Abstract
Xylella fastidiosa, the causal agent of several scorch diseases, is associated with leaf scorch symptoms in Chitalpa tashkentensis, a common ornamental landscape plant used throughout the southwestern United States. For a number of years, many chitalpa trees in southern New Mexico and Arizona exhibited leaf scorch symptoms, and the results from a regional survey show that chitalpa trees from New Mexico, Arizona, and California are frequently infected with X. fastidiosa. Phylogenetic analysis of multiple loci was used to compare the X. fastidiosa infecting chitalpa strains from New Mexico, Arizona, and trees imported into New Mexico nurseries with previously reported X. fastidiosa strains. Loci analyzed included the 16S ribosome, 16S-23S ribosomal intergenic spacer region, gyrase-B, simple sequence repeat sequences, X. fastidiosa-specific sequences, and the virulence-associated protein (VapD). This analysis indicates that the X. fastidiosa isolates associated with infected chitalpa trees in the Southwest are a highly related group that is distinct from the four previously defined taxons X. fastidiosa subsp. fastidiosa (piercei), X. fastidiosa subsp. multiplex, X. fastidiosa subsp. sandyi, and X. fastidiosa subsp. pauca. Therefore, the classification proposed for this new subspecies is X. fastidiosa subsp. tashke.
Xylella fastidiosa is a gram-negative bacterium that multiplies within the xylem and causes serious disease problems in many diverse plant species. X. fastidiosa is considered a “new world” pathogen and is mainly found within North, Central, and South America (30). In many native plant species this bacterium exists as an apparently benign endophyte, while in other instances proliferation of X. fastidiosa within the xylem leads to disease typified by symptoms, including leaf scorch, chlorosis, stunting, branch dieback, inedible fruit, and eventually the death of the plant (4, 15). X. fastidiosa is transmitted by xylem-feeding insect vectors such as sharpshooters, leafhoppers, and spittle bugs (35). Diseases caused by X. fastidiosa include Pierce's disease in grapes (7), citrus variegated chlorosis (CVC) (6), coffee leaf scorch (18), pecan leaf scorch (36), phony peach (41), plum leaf scald (32), and almond leaf scorch (25). X. fastidiosa has also been shown to be the causative agent of diseases found in landscape plants such as oleander leaf scorch (31), mulberry leaf scorch (14), and oak leaf scorch (3). In addition to the examples above proven through the completion of Koch's postulates, X. fastidiosa is known to be associated with leaf scorch type diseases in several other ornamental landscape species including crape myrtle, olive, day lily, and southern magnolia (12).
Chitalpa (Chitalpa tashkentensis Elias and Wisura) is an ornamental landscape plant that was developed for arid landscapes such as California, Arizona, Texas, and New Mexico. Chitalpa, originally bred in Russia and introduced into the United States in 1977, is an intergenic hybrid between desert willow (Chilopsis linearis Cav.) and Catalpa bignonioides Walt. (28). In the past, chitalpa trees across the Southwest were observed to display leaf scorch symptoms of unknown origin. X. fastidiosa was detected in many chitalpa trees that displayed leaf scorch symptoms in southern New Mexico (34). The first known occurrence of Pierce's disease in New Mexico was reported in 2007, and the strains of X. fastidiosa found in infected New Mexico grapes were very similar to those present in chitalpa trees from the same area (33). The common use of chitalpa as a landscape plant in the Southwest coupled with the recent discovery that it can harbor X. fastidiosa strains similar to those associated with Pierce's disease in New Mexico prompted a survey of chitalpa trees across the Southwest. The results of this survey show that chitalpa trees from New Mexico and Arizona are frequently infected with X. fastidiosa. Chitalpa plants imported into New Mexico nurseries from California were also found to contain similar strains of X. fastidiosa. A multilocus phylogenetic analysis was performed to further characterize these strains of X. fastidiosa. This analysis revealed that the X. fastidiosa isolates infecting chitalpa plants in New Mexico, Arizona, and imported into nurseries from California are highly related to each other and are distinct from the previously described subspecies fastidiosa (38).
MATERIALS AND METHODS
Collection of chitalpa samples.
Chitalpa trees exhibiting leaf scorch type symptoms from southern New Mexico, Arizona, and commercial nurseries in southern New Mexico were sampled during the summer and fall of 2006 and summer of 2007. Samples consisted of branches, stems, and leaves. The samples were placed in individual plastic bags and stored at 4°C.
ELISA of symptomatic chitalpa plants.
The presence of X. fastidiosa was first detected by enzyme-linked immunosorbent assay (ELISA). Two different methods were utilized for this assay. First, 0.5- to 1.0-g samples of leaf petioles and the midveins were placed in plastic samples bags with 3 to 5 ml of extraction buffer 3 (Agdia, Inc., Elkhart, IN) and crushed with a hammer at room temperature. Second, the sap was extracted from chitalpa branches by using a pressure chamber (Soilmoisture Equipment, Santa Barbara, CA). Sap was obtained between 20 and 40 bars of pressure. Crushed samples and extracted sap were then loaded into strips coated with X. fastidiosa-specific antibodies (X. fastidiosa PathoScreen kit; AgDia) and processed according to the manufacturer's instructions (Agdia). The results were analyzed by using a plate reader (Bio-Tek KC4, v.3.1) at 620 nm. All test plates included at least three negative controls, and samples were considered positive at greater than two times the average negative control.
Bacterial plating from chitalpa leaf tissue.
Leaves were surface sterilized by submerging in 70% ethanol for 2 min, followed by submerging the leaf in 30% bleach (1.5% sodium hypochlorite) for 2 min. The leaves were rinsed in sterile distilled water twice. Leaf sections from three separate leaves, consisting of mainly the petiole and main veins, were finely chopped on sterile filter paper and placed in an Eppendorf tube with 600 μl of sterile succinate-citrate-phosphate buffer. Leaf pieces were ground for 30 s with a homogenizer (43; http://nature.berkeley.edu/classes/es196/projects/2006final/zintzun%20.pdf). Then, 10 μl of this extract was added to 90 μl of sterile succinate-citrate-phosphate buffer and plated on XFD2 medium (1). The plates were incubated at 28°C and monitored for colony development for 6 weeks.
PCR detection of X. fastidiosa with total DNA, xylem fluid, and bacterial colonies.
