Abstract
Assessment of health risk and fecal bacterial loads associated with human fecal pollution requires reliable host-specific analytical methods and a rapid quantification approach. We report the development of quantitative PCR assays for quantification of two recently described human-specific genetic markers targeting Bacteroidales-like cell surface-associated genes. Each assay exhibited a range of quantification from 10 to 1 × 106 copies of target DNA. For each assay, internal amplification controls were developed to detect the presence or absence of amplification inhibitors. The assays predominantly detected human fecal specimens and exhibited specificity levels greater than 97% when tested against 265 fecal DNA extracts from 22 different animal species. The abundance of each human-specific genetic marker in primary effluent wastewater samples collected from 20 geographically distinct locations was measured and compared to quantities estimated by real-time PCR assays specific for rRNA gene sequences from total Bacteroidales and enterococcal fecal microorganisms. Assay performances combined with the prevalence of DNA targets in sewage samples provide experimental evidence supporting the potential application of these quantitative methods for monitoring fecal pollution in ambient environmental waters.
Waterborne diseases that originate from human fecal pollution remain a significant public health issue. As a result, a large number of methods have been developed to detect and quantify human fecal pollution (10, 12, 18, 20). The majority of these methods are based on real-time quantitative PCR (qPCR) assays designed to estimate the concentrations of 16S rRNA gene sequences from various subpopulations within the order Bacteroidales. This bacterial order constitutes a large proportion of the normal gut microbiota of most animals, including humans (3, 15, 27). Bacterial 16S rRNA genes are useful as markers because they have relatively low mutation rates (7) and are typically present in multiple operons, increasing template DNA levels available for detection (2, 11, 17, 29). While several studies have demonstrated the value of Bacteroides 16S rRNA gene-based qPCR assays, currently available assays cannot discriminate between several animal sources closely associated with humans, including cats, dogs, and/or swine (10, 12, 18, 20). Alternative qPCR assays targeting genes directly involved in host-specific interactions may be capable of increased discrimination of fecal pollution sources (22, 23) and are needed to complement existing qPCR-based approaches used to identify sources of human fecal pollution.
A recent metagenomic survey of a human fecal bacterial community using genome fragment enrichment has led to the identification of hundreds of candidate human fecal bacterium-specific DNA sequences (23). PCR assays targeting two gene sequences encoding a hypothetical protein potentially involved in remodeling of bacterial surface polysaccharides and lipopolysaccharides (assay 19) and a putative RNA polymerase extracytoplasmic function sigma factor (assay 22) from Bacteroidales-like microorganisms exhibited a high level of specificity (100%) for human fecal material (23). However, it remained to be determined whether these reported chromosomal DNA sequences are abundant and uniform enough within human populations to be detected once diluted in the environment. On the basis of these considerations, the next steps toward the application of these gene sequences for water quality monitoring applications were to design qPCR assays for their detection and then to use these assays to evaluate the overall abundance and distribution of these sequences in human populations relative to those of rRNA gene sequences from different currently recognized fecal indicator bacterial groups.
Here, we report the development of two qPCR assays for quantification of the human-specific DNA sequences targeted by previously reported PCR assays 19 and 22 (23). Method performance characteristics, including specificity, range of quantification (ROQ), limit of quantification, amplification efficiency, and analytical precision, were defined for each assay. An internal amplification control (IAC) was designed to monitor for the presence of inhibitors commonly associated with environmental sampling that can confound DNA target copy number estimations. Finally, the abundance of each DNA target in primary effluent wastewater samples representative of 20 geographically distinct human populations was measured by qPCR analysis. In addition, the abundances of these human-specific DNA genes in wastewater were compared to those of rRNA genes of Bacteroidales and enterococci, two general fecal indicator bacterial groups that have been widely used for water quality testing.
MATERIALS AND METHODS
Sample collection.
Individual fecal samples (n = 265) and wastewater samples (n = 20) were collected for analysis as previously described (23). Primary effluent wastewater samples were collected on-site from 20 different wastewater treatment facilities across the United States (Table 1). Facilities were selected based on population served and geographic location. Briefly, 500 ml of primary effluent was collected from each facility and immediately stored on ice. Samples were then packed in ice and shipped overnight to Cincinnati, OH, for laboratory testing. Twenty-five milliliters of primary effluent from each facility was filtered through a 0.2-μm-pore-size Supor-200 filters (Whatman), and each filter was placed in a sterile 1.5-ml microtube and stored at −80°C (<6 months) until DNA extraction and qPCR amplification.
TABLE 1.
