Abstract
Antibiotics such as chlortetracycline (CTC) have been used to promote growth of pigs for decades, but concerns over increased antibiotic-resistant infections in humans have prompted the development of alternative strategies. Developing alternatives to antibiotic growth promoters (AGPs) could be informed by information on the mechanisms of growth promotion, notably, how AGPs affect the microbial populations of the gastrointestinal tract. Pigs from three sows were aseptically delivered by cesarean section. Six piglets were distributed to each of two foster mothers until weaning, when piglets were fed a diet with or without 50 mg/kg CTC for 2 weeks. The ileal bacterial microbiota was characterized by using a cultivation-independent approach based on DNA extraction, PCR amplification, cloning, and sequencing of the 16S rRNA gene pool. The ileal and mucosal communities of these growing pigs were dominated by Lactobacillus bacteria, various members of the family Clostridiaceae, and members of the poorly known genus Turicibacter. Overall, CTC treatment resulted in three shifts: a decrease in Lactobacillus johnsonii, an increase in L. amylovorus, and a decrease in Turicibacter phylotypes. The composition of the microbiota varied considerably between individual pigs, as revealed by shared operational taxonomic units (OTUs) and similarity (SONS) analysis (θYC values). While the observed variation between untreated pigs obscured the possible effect of CTC, ∫-LIBSHUFF and SONS analyses of pooled libraries indicated a significant shift due to CTC in both the lumen and the mucosa, with some OTUs unique to either treated or control ileum. DOTUR analysis revealed little overlap between control and treated communities at the 3% difference level, indicating unique ileal communities in the presence of CTC.
Antibiotics have been used to promote animal growth for over 50 years. Antibiotic growth promoters (AGPs) such as tylosin, bacitracin, virginiamycin, and chlortetracycline (CTC) have been fed to pigs, chickens, and other animals to promote growth through increased feed intake, weight gain, and improved herd health (7, 36). Use of AGPs has come under increasing pressure with the growing consensus that their use leads to increased antibiotic-resistant infections in humans via generation of reservoirs of antibiotic-resistant bacteria that may enter the food chain through contamination (38, 46). The increasing concerns about antibiotic resistance have raised questions about whether the potential risks are worth the beneficial effects (44). Development of non-antibiotic-based alternative strategies to promote animal growth may benefit through increased understanding of AGP mechanisms of growth promotion.
The growth-promoting impact of antibiotics was first described in the 1940s, and their use soon became routine (29, 35). The gastrointestinal (GI) tract harbors a great diversity of bacteria at a very high density (27). The increased growth and feed efficiency promoted by AGPs may be due to alteration of the microbiota of the GI tract. Early hypotheses focused on the suppression of pathogenic bacteria (19), but the broad-spectrum antibiotics used as growth promoters do not target specific species. Suggested mechanisms of action have included suppression of subclinical infections, a decrease in the levels of growth-depressing bacterial metabolites, decreased consumption of nutrients by intestinal microbiota, and improvement of nutrient uptake due to a thinner intestinal wall (14, 48). Data on the effect of AGPs on pig intestinal microbiota are needed in order to determine the relative contributions of the various proposed mechanisms. Much of the evidence available points to the action of antibiotics on intestinal bacteria as the main component responsible for the growth effect on animals (17, 20, 36).
Traditional culture methods have provided some insights into pig GI microbiota, but culture-independent techniques utilizing analysis of rRNA genes have revealed a far greater diversity. Culture-independent methods have also helped to further our understanding of bacterial population dynamics and the complex interplay between the host and pathogenic and nonpathogenic bacteria. The construction of a large 16S rRNA bacterial clone library from the pig GI tract identified 375 phylotypes by using a similarity criterion of 97% (27). Studies utilizing denaturing gradient gel electrophoresis have shown the microbial variances between compartments of the pig intestinal tract, the effect of the diet on microbial communities of the colon, and the ileal microbiota changes produced by the use of several types of AGP (5, 28, 45). Each technique can hold its own bias or limitation, but combinations of fingerprinting and PCR techniques have led to a greater understanding of the composition of pig GI microbiota and their ecology (16, 49, 50).