Total DNA isolated from chitalpa plants or expressed xylem fluid obtained from the pressure chamber (see the ELISA methods described above) and bacterial colonies was used for PCR analysis. Total DNA was extracted from chitalpa plant samples by using the Qiagen Plant DNAeasy kit (Qiagen, Inc., Valencia, CA). The 272-1 and 272-2 external and internal primers for nested PCR were utilized to determine the presence of X. fastidiosa as previously described by Pooler et al. (29). Ten additional primer sets were also utilized for amplification, and the resulting products were sequenced; these included the 16S ribosome (V6 forward, 5′- AACGCGAAGAACCTTAC) and (23 reverse, 5′-GTGCCAAGGCATCCACC-3′), 16S-23S ribosomal internal transcribed spacer (ITS; 1493 forward, 5′-AGTCGTAACAAGGTAGCCGT-3′) and (23 reverse, 5′-GTGCCAAGGCATCCACC-3′) (19), gyrase subunit B (gyrase-B; forward [5′-AAGCGCCTCCGTGAGTTATC-3′] and reverse [5′-CCTTCACGCATATCATCACC-3′]), simple sequence repeat sequences ASSR12 (forward [5′-TGCTCATTGTGGCGAAGG-3′] and reverse [5′-CGCAACGTGCATTCATCG-3′]), OSSR9 (forward [5′-TAGGAATCGTGTTCAAACTG-3′] and reverse [5′-TTACTATCGGCAGCAGAC-3′]), and GSSR12 (forward [5′-TTACGCTGATTGGCTGCATTG-3′] and reverse [5′-GTCAAACACTGCCTATAGAGCG-3′]) (20) sequences specific to X. fastidiosa such as FY0076 (forward [5′-CGGGTCGTTCCTATCAACTT-3′] and reverse [5′-CCCTTCAACGATTCGGTCTA-3′]) (http://cropdisease.ars.usda.gov), HL (forward [5′-AAGGCAATAAACGCGCACTA-3′] and reverse [5′-GGTTTTGCTGACTGGCAACA-3′]) (10), and RST (rst31 [5′-GCGTTAATTTTCGAAGTGATTCGATT-3′] and rst33 [5′-CACCATTCGTATCCCGGTG-3′]) (24) and virulence-associated genes such as virulence-associated protein D (VapD) (forward [5′-CCATGGATCGCTGCCTAATCG-3′] and reverse [5′-GGATCCCTAATCATCAGGATTTGG-3′]). The components for the PCR included 1× PCR buffer (100 mM Tris-HCl, 500 mM KCl [pH 8.3]), 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 0.1 ng of each primer, and 2 U of Taq polymerase. Templates consisted of 1 μl of the total chitalpa DNA, 1 μl of 1:100 dilution of xylem fluid, or a small portion of the bacterial colony for whole-cell PCR. The reaction conditions were as follows: an initial denaturation step of 95°C for 2 min; 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 2 min; with a final elongation step of 72°C for 5 min. The products were resolved on 1% agarose gels, stained with ethidium bromide, and visualized under UV light with a Kodak Image 2000R Station (Eastman Kodak Company, Rochester, NY).
DNA sequencing and sequence analysis.
The PCR products were either gel purified using Qiaex (Qiagen) or treated with ExoSapIt according to the manufacturer's instructions (USB, Cleveland, OH) and directly sequenced using a BigDye Terminator version 3.1 kit (ABI, Foster City, CA). Sequencing reactions were purified by using Performa DTR gel filtration cartridges (Edge BioSystem, Gaithersburg, MD) and run on an ABI 3100 automated sequencer (NMSU-LiCor facility). The sequences were analyzed by using the sequence scanner software (Bio-Rad, Hercules, CA). Sequence similarities were searched by using the BLAST at the National Center for Biotechnology Information website (2). Multiple sequence alignments were generated by using Geneious Pro v4.0.4 (8). Maximum-likelihood trees were generated by using PAUP 4.0 and bootstrapped 1,000 times (40). Sequences obtained during the present study were deposited in GenBank (Table 1).
TABLE 1.
GenBank accession numbers for the X. fastidiosa sequences obtained during this study
| X. fastidiosa isolate | Locus | GenBank accession no. |
|---|---|---|
| NM02 | 16S ribosome | FJ755926 |
| AZ03 | 16S ribosome | FJ755927 |
| AZ04 | 16S ribosome | FJ755928 |
| CA01 | 16S ribosome | FJ755929 |
| NM01 | 16S-23S ITS | EU714194 |
| NM02 | 16S-23S ITS | EU714192 |
| NM05 | 16S-23S ITS | EU714193 |
| NM grape 1 | 16S-23S ITS | EU714195 |
| AZ01 | 16S-23S ITS | EU714188 |
| AZ03 | 16S-23S ITS | EU714189 |
| AZ04 | 16S-23S ITS | EU714187 |
| CA01 | 16S-23S ITS | EU714190 |
| CA02 | 16S-23S ITS | EU714191 |
| NM01 | Gyrase-B | EU714196 |
| NM02 | Gyrase-B | EU714197 |
| AZ03 | Gyrase-B | EU714198 |
| AZ04 | Gyrase-B | EU714199 |
| NM01 | RST (RNA pol Sigma-70) | EU714203 |
| NM02 | RST (RNA pol Sigma-70) | EU714204 |
| AZ01 | RST (RNA pol Sigma-70) | EU714200 |
| AZ03 | RST (RNA pol Sigma-70) | EU714201 |
| AZ04 | RST (RNA pol Sigma-70) | EU714202 |
| NM01 | HL (hypothetical protein) | EU714205 |
| NM02 | HL (hypothetical protein) | EU714206 |
| AZ03 | HL (hypothetical protein) | EU714207 |
| AZ04 | HL (hypothetical protein) | EU714208 |
| NM grape 1 | HL (hypothetical protein) | EU714209 |
| NM01 | ASSR (hypothetical protein) | EU714219 |
| NM02 | ASSR (hypothetical protein) | EU714220 |
| AZ03 | ASSR (hypothetical protein) | EU714221 |
| AZ04 | ASSR (hypothetical protein) | EU714222 |
| NM01 | GSSR (noncoding) | EU714210 |
| NM02 | GSSR (noncoding) | EU714211 |
| AZ03 | GSSR (noncoding) | EU714212 |
| AZ04 | GSSR (noncoding) | EU714213 |
| NM grape 1 | GSSR (noncoding) | EU714214 |
| NM01 | OSSR (exopolyphosphatase) | EU714225 |
| NM02 | OSSR (exopolyphosphatase) | EU714226 |
| AZ03 | OSSR (exopolyphosphatase) | EU714223 |
| AZ04 | OSSR (exopolyphosphatase) | EU714224 |
| NM01 | 272 (hypothetical protein) | EF109036.1 |
| NM02 | 272 (hypothetical protein) | EF109937.1 |
| AZ03 | 272 (hypothetical protein) | EU714227 |
| AZ04 | 272 (hypothetical protein) | EU714228 |
| NM01 | FY0076 (phage primase) | EU714215 |
| NM02 | FY0076 (phage primase) | EU714216 |
| AZ03 | FY0076 (phage primase) | EU714217 |
| AZ04 | FY0076 (phage primase) | EU714218 |
| NM01 | VapD | EU714229 |
| NM02 | VapD | EU714230 |
| AZ04 | VapD | EU714231 |
| AZ05 | VapD | EU714232 |
Pathogenicity studies.