Primary effluent wastewater sample information
| Facility | Location | Population served | Inflow (mgd)a |
|---|---|---|---|
| Sacramento RWTP | Sacramento, CA | 1,200,000 | 168 |
| Clarksburg WWTP | Clarksburg, WV | 24,498 | 8.65 |
| Lincoln Northeast WWTF | Lincoln, NE | 55,000 | 5 |
| Lower East Fork WWTP | Milford, OH | 55,000 | 6.53 |
| West Point WWTP | Seattle, WA | 1,400,000 | 98.1 |
| Crystal Lake WWTP No.2 | Crystal Lake, IL | 38,600 | 5.8 |
| Little Falls WWTP | Little Falls, NY | 49,000 | 5.14 |
| Wildcat Hill WWTP | Flagstaff, AZ | 60,000 | 3.3 |
| Northwest Bergen County WWTP | Waldwick, NJ | 102,448 | 10 |
| Moorehead WWTP | Moorehead, KY | 20,454 | 2.5 |
| Buffalo WWTP | Buffalo, MO | 6,000 | 0.72 |
| Saginaw WWTP | Saginaw, MI | 57,523 | 25 |
| Bonner Springs WWTP | Bonner Springs, KS | 7,500 | 0.53 |
| Frankurt Sewer Department | Frankfurt, KY | 48,000 | 6.69 |
| Old Town PCF | Old Town, ME | 9,500 | 1.2 |
| Rutland WWTP | Rutland, VT | 22,000 | 5.85 |
| Maui County Kahului WWTF | Kahului, HI | 41,720 | 4.3 |
| City of St. Peter WWTP | St. Peter, MN | 10,850 | 1.1 |
| Las Vegas WWTP | Las Vegas, NV | 815,207 | 68 |
| Marshall St. Advanced WWTP | Clearwater, FL | 65,000 | 5.3 |
| Total | 4,088,300 | 431.7 |
Inflow indicates the average rate of sewage influent at each treatment facility, reported in millions of gallons per day (mgd).
Individual fecal samples were collected over a 12-month period at various locations across the United States from 22 different animal species likely to affect watersheds or beaches, including Homo sapiens (human, n = 16), Lama pacos (alpaca; n = 2), an Anser sp. (Canadian goose; n = 12), Felis catus (cat; n = 10), Gallus gallus (chicken; n = 10), Bos taurus (cow; n = 80), Odocoileus virginianus (white-tail deer; n = 15), Odocoileus hemionus (mule deer; n = 5), Cervus elaphus (elk; n = 5), Alces alces (moose; n = 1), Antilocapra american (pronghorn; n = 4), Canis familiaris (dog; n = 10), an Anas sp. (duck; n = 12), Capra aegagrus (goat; n = 7), Equus caballus (horse; n = 12), a Pelecanus sp. (pelican; n = 5), Sus scrofa (pig; n = 22), a Laridae sp. (gull; n = 12), Ovis aries (sheep; n = 10), Zalophus californianus (sea lion; n = 5), a Delphinidae sp. (marine dolphin; n = 3), and a Meleagris sp. (turkey; n = 7). Each fecal sample was collected from a different individual to maximize the opportunity to observe false-positive amplifications.
DNA extraction of fecal and primary effluent wastewater samples.
All DNA extractions were performed with a FastDNA kit for soils (Q-Biogene, Carlsbad, CA) as described previously (23), with the exception that a FastPrep-24 instrument (MP, Solon, OH) at a setting of 6 m/s for 120 s was used for cell lysis. DNA extraction yields were determined with a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). Filtration and extraction controls, with purified water substituted for primary effluent, were performed each day samples were received or extracted to monitor for potential contamination.
Oligonucleotides and primers.
TaqMan probe and primer assays targeting the rRNA genes of Bacteroidales (GenBac3) and Enterococcus (Entero1) are reported elsewhere (8, 13). qPCR probe and primer sequences for the putative human-specific HumM2 and HumM3 assays (Table 2) were designed with Primer Express software (Applied Biosystems, Foster City, CA) based on the previously reported end point PCR assays HumM19 and HumM22, respectively (23). Primers and TaqMan probes were designed using the default parameters of the Primer Express software (version 1.5; Applied Biosystems). Fluorogenic probes were 5′ labeled with 6-carboxyfluorescein (FAM) or VIC and 3′ labeled with 6-carboxytetramethylrhodamine (TAMRA). Optimal primer and probe reaction concentrations were determined according to a standard Applied Biosystems protocol (1). The HumM2 and HumM3 assay primer and probe sets (Table 2) were tested for specificity with animal fecal and wastewater sample composites (5 ng DNA template per PCR assay).
TABLE 2.
Oligonucleotides, primers, and probes
| Assay and sequence type | Primer or probe sequence (5′ to 3′) | Size (bp) | Source or reference |
|---|---|---|---|
| GenBac3 | 129 | 20 | |
| Forward | GGGGTTCTGAGAGGAAGGT | ||
| Reverse | AGTAGCGGAAGGATGACGG | ||
| Probe | FAM-CAATATTCCTCACTGCTGCCTCCCGTA-TAMRA | ||
| Entero1 | 92 | 13 | |
| Forward | AGAAATTCCAAACGAACTTG | ||
| Reverse | AATGATGGAGGTAGAGCAC | ||
| Probe | FAM-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-TAMRA | ||
| HumM2 | 101 | This study | |
| Hum2F | CGTCAGGTTTGTTTCGGTATTG | ||
| Hum2R | TCATCACGTAACTTATTTATATGCATTAGC | ||
| Probe | FAM-TATCGAAAATCTCACGGATTAACTCTTGTGTACGC-TAMRA | ||
| HumM3 | 83 | This study | |
| Hum3F | GTAATTCGCGTTCTTCCTCACAT | ||
| Hum3R | GGAGGAAACAAGTATGAAGATAGAAGAATTAA | ||
| Probe | FAM-AGGTCTGTCCTTCGAAATAGCGGT-TAMRA | ||
| Human IAC | 258 | 30; this study | |
| Frag1 | GATCATGAGTTCACATGTCCGAGTAATTCGCGTTCTTCCTCACATACGTCAGGTTTGTTTCGGTATTG AGTTAGGAACAGGCGGCGACGAATGTTAATCTTCTATCTTC | ||
| Frag2 | TCCGGTGATGTCTCGAGAGTGTCTCATCACGTAACTTATTTATATGCATTAGCGGTGAAGGTCTGGGAGGAAACAAGTATGAAGATAGAAGAATTAACATTCGTCGCCGC | ||
| Frag3 | AGTTAGGAACAGGCGGCGACGAATGTTAATTCTTCTATCTTCATACTTGTTTCCTCCCAGACCTTCACCGCTAATGCATATAAATAAGTTACGTGATGAGACACTCTCGA | ||
| Frag4 | CCGTCATCCTTCACGCTACTGATGTCTGCATGGTATATGTTGAGTGCAATGGGATTTTATCCGGTGAATCCGGTGATGTCTCGAGAGTGTCTCATCACGTAACTTATTTA | ||
| Probe | VIC-TAGGAACAGGCGGCGACGA-TAMRAa |
The TaqMan probe was modified from the previously reported UT probe (30).
DNA preparations from pure bacterial cultures.
American Type Culture Collection (ATCC) bacterial strains were used to prepare DNA standards for the Bacteroidales and Enterococcus qPCR assays. Enterococcus faecalis (ATCC 29212) was cultured as previously described (8). Bacteroides thetaiotaomicron (ATCC 29741) cells were grown in chopped-meat carbohydrate broth (Remel, Lenexa, KS) in accordance with the manufacturer's instructions. Both cultures were harvested by centrifugation at 8,000 × g for 5 min, washed twice using sterile phosphate-buffered saline (Sigma, St. Louis, MO), and stored in aliquots at −40°C. The cell concentrations of each organism in the final washed suspensions were determined by bright-field microscopy at ×40 magnification in disposable hemocytometer chambers (no. CP2-002; Nexcelom Bioscience, Lawrence, MA). DNA was isolated from the cell suspensions by using a bead-beating extraction approach (8) and incubated for 1 hour at 37°C with 0.017 μg/μl RNase A (Gentra Systems). DNA purification was performed using a silica column adsorption kit (DNA-EZ; GeneRite, Kendall Park, NJ). DNA concentrations of cell extracts were determined by spectrophotometric absorbance readings at 260 nm (A260), and the purity of the DNA preparations was determined by A260/A280 ratios.
Construction of IAC and plasmid DNA standards.
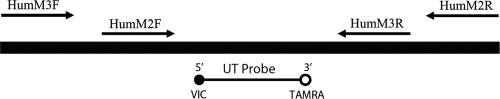
A plasmid DNA construct was developed to function as an IAC DNA target that can be spiked into DNA extracts to monitor for PCR inhibition and also as a plasmid DNA standard for calculation of HumM2 and HumM3 qPCR assay calibration curves. The IAC construct was designed to contain a single site for hybridization of a unique TaqMan VIC-labeled probe sequence flanked by multiple primer binding sequences (Table 2 and Fig. 1). To build the human assay IAC construct, long oligonucleotides (>100 bp) (Table 1) containing multiple primer sequences (23) were designed such that their 3′ ends overlapped. The overlapping fragments were then combined into a single DNA molecule by using overlap extension PCR (9). The IAC construct was inserted into a plasmid vector, purified, linearized, quantified, and diluted to generate samples ranging from approximately 10 to 1 × 106 molecules of template DNA as described previously (21).