PCR-positive X. fastidiosa bacterial colonies from the NM02 and NM grape 1 samples were grown in liquid XFD2 medium at 28°C with shaking for 3 weeks. The bacteria were centrifuged to obtain a pellet, and the pellet was resuspended in succinate-citrate buffer (106 bacterial cells/ml). The resuspended bacterial cells (∼200 μl) were injected into the stems of Nicotiana benthamiana plants by using a 28-gauge needle. Control plants consisted of plants that were inoculated with the citrate-succinate buffer in a similar fashion and plants that were not inoculated. Two months later, the plants were evaluated for symptoms, and the leaves above the inoculation site were tested for X. fastidiosa by PCR with the 272-1 and 272-2 primers. Leaves from above the inoculation site were also used for plating bacterial colonies (as described above).
RESULTS
Identification of X. fastidiosa in C. tashkentensis by ELISA and PCR.
Many chitalpa plants exhibiting classic symptoms of leaf scorch were observed in New Mexico and Arizona during 2006, 2007, and 2008. The common characteristics associated with this leaf scorch were leaf chlorosis, leaf necrosis, defoliation, and branch dieback (Fig. 1). The leaves first displayed chlorosis followed by necrosis in ringlike patterns beginning at the leaf tip (Fig. 1A). Leaf tip and marginal necrosis was also a common symptom on these trees (Fig. 1B). Samples from symptomatic chitalpa plants were collected from southern New Mexico and southern Arizona and analyzed for the presence of X. fastidiosa. Samples were also collected from three nurseries in southern New Mexico that had directly imported chitalpa plants from California. ELISA analysis of chitalpa samples collected from New Mexico, Arizona, and nurseries identified 36 of 67 independent plants as positive for X. fastidiosa (Table 2). Four chitalpa samples were borderline by ELISA analysis, while the remaining 25 samples were negative for X. fastidiosa.
FIG. 1.
C. tashkentensis in Southern New Mexico exhibiting leaf scorch symptoms. (A and B) Leaf chlorosis and necrosis from symptomatic chitalpa trees. (C) Flowers of the chitalpa tree. (D) Symptomatic chitalpa tree exhibiting branch dieback.
TABLE 2.
Data from Southwest chitalpa samplesa
| Chitalpa sample | Sequence identification | Assay result
|
||
|---|---|---|---|---|
| ELISA | PCR | Bacterial colony | ||
| New Mexico | ||||
| FG1 | + | + | + | |
| FG4 | + | No test | Not plated | |
| MA1 | NM01 | + | + | + |
| WO1 | + | + | + | |
| WO1a | + | + | - | |
| WO1b | + | No test | Not plated | |
| WO2a | + | + | - | |
| WO2b | + | No test | Not plated | |
| PB1 | NM02 | + | + | + |
| CBH | NM03 | + | + | + |
| CB624 | NM04 | + | + | - |
| CB418 | - | - | - | |
| CH33 | + | + | - | |
| MA2 | NM05 | + | + | + |
| MA3 | + | + | + | |
| VP | + | No test | Not plated | |
| MA6 | NM06 | + | + | - |
| HD3 | NM07 | + | + | - |
| BM1 | - | - | - | |
| BM2 | - | - | - | |
| SW1 | NM08 | + | + | + |
| SW5 | NM09 | + | + | - |
| MO1 | NM10 | + | + | - |
| FG2 | NM11 | + | + | - |
| FG3 | NM12 | + | + | - |
| WO2 | NM13 | + | + | + |
| LVC1 | + | - | - | |
| LVC2 | - | + | - | |
| CV1 | + | No test | Not plated | |
| CV2 | Borderline | - | Not plated | |
| CV3 | - | - | Not plated | |
| HD3 | + | + | - | |
| Arizona | ||||
| SV1 | AZ01 | + | + | - |
| SV2 | AZ02 | + | + | - |
| SV3 | AZ03 | + | + | - |
| SV4 | AZ04 | + | + | - |
| SV5 | AZ05 | + | + | - |
| SV6 | AZ06 | + | + | - |
| SV7 | AZ07 | + | + | - |
| SV8 | - | - | Not plated | |
| SV9 | - | - | Not plated | |
| SV10 | - | - | Not plated | |
| SV11 | AZ11 | + | + | - |
| TU1 | - | - | Not plated | |
| TU2 | - | - | Not plated | |
| TU3 | - | - | Not plated | |
| TU4 | - | - | Not plated | |
| TU5 | - | - | Not plated | |
| TU6 | + | + | + | |
| TU7 | - | - | Not plated | |
| TU8 | - | - | Not plated | |
| TU9 | - | - | Not plated | |
| TU10 | - | - | Not plated | |
| TU11 | - | - | Not plated | |
| TU12 | - | - | Not plated | |
| TU13 | - | - | Not plated | |
| TU14 | - | - | Not plated | |
| Nursery samples | ||||
| 701 | CA1 | - | + | Not plated |
| 699 | - | - | Not plated | |
| 700 | - | - | Not plated | |
| 702 | - | - | Not plated | |
| 703 | - | - | Not plated | |
| 81 | CA2 | Borderline | + | Not plated |
| 639 | CA3 | - | + | + |
| 79 | + | + | Not plated | |
| 80 | Borderline | - | Not plated | |
| 59 | - | - | Not plated | |
| 63 | - | + | Not plated | |
The PCR result was determined to be positive or negative by the presence of a product at the correct size on an agarose gel. The bacterial colony column refers to samples that yielded X. fastidiosa colonies when cultured.
Total DNA or xylem fluid isolated from the chitalpa samples was also analyzed for X. fastidiosa by PCR amplification. PCR analysis of the chitalpa samples using previously described nested primers (29) specific for X. fastidiosa produced the expected 450-bp amplicon for 33 of the 62 samples tested by PCR (Table 2). The ELISA and PCR data were in agreement for all samples except three: LVC1, LVC2, and CA1 (Table 2). The LVC2 and CA1 chitalpa samples were found to be negative for X. fastidiosa by ELISA analysis but were positive when tested by PCR. This discrepancy is likely due to the difference in sensitivity between ELISA and PCR techniques for detecting X. fastidiosa. PCR is much more sensitive and detects dead or alive cells. The chitalpa sample LVC1 was positive for X. fastidiosa by ELISA, while the PCR amplification for the pathogen was negative. The presence of the X. fastidiosa varied spatially within the plants (data not shown), and this spatial variation may account for this discrepancy.
Culturing of X. fastidiosa from infected chitalpa plants.
X. fastidiosa was cultured from infected chitalpa trees as previously described (43). Twenty days after plating on XFD2 medium (1), small white colonies were visible. Colonies were obtained from 11 of the symptomatic chitalpa trees (Table 2). These colonies were confirmed to be X. fastidiosa by direct sequencing of amplicons produced from whole-cell PCR with the nested PCR primers performed as described above (data not shown).
Pathogenicity studies of X. fastidiosa colonies in N. benthamiana.
PCR positive X. fastidiosa bacterial colonies from NM02 and NM grape were grown in XFD2 liquid media and needle inoculated into the stems of N. benthamiana plants. Control plants consisted of noninjected N. benthamiana and N. benthamiana plants injected with succinate-citrate buffer. At 2 months after inoculation the plants were evaluated for symptoms. Five of the fourteen plants inoculated with NM02 X. fastidiosa isolate were dead. Of the remaining nine plants, eight exhibited symptoms such as chlorosis and necrosis of the leaves. One plant was asymptomatic. The plants inoculated with the NM grape X. fastidiosa resulted with one plant dead and five of the remaining six plants exhibited chlorotic symptoms on the leaves. The leaves above the inoculation site were tested for the presence of X. fastidiosa by PCR with the nested 272-1 and 272-2 primers. Six of the eight living symptomatic plants inoculated with NM02 were positive for X. fastidiosa by PCR. Five plants inoculated with the NM grape isolate were PCR positive. None of the control plants died or exhibited symptoms associated with X. fastidiosa. The control plants were also negative for X. fastidiosa by PCR. Bacterial plating from the plants resulted in bacterial colonies that were PCR positive for X. fastidiosa from the plants inoculated with the NM grape isolate. No bacterial colonies resulted from plating the NM02-inoculated plant samples.
Phylogenetic analysis of X. fastidiosa 16S ribosome including the 16S-23S ITS.
The 1,200-bp fragment amplified from the 16S ribosome including 16S-23S rRNA ITS region from the New Mexico, Arizona, and California X. fastidiosa samples were analyzed by sequence analysis. BLAST analysis of the 16S ribosome showed that these sequences are 99% similar to several strains of X. fastidiosa. BLAST analysis of the ITS regions showed that the New Mexico, Arizona, and California sequences are 99% similar to citrus CVC-9a5c (AE003849.1), the almond-Dixon (AF073251.1), and the oleander-OLS (AF073213.1) and 98% similar to the grape Temecula-1 (AE009442.1).
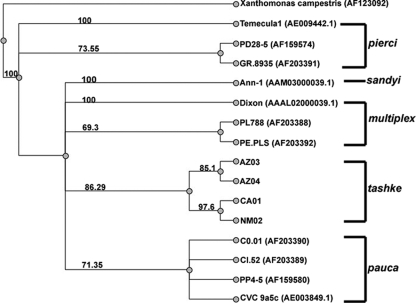
Previous work by Schaad et al. (38) and Schuenzel et al. (39) proposed that all X. fastidiosa strains can be subdivided into four main subspecies which include fastidiosa (pierci), sandyi, multiplex, and pauca based upon their ITS similarities, DNA relatedness, growth, serology, and multiple locus sequence analysis. Multiple sequence alignments were constructed using the 16S ribosome and ITS consensus sequences from representative members of four subspecies fastidiosa (piercei), sandyi, multiplex, and pauca (obtained directly from the Schaad et al. [38] or GenBank). A phylogenetic tree was generated from this alignment by using the maximum-likelihood method, which was bootstrapped 1,000 times and includes Xanthomonas campestris as the outgroup (Fig. 2). This 16S ribosome and ITS-based tree indicates that all of the chitalpa X. fastidiosa (California, Arizona, and New Mexico) sequences are more closely related to each other than any other described X. fastidiosa strain. All of these sequences cluster together on a single well-supported branch that is separate from the other distinct subspecies branches. Based on this analysis of the 16S ribosome and ITS sequences, the X. fastidiosa chitalpa group is equally related to the multiplex and pauca genotypes and less related to the pierci and sandyi groups.
FIG. 2.
Phylogram of the 16S ribosome and ITS region. Included in this phylogenetic tree are the isolates from chitalpa obtained from New Mexico, Arizona, and California. The sequences for the subgroups pierci, multiplex, and pauca were taken from Schaad et al. (38), and the sequences obtained from GenBank are noted with the GenBank accession numbers. The maximum-likelihood method using PAUP 4.0 was utilized to make the tree. The tree was bootstrapped 1,000 times, and bootstrapping consensus percentages are shown on the tree with Xanthomonas as the outgroup.
Similar phylogenetic trees were also obtained using alternative methods such as discrete character parsimony algorithm (version 3.67), along with nearest-neighbor methods, including Hasegawa-Kishino-Yano, Tamuri-Nei, and Jukes-Cantor (data not shown).
Multilocus phylogenetic analysis.