FIG. 1.
Diagram of human-specific plasmid DNA IAC composite construct. The IAC (258 bp) consists of a VIC-labeled universal probe binding site (30) flanked by primer sequences for HumM2 (101 bp) and HumM3 (83 bp) qPCR assays.
qPCR assays and quantification.
The four qPCR assays used in this study were HumM2, HumM3, GenBac3, and Entero1 (Table 2). Amplification was performed with a 7900HT fast real-time sequence detector (Applied Biosystems). Reaction conditions and thermal cycling parameters for GenBac3 and Entero1 are described elsewhere (24). For HumM2 and HumM3, reaction mixtures (25 μl) contained 1× TaqMan universal PCR master mix with AmpErase uracil-N-glycosylase (Applied Biosystems), 0.2 mg/ml bovine serum albumin (Sigma), 1 μM of each primer, 80 nM FAM- or VIC-labeled TaqMan probe (Applied Biosystems), and either 1 to 100 ng genomic DNA (fecal and wastewater samples) or 10 to 1 × 106 target gene copies (human IAC plasmid DNA). Reaction mixtures for multiplex applications were the same as those described above, with additions of both 80 nM of VIC- or tetrachloro-6-carboxyfluorescein (TET)-labeled TaqMan probes for IAC plasmid DNA and 80 nM of FAM-labeled TaqMan probe for native DNA targets. IAC spike concentrations were either 25 or 50 copies. All reactions were performed in triplicate using MicroAmp optical 96-well reaction plates with MicroAmp optical caps (Applied Biosystems). The thermal conditions were 50°C for 2 min to activate uracil-N-glycosylase, followed by 10 min of incubation at 95°C to activate AmpliTaq Gold enzyme, and the temperature profile then followed a 40-cycle pattern with a short denaturation at 95°C for 15 s and a combined annealing and primer extension phase at 60°C for 1 min. Data were initially analyzed with Sequence Detector Software (version 2.2.2) at threshold determination levels of 0.08 for human-specific assays (HumM2 and HumM3) and 0.03 for general fecal indicator bacterial assays (GenBac3 and Entero1). Threshold cycle (CT) values were exported to Microsoft Excel for further statistical analysis. A minimum of three no-template amplifications, with purified water substituted for template DNA, were performed for each 96-well qPCR experiment to monitor for potential contamination.
Calculations and statistical analysis.
The specificities of HumM2 and HumM3 were determined as specificity = d/(b + d), where b represents false positives and d represents true negatives. Master calibration curves, unknown DNA concentration estimates, and credible intervals were determined using a Markov Chain Monte Carlo approach (25). Bayesian calculations were performed using the publicly available software program WinBUGS version 1.4.1 (http://www.mrc-bsu.cam.ac.uk/bugs) (14). The WinBUGS program code and the resulting data output used to develop master calibration curves for HumM2 and HumM3 DNA standards are available (not shown). An analysis of covariance model was used to compare the intercepts and slopes of individual standard curves used to calculate the master calibration equations. One-way analysis-of-variance (ANOVA) tests comparing CT values for reactions with known amounts of standard target DNA were used to define the ROQ for each assay. The precision of CT measurements determined from DNA standards was expressed as a percent coefficient of variation (CV; standard deviation expressed as a percentage of the mean). A one-way random-effect ANOVA model (with location as a random factor) was used to test the hypotheses that the variability between untreated wastewater sample locations was zero. A paired two-sample t test was used to compare the overall mean difference between HumM2, HumM3, Entero1, and GenBac3 qPCR mean CT values for wastewater samples.
RESULTS
Master calibration curves and ROQ.
Overall fitted curves representing multiple independent runs of the DNA standards were compared using analysis of covariance tests. Independent fitted curves for each qPCR assay demonstrated a significant difference in intercepts (P < 0.05) but no difference in slopes (P > 0.05). Calibration curve equations and performance characteristics of the four qPCR assays are shown in Table 3. Calibration curves for GenBac3 and Entero1 general fecal indicator bacterial assays were generated from eight independent runs using genomic DNA standards extracted from cultured cell suspensions whereas, HumM2 and HumM3 fitted curves were generated from 12 independent fitted curves, each using plasmid DNA standards. ROQs spanned the entire range of standard concentrations tested for all qPCR assays, including 10 to 1 × 106 copies for human-specific assays and 40 to 4 × 104 copies for general fecal indicator bacterial assays. The precision of CT measurements across defined ROQs for all assays was less than 3% CV, and amplification efficiencies ranged from 1.87 to 1.99 (Table 3). No-template controls indicated the absence of contamination in 98.9% of qPCR experiments, and all extraction blanks tested negative for the presence of extraneous DNA molecules.