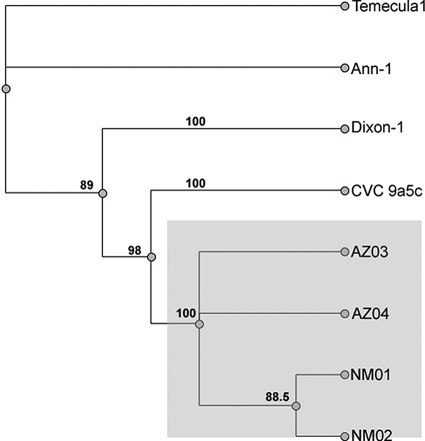
Analysis of the 16S ribosome and ITS region suggested that the chitalpa from the New Mexico and Arizona groups are highly related to each other and distinct from any other previously described strains of X. fastidiosa. To increase the resolution of the phylogenetic analysis nine additional loci were examined for the New Mexico and Arizona groups. These loci include gyrase-B; the simple sequence repeat sequences ASSR (codes for a conserved hypothetical protein), OSSR (exopolyphosphatase), and GSSR (noncoding region) (20); sequences specific to X. fastidiosa, including FY0076 (this includes the coding region for a phage-related protein) (http://cropdisease.ars.usda.gov), HL (hypothetical protein) (10), and RST (sigma-70 factor) (24); and the virulence-associated D (VapD) gene. The corresponding sequences for X. fastidiosa strains from CVC 9a5c, Temecula-1, Almond (Dixon), and Oleander (Ann-1) were obtained from GenBank. These loci were analyzed individually and then linked together into one concatameric sequence. Since sequences for all of these loci do not exist for other related species this analysis was performed with the four fully sequenced X. fastidiosa strains (CVC 9a5c, Temecula-1, Oleander, and Almond) without an outgroup. The tree generated from alignment of these concatameric sequences affirms the results obtained from 16S ribosome and ITS-based analysis further, indicating that the New Mexico and Arizona isolates are more closely related to each other than to other reported X. fastidiosa isolates (Fig. 3). Also similar to the 16S ribosome and ITS results, the concatemer-based results indicate that the X. fastidiosa isolates from chitalpa (New Mexico and Arizona isolates) cluster into a unique clade that is separate from those containing other recognized subspecies. Interestingly, the total concatameric sequences from the chitalpa strains are most similar to the CVC 9a5c isolate on this tree (Fig. 3).
FIG. 3.
Concatameric phylogram. Sequences from eight different loci were linked for each X. fastidiosa isolate. These concatamers were used to make nucleotide alignments and phylogenetic trees using Geneious Pro v4.0.4. The maximum-likelihood method using PAUP 4.0 was utilized to make the tree. The tree was bootstrapped 1,000 times, and bootstrapping consensus percentages are shown on the tree. The concatamers included OSSR, ASSR, GSSR, gyrase-B, 272-1 (nested), RST, HL, and FY0076 sequences that were separated by four N′s to keep spatial alignment. Isolates included the New Mexico and Arizona isolates, as well as the four completely sequenced strains of X. fastidiosa CVC 9a5c (GenBank accession AE003849.1), Temecula-1 (AE009442.1), Oleander (Ann-1) (AAM03000039.1), and almond (Dixon) (AAAL02000003.1).
VapD.
Virulence-associated proteins are utilized by a wide range of pathogenic bacteria. In particular, VapD is found in Dichelobacter, Haemophilus, and Helicobacter (16, 22). The X. fastidiosa CVC 9a5c strain was found to contain a VapD gene on pXF51 (22). The VapD protein from CVC 9a5c is similar to a VapD found in Riemerella anatipestifer and Actinobacillus actinomycetemcomitans (5). The function of this protein is unknown, although microarray analysis with the CVC 9a5c strain indicates that it may be turned on during heat stress (27). In silico analysis of sequenced X. fastidiosa strains identified a putative chromosomal vapD gene in the oleander strain (Ann-1). However, none of the other X. fastidiosa strains with complete sequences available contain an apparent homologue, nor have any other vapD sequences been reported for other X. fastidiosa isolates. Analysis of the putative chromosomal vapD gene within the Ann-1 genome has a contiguous open reading frame of 103 amino acids that are 98% identical to the CVC 9a5c VapD amino acid sequence. However, there is a significant gap within the N-terminal coding sequence compared to the CVC 9a5c vapD gene. The open reading frame begins with 14 amino acids that differ from the CVC 9a5c VapD by only one amino acid. This is then followed by a gap of 31 nucleotides that causes a frameshift within the coding sequence. The coding sequence for the 103-amino-acid ORF follows this gap and is thus out of frame with the sequences coding for the apparent N-terminal amino acids of the VapD protein. Thus, it is not clear whether the vapD-related sequence in the Ann-1 strain represents a real gene. It is possible that the Ann-1 strain contains a nonfunctional vapD gene that was acquired through horizontal exchange and has since become inactivated through a frame shifting deletion in the N-terminal coding region. Alternatively, the Ann-1 strain may be utilizing a truncated version of VapD where the N-terminal 14 amino acids are bypassed. The following analysis involving the putative oleander Ann-1 vapD was performed with the second contiguous open reading frame. The vapD from oleander (Ann-1) and CVC 9a5c X. fastidiosa share a nucleotide identity of 93% and share 98% identity at the amino acid level.
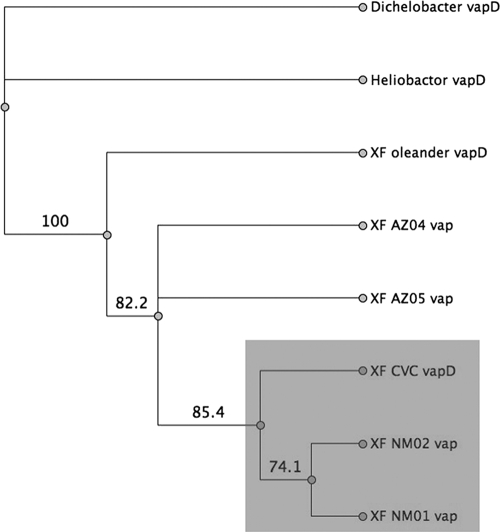
VapD PCR products were amplified from the New Mexico and Arizona isolates using primers designed from the CVC 9a5c vapD sequence. Sequencing of the amplicons showed that they contained one contiguous open reading frame coding for VapD-related protein sequences. The vapD sequences from the New Mexico and Arizona strains did not contain the frame-shifting deletion observed in the Ann-1 genomic vapD sequence. BLAST analysis indicates that the vapD sequences from the New Mexico isolates are 96% identical to vapD from CVC 9a5c and 94% identical to the partial vapD from the Oleander (Ann-1) strain, while the Arizona vapD PCR products are 94% identical to the CVC 9a5c and 91% identical to the partial Oleander (Ann-1) vapD at the nucleotide level. BLAST analysis of the deduced amino acid sequences for the NM VapD protein indicate that they share 99% identity to the CVC 9a5c VapD and 97% identity to the partial oleander (Ann-1) VapD. The Arizona VapD protein products share 86% identity with the CVC 9a5c VapD and 84% identity with the partial oleander (Ann-1) VapD protein. The deduced VapD amino acid sequences from the New Mexico and Arizona isolates were compared to those from the CVC 9a5c and Oleander (Ann-1) strains and VapD sequences from Dichelobacter (GenBank accession no. YP_001209) and Helicobacter (GenBank accession no. NP_207759), with the Dichelobacter VapD sequence serving as the outgroup. This alignment compared the entire VapD protein sequence including the gap at the N-terminal end of the oleander (Ann-1) VapD. Interestingly, the New Mexico VapD proteins cluster together and are most closely related to the VapD from the CVC 9a5c strain, whereas the Arizona VapD protein sequences cluster together on a separate branch (Fig. 4).