TABLE 3.
Calibration curve equations and performance characteristics of qPCR assays
| Assay | Calibration equation | Amplification efficiencya | ROQ (no. of copies) for target DNA | % CV across ROQ | Methodb |
|---|---|---|---|---|---|
| Entero1 | Y = 38.0 − 3.42X | 1.96 | 40 to 4 × 104 | 2.24 | Multiplex |
| GenBac3 | Y = 38.1 − 3.34X | 1.99 | 40 to 4 × 104 | 2.92 | Simplex |
| HumM2 | Y = 41.8 − 3.67X | 1.87 | 10 to 1 × 106 | 2.46 | Simplex |
| HumM3 | Y = 41.9 − 3.66X | 1.88 | 10 to 1 × 106 | 2.40 | Simplex |
Amplification efficiency = 10(1/−slope).
Either a simplex approach or a multiplex strategy where the target DNA was simultaneously detected with an IAC.
Evaluation of multiplex host-specific qPCR application.
A composite synthetic internal control was developed for each host-specific assay to monitor fecal and wastewater DNA extracts for potential PCR inhibition. The IAC construct was designed with the intention of allowing target DNA and an IAC to be coamplified with the same set of primers, under the same reaction conditions, in the same PCR tube. The target DNA and IAC product could then be detected and quantified simultaneously with different fluorescently labeled TaqMan probes, provided that (i) there is no significant difference (P > 0.05) between simplex and multiplex standard curve intercepts and slopes and (ii) a fixed amount of IAC could be quantified across a range of genomic DNA standard concentrations (21). An IAC spike of 50 copies was undetectable at human fecal DNA concentrations ranging from 1 to 100 ng for the HumM2 assay, while a significant difference between simplex and multiplex curve intercepts and slopes was observed for the HumM3 assay (P < 0.05), suggesting that neither of these assays is reliable as a multiplex reaction (data not shown). The failure of both assays to perform in a multiplex environment is most likely due to competition between genomic target DNA (FAM labeled) and the IAC spike (VIC labeled). Thus, only the HumM3 IAC could be used to monitor for PCR inhibition, and only in a simplex application.
Monitoring for PCR inhibition in DNA extracts.
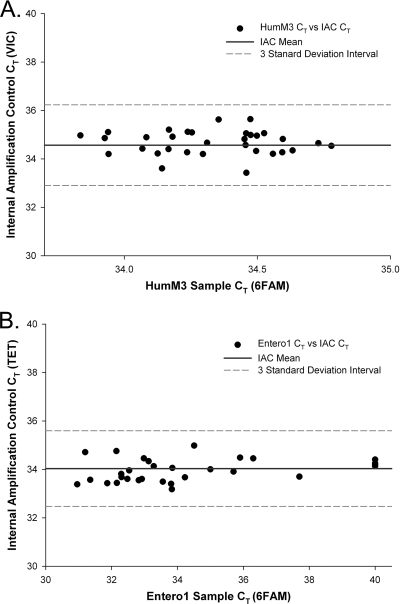
DNA isolation from wastewater and fecal samples may not remove all substances that can interfere with qPCR, and the degree of interference may vary between samples. Therefore, internal controls designed to evaluate the suitability of isolated DNA for quantitative analysis were included for each DNA extract. All fecal DNA extracts were screened for inhibition of the HumM3 IAC assay. The criterion for concluding that there was no significant PCR inhibition of the HumM3 IAC assay by these samples was established as a CT of 34.6 ± 1.65, based on repeated experiments measuring the simplex mean CT and standard deviation values for control reaction mixtures containing 50 copies of IAC in buffer (Fig. 2A). Wastewater DNA extracts were also tested using the previously reported multiplex Entero1 application with a 25-copy IAC spike. The criterion for concluding that there was no significant PCR inhibition in these assays was defined as a CT of 34.0 ± 1.41 (Fig. 2B). IAC analyses indicated the absence of PCR inhibitors in all fecal and untreated wastewater DNA extracts on the basis of both of these criteria.
FIG. 2.
Results of qPCR IAC inhibition tests for fecal and wastewater DNA extracts. Scatter plots show IAC (VIC or TET probes) and genomic DNA (FAM probe) CT values from analyses of fecal DNA extracts with HumM3 (A) and of wastewater DNA extracts with Entero1 (B). Confidence intervals (dashed lines) represent 3 standard deviations from the mean IAC CT (solid lines; HumM3 CT = 34.6 and Entero1 CT = 34.0) established from repeated control experiments.