FIG. 4.
Protein VapD phylogram. VapD translated sequences from the New Mexico and Arizona X. fastidiosa isolates were aligned with the VapD protein sequences from CVC 9a5c (GenBank accession no. NP_061707) and Oleander (Ann-1) (GenBank accession no. YP_001209) strains obtained from GenBank (www.ncbi.nlm.nih.giv). VapD from Helicobacter pylori (GenBank accession no. NP_207759) and Dichelobacter (GenBank accession no. YP_001209) were also included in the alignment, and Dichelobacter was used as the outgroup for the phylogenetic tree. The Jukes-Cantor neighbor-joining method was used, and results were bootstrapped 1,000 times. Initial protein alignments were completed by using the Global Alignment method with a Blossum62 as the cost matrix.
DISCUSSION
Environmental pressures placed on chitalpa trees, such as limited water and high temperatures, have previously been proposed to be the cause of the leaf scorch symptoms. However, the symptoms observed on these chitalpa trees—chlorosis, leaf necrosis, defoliation, and branch dieback—were reminiscent of symptoms described for plants infected with X. fastidiosa. Samples from chitalpa trees in New Mexico, Arizona, and imported into nurseries were collected and tested positive for the presence of X. fastidiosa by ELISA and PCR assays. Although the link between X. fastidiosa and leaf scorch symptoms is unclear, it should be noted that most of the newly imported trees exhibited mild leaf scorch symptoms. Interestingly, the mild symptoms in these trees correlated with lower ELISA readings compared to some of the severely symptomatic trees sampled in the field. The data indicate that X. fastidiosa-infested chitalpa are widely distributed across the southwestern United States, being present at every location sampled, and that similar X. fastidiosa strains are also present in nursery stock being distributed across the southwestern United States.
The pathogenicity study indicated that cultures of the X. fastidiosa strain associated with chitalpa and the Pierce's disease outbreak in New Mexico are pathogenic in N. benthamiana. In a previous study, Nicotiana tabacum was shown to be a suitable host for X. fastidiosa assays, in particular with Pierce's disease strains and almond leaf scorch disease strains (9). Pathogenicity testing was performed on the New Mexico strains of X. fastidiosa by inoculation of liquid cultures of chitalpa or New Mexico-Pierce's disease strains into a different species of tobacco, N. benthamiana. Plants inoculated with these strains developed symptoms consistent with X. fastidiosa infection. Interestingly, further examination of chitalpa trees and the X. fastidiosa cultures showed that they contained both X. fastidiosa and an endophytic Methylobacterium sp. that apparently cocultured with X. fastidiosa (data not shown). An interaction between Methylobacterium and X. fastidiosa was first observed in citrus plants infected with CVC (17). Since our pathogenicity studies were conducted with mixed cultures we are unable to directly establish that the chitalpa strains of X. fastidiosa are pathogenic. However, inoculation of pure Methylobacterium cultures into N. benthamiana resulted in no symptoms observed with the plants (data not shown). Therefore, the observed disease cannot be attributed to the cocultured Methylobacterium. Although the pathogenicity study indicates that this particular X. fastidiosa strain is likely pathogenic in tobacco, we have not yet completed Koch's postulates. The origin of X. fastidiosa in the southwest chitalpa trees is unknown. The observation that chitalpa across the Southwest are colonized by similar strains of X. fastidiosa, which are distinct from those previously reported, suggests that X. fastidiosa was introduced into chitalpa prior to the trees being distributed across the Southwest. This could have occurred through one of the parents that was used to make the chitalpa hybrid or from an area where chitalpa are produced and distributed. Limited testing for X. fastidiosa in local desert willow and catalpa trees resulted in only one catalpa tree that was positive for X. fastidiosa, while no desert willow trees tested positive for X. fastidiosa (data not shown). The association of this new X. fastidiosa strain with the first outbreak of Pierce's disease in New Mexico indicates that the frequently infected chitalpa trees may be a reservoir for this disease. Although stable populations of known insects known to vector Pierce's disease have never been observed in New Mexico, the presence of known X. fastidiosa vector populations in neighboring states suggests that appropriate vectors may exist, at least transiently or in low numbers, in this area. Bluegreen sharpshooters were caught in traps near nurseries in central New Mexico, and bluegreen and smoke-tree sharpshooters were also trapped in southwestern New Mexico, suggesting that these insects enter New Mexico at least occasionally (N. Goldberg and C. Sutherland, unpublished data). Also, there are native sharpshooters present in New Mexico. These sharpshooters are not specifically noted as vectors for X. fastidiosa transmission, but they are xylem feeders and are potentially capable of vectoring X. fastidiosa.
Phylogenetic analysis of 10 different loci of the X. fastidiosa isolates from the southwest chitalpa samples revealed that these X. fastidiosa isolates are more genetically related to each other than to other previously described strains of X. fastidiosa (Fig. 2, 3, and 4). Analysis of the 16S-23S ribosomal ITS region resulted in five distinct clusters corresponding to the four previously established subspecies (pierci, pauca, sandyi, and multiplex) and a new separate fifth cluster containing all of the chitalpa isolates (Fig. 2). Previous work classifying X. fastidiosa subspecies suggested that the ITS sequence can fully differentiate strains into their specific subspecies (38, 39). These strains clustered as previously reported with the chitalpa and NM grape X. fastidiosa isolates clustering on a separate unique branch. Based on this analysis, the chitalpa isolates appear to be a separate unique subspecies.
Multiple locus analysis was utilized to further evaluate phylogenetic relationships between different isolates and strains of X. fastidiosa. Multilocus approaches used by a number of other groups included RAPD [random(ly) amplified polymorphic DNA] analysis, simple sequence repeats, and single nucleotide polymorphism analysis (11, 13, 20, 23, 37, 42). Concatameric analysis of sequences from multiple loci is often utilized to obtain a more robust and comprehensive view of phylogenetic relationships and was used to identify sandyi as a separate subspecies of X. fastidiosa (21, 26, 39, 42). Sequences from multiple loci, including gyrase-B, simple sequence repeats, and specific X. fastidiosa genes, were linked together into one concatameric sequence for phylogenetic analysis. Comparison of the concatameric sequences with the four completely sequenced strains of X. fastidiosa (CVC 9a5c, Temecula-1, Oleander [Ann-1], and Almond [Dixon]) further indicated that the New Mexico and Arizona isolates are most similar to each other and then to the CVC 9a5c strain. This analysis supports that the New Mexico and Arizona strains compose a new subgroup that is independent from the previously described fastidiosa (piercei), sandyi, multiplex, and pauca subspecies.
In addition to looking at genomic elements common to all characterized X. fastidiosa isolates, genes that may have had the opportunity for easy horizontal transfer, including the vapD gene were also considered. This gene has only been described in the X. fastidiosa strain CVC 9a5c strain, where it is hypothesized to have transferred into CVC 9a5c by phage mediated horizontal transfer (22). The New Mexico and Arizona isolates were found to contain vapD sequences and the New Mexico vapD sequences were most closely related to the CVC 9a5c vapD (Fig. 4). The presence of vapD sequences and their similarity to CVC 9a5c independently suggests the relatedness of the New Mexico isolates to the CVC strain, as implied by the analysis of other loci (Fig. 3). However, until further elucidation of the presence of other vapD sequences in X. fastidiosa strains occurs, this analysis is limited.
The phylogenetic analysis reported here indicates that these isolates comprise a unique subspecies that is different than the four previously defined. We suggest calling this subspecies X. fastidiosa subsp. tashke due to its discovery in and wide association with C. tashkentensis.
Acknowledgments
We thank Richard Heerema for the use of the pressure chamber. We also thank Rio Stamler and Jenna Painter for technical support.
This study was supported by USDA grant 2006-06129 and NIGMS grant S06 GM08136.
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Almeida, R. P., R. Mann, and A. H. Purcell. 2004. Xylella fastidiosa cultivation on a minimal solid defined medium. Curr. Microbiol. 48:368-372. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Barnard, E. L., E. C. Ash, D. L. Hopkins, and R. J. McGovern. 1998. Distribution of Xylella fastidiosa in oaks in Florida and its association with growth decline in Quercus laevis. Plant Dis. 82:569-572. [DOI] [PubMed] [Google Scholar]
- 4.Buzombo, P., J. Jaimes, V. Lam, K. Cantrell, M. Harkness, D. McCullough, and L. Morano. 2006. An American hybrid vineyard in the Texas Gulf Coast: analysis within a Pierce's disease hot zone. Am. J. Enol. Viticulture 57:347-355. [Google Scholar]
- 5.Catani, C. F., A. R. Azzoni, D. P. Paula, S. F. S. Tada, L. K. Rosselli, A. P. de Souza, and T. Yano. 2004. Cloning, expression, and purification of the virulence-associated protein D from Xylella fastidiosa. Protein Expr. Purif. 37:320-326. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C. J., M. Garnier, L. Zreik, V. Rossetti, and J. M. Bove. 1993. Culture and serological detection of the xylem-limited bacterium causing citrus variegated chlorosis and its identification as a strain of Xylella fastidiosa. Curr. Microbiol. 27:137-142. [DOI] [PubMed] [Google Scholar]
- 7.Davis, M. J., A. H. Purcell, and S. V. Thomson. 1978. Pierce's disease of grapevines isolation of the casual bacterium. Science 199:75-77. [DOI] [PubMed] [Google Scholar]
- 8.Drummond, A. J., K. M. Heled, R. Moir, T. Thierer, B. Ashton, A. Wilson, and S. Stones-Havas. 2006. Geneious, v2.5. Biomatters, Ltd., Auckland, New Zealand.
- 9.Francis, M., E. L. Civerolo, and G. Bruening. 2008. Improved bioassay of Xylella fastidiosa using Nicotiana tabacum cultivar SRI. Plant Dis. 92:14-20. [DOI] [PubMed] [Google Scholar]
- 10.Francis, M., H. Lin, J. Cabrera-La Rosa, H. Doddapaneni, and E. L. Civerolo. 2006. Genome-based PCR primers for specific and sensitive detection and quantification of Xylella fastidiosa. Eur. J. Plant Pathol. 115:203-213. [Google Scholar]
- 11.Hendson, M., A. H. Purcell, D. Chen, C. Smart, M. Guilhabert, and B. Kirkpatrick. 2001. Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa. Appl. Environ. Microbiol. 67:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez-Martinez, R., H. S. Costa, D. A. Cooksey, and F. Wong. 2006. Documentation and characterization of Xylella fastidiosa strains in landscape hosts. Pierce's Dis. Res. Symp. 2006:191-197. [Google Scholar]
- 13.Hernandez-Martinez, R., K. A. de la Cerda, H. S. Costa, D. A. Cooksey, and F. P. Wong. 2007. Phylogenetic relationships of Xylella fastidiosa strains isolated from landscape ornamentals in southern California. Phytopathology 97:857-864. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Martinez, R., T. R. Pinckard, H. S. Costa, D. A. Cooksey, and F. P. Wong. 2006. Discovery and characterization of Xylella fastidiosa strains in southern California causing mulberry leaf scorch. Plant Dis. 90:1143-1149. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins, D. L. 1989. Xylella fastidiosa: xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 27:271-290. [Google Scholar]