Specificity of host-specific qPCR assays.
The specificities of the HumM2 and HumM3 assays were tested with a reference collection of fecal samples from hundreds of nontarget animals (Table 4). HumM2 and HumM3 assays exhibited specificity values of 99.2% and 97.2%, respectively. HumM2 elicited false positives with two chicken fecal samples (CT values of 29.3 ± 0.16 and 29.1 ± 0.14), while HumM3 cross-reacted with a single elk sample (CT of 33.6 ± 0.35) and six sheep samples (CT values ranging from 24.4 ± 0.05 to 36.9 ± 0.73). Both assays successfully detected respective DNA targets in all human fecal and primary effluent wastewater DNA extracts (Table 4).
TABLE 4.
Specificity of HumM2 and HumM3 qPCR assaysa
| Animal source | No. of animals | Avg CT (SD) for:
|
|
|---|---|---|---|
| HumM2 | HumM3 | ||
| Alpaca | 2 | ||
| Cow | 80 | ||
| Goat | 7 | ||
| Sheep | 10 | 34.1 (0.03) | |
| Horse | 12 | ||
| Pig | 22 | ||
| Antelope | 4 | ||
| Whitetail deer | 15 | ||
| Mule deer | 5 | ||
| Moose | 1 | ||
| Elk | 5 | 35.7 (0.24) | |
| Canadian goose | 12 | ||
| Duck | 12 | ||
| Pelican | 5 | ||
| Gull | 12 | ||
| Turkey | 7 | ||
| Chicken | 10 | 32.1 (0.30) | |
| Marine dolphin | 3 | ||
| California sea lion | 5 | ||
| Cat | 10 | ||
| Dog | 10 | ||
| Human | 16 | 29.7 (0.03) | 30.3 (0.10) |
| Wastewater | 20 | 31.8 (0.54) | 32.8 (1.01) |
| Total | 285 | ||
CT values were generated from 1 ng of total DNA.
Quantification of fecal bacterial genes in untreated wastewater.
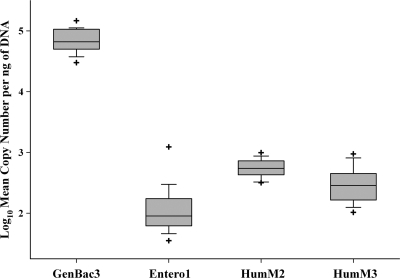
Primary effluent wastewater samples were collected from 20 different geographic locations to characterize target DNA variability between localities and to compare the relative abundance of each target DNA to those of enterococci and general Bacteroidales 16S rRNA genes. A one-way random effect ANOVA model indicated that there is significant variability (P < 0.05) in CT values among all locations for each assay. Variance (σ2) between wastewater sample locations ranged from 0.30 for HumM2, 1.06 for HumM3, and 1.65 for Entero1 to 0.45 for GenBac3. Target DNA relative abundances for each assay were compared by normalizing data sets to 1 ng of template DNA and plotting log10 mean copy number estimates for each wastewater sample by a qPCR assay. A box-and-whisker diagram was used to display differences between wastewater sample DNA target estimates for each qPCR assay, including the smallest observation, lower quartile (25th percentile), median, upper quartile (75th percentile), largest observation, and outliers (Fig. 3).
FIG. 3.
Box-and-whisker diagram depicting the relative abundances of gene targets from HumM2, HumM3, Entero1, and GenBac3 qPCR assays for all primary effluent sewage sample locations. Estimated gene target concentrations are reported as log10 mean copy numbers per ng of total DNA. The boundary of the box closest to zero indicates the 25th percentile, the line within the box represents the median, and the boundary of the box farthest from zero indicates the 75th percentile. Whiskers above and below the box indicate the 10th and 90th percentiles, respectively. “+” denotes outlier measurements.
DISCUSSION
Human-specific qPCR.
We report on two qPCR assays that detect predominantly human fecal DNA when tested against a panel of samples representing agriculturally important animals, such as cattle, poultry, and swine, as well as many wildlife species. These qPCR assays were designed to target the same gene sequences as two end point PCR assays (assays 19 and 22) that were previously reported to be 100% human specific based on a fecal reference collection consisting of 160 individual samples representing 11 different animal species (23). The slight decrease in the specificity of the real-time qPCR assays compared to the level for the end point PCR assays may be due to the larger nontarget fecal sample reference library used to establish specificity values or factors associated with the TaqMan qPCR approach, such as constraints in primer design, PCR reagent chemistry, thermal cycling settings, and an increased number of amplification thermal cycles. Regardless of the reason, the HumM2 and HumM3 qPCR assays exhibit extremely high levels of specificity exceeding 97.5%.