- 16.Katz, M. E., R. A. Strugnell, and J. I. Rood. 1992. Molecular characterization of a genomic region associated with virulence in Dichelobacter nodosus. Infect. Immun. 60:4586-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lacava, P. T., W. L. Araujo, J. Marcon, W. Maccheroni, Jr., and J. L. Azevedo. 2004. Interaction between endophytic bacteria from citrus plants and the phytopathogenic bacteria Xylella fastidiosa, causal agent of citrus-variegated chlorosis. Lett. Appl. Microbiol. 39:55-59. [DOI] [PubMed] [Google Scholar]
- 18.Li, W. B., W. D. Pria, C. Teixeira, V. S. Miranda, A. J. Ayres, C. F. Franco, M. G. Costa, C. X. He, P. I. Costa, and J. S. Hartung. 2001. Coffee leaf scorch caused by a strain of Xylella fastidiosa from citrus. Plant Dis. 85:501-505. [DOI] [PubMed] [Google Scholar]
- 19.Li, X., and S. H. deBoer. 1995. Selection of polymerase chain reaction primers from an RNA intergenic spacer region for specific detection of Clavibacter michiganensis subsp. spedonicus. Phytopathology 85:837-842. [Google Scholar]
- 20.Lin, H., E. L. Civerolo, R. Hu, S. Barros, M. Francis, and M. A. Walker. 2005. Multilocus simple sequence repeat markers for differentiating strains and evaluating genetic diversity of Xylella fastidiosa. Appl. Environ. Microbiol. 71:4888-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manso-Silvan, L., X. Perrier, and F. Thiancourt. 2007. Phylogeny of the Mycoplasma mycoides cluster based on analysis of five conserved protein coding sequences and possible implications for the taxonomy of the group. Int. J. Syst. Evol. Microbiol. 57:2247-2258. [DOI] [PubMed] [Google Scholar]
- 22.Marques, M. V., A. M. da Silva, and S. L. Gomes. 2001. Genetic organization of plasmid pXF51 from the plant pathogen Xylella fastidiosa. Plasmid 45:184-199. [DOI] [PubMed] [Google Scholar]
- 23.Martinati, J. C., F. T. Pacheco, V. F. de Miranda, and S. M. Tsai. 2005. Phylogenetic relationships of Xylella fastidiosa strains based on 16S-23S rDNA sequences. Curr. Microbiol. 50:190-195. [DOI] [PubMed] [Google Scholar]
- 24.Minsavage, G. V., C. M. Thompson, D. L. Hopkins, R. M. V. B. C. Leite, and R. E. Stall. 1994. Development of a polymerase chain-reaction protocol for detection of Xylella fastidiosa in plant tissue. Phytopathology 84:456-461. [Google Scholar]
- 25.Mircetich, S. M., S. K. Lowe, W. J. Moller, and G. Nyland. 1976. Etiology of almond leaf scorch disease and transmission of the casual agent. Phytopathology 66:17-24. [Google Scholar]
- 26.Nightingale, K. K., K. Windham, and M. Wiedmann. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunes, L. R., Y. B. Rosato, N. H. Muto, G. M. Yanai, V. S. da Silva, D. B. Leite, E. R. Goncalves, A. A. de Souza, H. D. Coletta, M. A. Machado, S. A. Lopes, and R. C. C. Oliveira. 2003. Microarray analyses of Xylella fastidiosa provide evidence of coordinated transcription control of laterally transferred elements. Genome Res. 13:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen, R. T., and T. G. Ranney. 2006. Susceptibility of Catalpa, Chilopsis, and hybrids to powdery mildew and catalpa sphinx larvae. Hortscience 41:1629-1634. [Google Scholar]
- 29.Pooler, M. R., I. S. Myung, J. Bentz, J. Sherald, and J. S. Hartung. 1997. Detection of Xylella fastidiosa in potential insect vectors by immunomagnetic separation and nested polymerase chain reaction. Lett. Appl. Microbiol. 25:123-126. [DOI] [PubMed] [Google Scholar]
- 30.Purcell, A. H. 1997. Xylella fastidiosa: a regional problem or a global threat. J. Plant Pathol. 79:99-105. [Google Scholar]
- 31.Purcell, A. H., S. R. Saunders, M. Hendson, M. E. Grebus, and M. J. Henry. 1999. Causal role of Xylella fastidiosa in oleander leaf scorch disease. Phytopathology 89:53-58. [DOI] [PubMed] [Google Scholar]
- 32.Raju, B. C., J. M. Wells, G. Nyland, R. H. Brlansky, and S. K. Lowe. 1982. Plum leaf scald isolation and culture and pathogenicity of the casual agent. Phytopathology 72:1460-1466. [Google Scholar]
- 33.Randall, J. J., M. Radionenko, J. M. French, N. P. Goldberg, and S. F. Hanson. 2007. First report of Pierce's disease in New Mexico. Plant Health Prog. doi: 10.1094/PHP-2007-1002-01-RS. [DOI]
- 34.Randall, J. J., M. Radionenko, J. M. French, M. W. Olsen, N. P. Goldberg, and S. F. Hanson. 2007. Xylella fastidiosa detected in New Mexico in chitalpa, a common landscape ornamental plant. Plant Dis. 91:329. [DOI] [PubMed] [Google Scholar]
- 35.Redak, R. A., A. H. Purcell, J. R. Lopes, M. J. Blua, R. F. Mizell III, and P. C. Andersen. 2004. The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu. Rev. Entomol. 49:243-270. [DOI] [PubMed] [Google Scholar]
- 36.Sanderlin, R. S., and K. I. Heyderich-Alger. 2000. Evidence that Xylella fastidiosa can cause leaf scorch disease of pecan. Plant Dis. 84:1282-1286. [DOI] [PubMed] [Google Scholar]
- 37.Scally, M., E. L. Schuenzel, R. Stouthamer, and L. Nunney. 2005. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl. Environ. Microbiol. 71:8491-8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaad, N. W., E. Postnikova, G. Lacy, M. Fatmi, and C. J. Chang. 2004. Xylella fastidiosa subspecies: X. fastidiosa subsp. piercei, subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., and X. fastidiosa subsp. pauca subsp. nov. Syst. Appl. Microbiol. 27:763-764. [DOI] [PubMed] [Google Scholar]
- 39.Schuenzel, E. L., M. Scally, R. Stouthamer, and L. Nunney. 2005. A multigene phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa. Appl. Environ. Microbiol. 71:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 41.Wells, J. M., B. C. Raju, and G. Nyland. 1983. Isolation culture and pathogenicity of the bacterium causing phony disease of peach Prunus persica. Phytopathology 73:859-862. [Google Scholar]
- 42.Wickert, E., M. A. Machado, and E. G. Lemos. 2007. Evaluation of the genetic diversity of Xylella fastidiosa strains from citrus and coffee hosts by single-nucleotide polymorphism markers. Phytopathology 97:1543-1549. [DOI] [PubMed] [Google Scholar]
- 43.Wistrom, C., and A. H. Purcell. 2005. The fate of Xylella fastidiosa in vineyard weeds and other alternate hosts in California. Plant Dis. 89:994-999. [DOI] [PubMed] [Google Scholar]