Master calibration curves were used in this study due to the large numbers of fecal and wastewater samples processed and the need to maximize the number of samples in each experiment setup and reduce expenses. Each master curve was compiled from up to 12 independent runs in order to reflect sources of intra- and interrun variability. Master calibration curves were acceptable in this study because (i) there were no significant differences in the slopes of fitted curves between independent runs (P > 0.05), (ii) the analytical precision (percent CV) over the ROQ between runs averaged less than 3%, and (iii) the Bayesian approach accounts for run-to-run variability with a 95% credible interval when generating fitted calibration curves (25).
Abundance of host-specific and fecal indicator genes.
Little is known regarding the abundance and geographical distribution of human-specific genes in sewage. In this study, we tested 20 primary effluent wastewater samples collected from different geographic locations in the United States, ranging from Hawaii to Florida. The wastewater samples were representative of approximately 4.1 million individuals, responsible for generating an average of 5,180 million gallons of raw sewage per year, and were ideal for estimating the abundances of host-specific gene targets in different human populations. Host-specific and general fecal indicator bacterial qPCR assays successfully detected respective genetic targets from 1 ng of DNA for 100% of the wastewater samples regardless of locality. The general Bacteroidales assay (GenBac3) detected the highest target gene concentrations in all samples, which supports previous research reporting that Bacteroidales often makes up a large portion of the human fecal bacterial community (6, 16, 28). The HumM2 and HumM3 gene targets were the next-most-abundant markers and more prevalent than the enterococcal 23S rRNA genes (Fig. 3). Enterococci are routinely detected in feces-polluted waters (26). The observation that host-specific gene targets are more abundant than enterococcal 23S rRNA genes suggests that detectable quantities of HumM2 and HumM3 gene targets may be present in ambient waters.
All qPCR assays exhibited less than 3.9% dispersion of CT values from an overall wastewater sample mean [(one-way random-effect ANOVA qPCR standard deviation/mean) × 100] regardless of gene target. In addition, a significant difference (P < 0.05) was observed in concentrations of all qPCR gene targets between wastewater geographic locations. Fluctuations in relative gene target concentrations between wastewater samples could result from differences in local population diet, age, and/or health but could also reflect uncertainty associated with single sample events. Regardless of the reason, low dispersion percentages (<3.9%) suggest that the human-specific gene targets can be detected with a level of confidence similar to those for 16S rRNA general Bacteroidales and 23S rRNA enterococcal gene targets.
Implications for MST.
Recreational and drinking source waters continue to be impacted by human fecal pollution and can impose a direct threat to human health (4, 5, 19). In addition to human waste, many other agricultural and wildlife animal sources can contribute to the total fecal load. Most microbial source tracking (MST) methods attempt to identify specific fecal sources to help local authorities prioritize polluted areas for restoration. Recent advances in PCR-based methods now allow for the estimation of host-specific DNA target concentrations. These quantitative approaches can extend the utility of MST applications by supplying information regarding the concentration of host-specific fecal pollution sources. To date, no qPCR-based method has been found to be 100% specific for human fecal pollution (10, 12, 18, 20). Animals that cohabitate with humans, such as cats and dogs, and animals that share similar digestive physiologies, such as pigs, are the most problematic. Fecal pollution originating from pets can confound MST studies where cat and dog waste is mixed with sewage and/or runoff after rain events. A similar problem can arise in watersheds affected by swine sources of fecal pollution. HumM2 and HumM3 are the first qPCR assays available that can discriminate between all three of these sources of fecal pollution. In addition, these assays can quantify as few as 10 copies of target DNA per reaction with a high degree of precision. DNA targets of these assays were widely distributed among 20 different human populations and more abundant than fecal enterococci in almost all wastewater samples tested.
To explore the potential of the HumM2 and HumM3 qPCR assays for environmental monitoring, each assay underwent preliminary testing with DNA isolated from river, stream, and storm water samples (n = 6). All six samples contained general Bacteroidales target sequences (GenBac3 CT values ranging from 34.3 ± 1.26 to 26.2 ± 0.10), suggesting the presence of fecal pollution. Two of these samples, both collected from locations situated within 100 m downstream of a wastewater discharge pipe, generated CT values for both host-specific assays (CT values ranging from 35.8 ± 0.46 to 34.7 ± 0.32). These preliminary results, combined with the high levels of specificity and broad distribution of their DNA targets in wastewater samples, suggest that the HumM2 and HumM3 assays may have future utility in MST applications. However, to realize the full potential of these qPCR assays, several issues remain to be addressed. Future studies characterizing the survival of target DNA molecules through the wastewater treatment process and in the environment are needed to generate reliable estimates of the impact of these sources on ambient water samples. Research projects focusing on the relevance of each qPCR assay to current culture-based and qPCR-based fecal indicator methods (such as Escherichia coli and enterococci) are also critical for successful MST applications. Finally, epidemiological studies are necessary to establish any links between the prevalence of host-specific DNA targets and relevant public health risks.
Acknowledgments
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed or partially funded and collaborated in the research described herein.
This research has been subjected to the agency's peer and administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print on 10 July 2009.
REFERENCES
- 1.ABI. 2002. TaqMan universal PCR master mix protocol. Applied Biosystems, Foster City, CA.
- 2.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, A. F., M. Lindberg, H. Jackobsson, F. Backhed, P. Nyren, and L. Engstrand. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balarajan, R., V. Soni Raleigh, P. Yuen, D. Wheeler, D. Machin, and R. Caftwright. 1991. Health risks associated with bathing in sea water. BMJ 303:1444-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dufour, A. P. 1984. Health effects criteria for fresh recreational waters. U.S. Environmental Protection Agency, Cincinnati, OH.
- 6.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimont, F., and P. A. Grimont. 1986. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann. Inst. Pasteur Microbiol. 137B:165-175. [DOI] [PubMed] [Google Scholar]
- 8.Haugland, R. A., S. C. Siefring, L. J. Wymer, K. P. Brenner, and A. P. Dufour. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559-568. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kildare, B. J., C. M. Leutenegger, B. S. McSwain, D. G. Bambic, V. B. Rajal, and S. Wuertz. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701-3715. [DOI] [PubMed] [Google Scholar]
- 11.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Layton, A., L. McKay, D. Williams, V. Garrett, R. Gentry, and G. Sayler. 2006. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72:4214-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig, W., and K.-H. Schleifer. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556-562. [DOI] [PubMed] [Google Scholar]
- 14.Lunn, D. J., A. Thomas, N. Best, and D. Spiegelhalter. 2000. WinBUGS—a Bayesian modeling framework: concepts, structure, and extensibility. Stat. Comput. 10:325-337. [Google Scholar]
- 15.Matsuki, T., K. Watanabe, J. Fujimoto, T. Takada, and R. Tanaka. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore, W. E. C., and I. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson, K. E., R. D. Fleischmann, R. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okabe, S., N. Okayama, O. Savichtcheva, and T. Ito. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890-901. [DOI] [PubMed] [Google Scholar]
- 19.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seurinck, S., T. Defoirdt, W. Verstraete, and S. Siciliano. 2005. Detection and quantification of human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol. 7:249-259. [DOI] [PubMed] [Google Scholar]
- 21.Shanks, O. C., E. Atikovic, A. D. Blackwood, J. Lu, R. T. Noble, J. Santo Domingo, S. Siefring, M. Sivaganesan, and R. P. Haugland. 2008. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 74:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shanks, O. C., J. W. Santo Domingo, R. Lamendella, C. A. Kelty, and J. E. Graham. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanks, O. C., J. W. Santo Domingo, J. Lu, C. A. Kelty, and J. E. Graham. 2007. Identification of bacterial DNA markers for the detection of human fecal pollution in water. Appl. Environ. Microbiol. 73:2416-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siefring, S. C., M. Varma, E. Atikovic, L. J. Wymer, and R. A. Haugland. 2008. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J. Water Health 6:225-237. [DOI] [PubMed] [Google Scholar]
- 25.Sivaganesan, M., S. Seifring, M. Varma, R. A. Haugland, and O. C. Shanks. 2008. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinformatics 9:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.U.S. Environmental Protection Agency. 2000. Improved enumeration methods for recreational water quality indicators: enterococci and Escherichia coli. Report EPA-821-R-97-004. U.S. Environmental Protection Agency, Cincinnati, OH.
- 27.Wang, R. F., M. L. Beggs, B. D. Erickson, and C. E. Cerniglia. 2004. DNA microarray analysis of predominant human intestinal bacteria in fecal samples. Mol. Cell. Probes 18:223-234. [DOI] [PubMed] [Google Scholar]
- 28.Wood, J., K. P. Scott, G. Avgustin, C. J. Newbold, and H. J. Flint. 1998. Estimation of the relative abundance of different Bacteroides and Prevotella ribotypes in gut samples by restriction enzyme profiling of PCR-amplified 16S rRNA gene sequences. Appl. Environ. Microbiol. 64:3683-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, Y., D. Zhang, L. Wenquan, J. Chen, Y. Peng, and W. Cao. 2003. A novel real-time quantitative PCR method using attached universal template probe. Nucleic Acids Res. 31:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]