Studies on the effect of antibiotics on intestinal microbiology have focused on colonic or fecal microbiota because bacterial densities are highest (14) and sampling is noninvasive, allowing temporal studies. Yet, nutrient uptake occurs primarily in the small intestine, the region where bacterial activity would therefore have the greatest influence on growth (14). Demands on the GI tract to respond to bacteria by increased mucus production occur primarily in the small intestine (13). The main growth-promoting effect of antibiotics is therefore more likely to occur in the small intestine, specifically in the ileum, where bacterial numbers have reached a high density. One study showed that AGPs, including bacitracin, CTC, and tylosin, caused a shift in the ileal microbial profile of pigs (5). In that study, only one pig was used per treatment, so the basal variation in microbiota between individuals was not taken into account.
The objective of this study was to examine how the AGP CTC affects the microbial community of the porcine ileum. To account for variation in the intestinal microbiota as influenced by both antenatal and postnatal environment, pigs from three separate sows were aseptically delivered by cesarean (C) section and distributed to two foster mothers until weaning, when piglets were fed a diet either with or without the AGP CTC. A cultivation-independent approach based on DNA extraction, PCR amplification, and cloning and sequencing of the 16S RNA gene was taken to characterize the pig ileal microbiota.
MATERIALS AND METHODS
Animals and treatments.
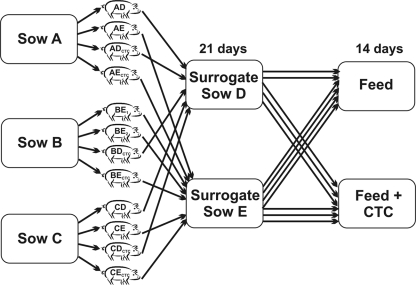
A single SP-1 (Duroc × Yorkshire) boar and three line 13 (Landrace × Yorkshire) healthy sows (AusGene International Inc., Gridley, IL) selected from the University Swine Unit at South Dakota State University were mated. Sows tested negative for porcine respiratory and reproductive syndrome virus and rotavirus. All animal protocols were approved by the South Dakota State University (SDSU) Institutional Animal Care and Use Committee (IACUC). Half-sib piglets from these sows were delivered 2 to 3 days before the expected parturition date by C section. In total, 12 piglets, 4 from each of the three sows, were used for this study (Fig. 1). The sows were named A, B, and C, respectively, and were ear notched accordingly. By using prostaglandin to induce labor, the parturition of two sows required as surrogate mothers (D and E) was synchronized to precede procurement of the experimental piglets. Two pigs each originating from sows A, B, and C were then fostered onto each of the two surrogate sows (D and E). The two foster mothers and their surrogate litters were kept in separate isolation rooms, thus allowing colonization of the pigs' intestines with the normal intestinal microbiota derived from the specific foster mothers. After 21 days, piglets were weaned and regrouped as control (antibiotic-free) and antibiotic-fed groups (n = 6). The fostered control and antibiotic-fed groups were formed by including piglets from each foster sow. Fostering on nurse sows and redivision at weaning were done in an attempt to nullify the effect of the antenatal environment on the intestinal microbiota composition of experimental piglets. All three groups of animals were fed a weanling diet ad libitum (see Table S2 in the supplemental material), except for the addition of CTC at 50 mg/kg to the feed of the antibiotic-fed group, as described previously (15). Piglets were named for their mother (A, B, or C), their foster mother (D or E), and the feed type (control or CTC). A piglet from sow A fostered on sow D and fed CTC was named piglet ADCTC. During fostering, an error occurred so that three piglets from the sow B litter were placed with surrogate sow E instead of two each with sows D and E.
FIG. 1.
Diagram outlining the sources and treatment of piglets used in this study. Piglets were delivered by C section from one of three sows (A, B, or C) and placed with one of two foster sows (D or E) for 21 days, after which they were weaned and fed a diet either with or without 50 mg/kg CTC.
Sample collection.
Piglets were sacrificed at 35 (piglets A), 36 (piglets B), and 37 (piglets C) days postnatally. Piglets from each group were euthanized by using a standard protocol approved by the SDSU IACUC. The abdomen was opened immediately, mesenteries were removed, and the ileum bearing a continuous Peyer's patch was located. A distal segment (20 cm) of the ileum was removed, and the ingesta (lumen material) was removed by gentle squeezing. The ileal segment was filled with cold, sterile phosphate buffer (10 mM, pH 7.0) and massaged gently to suspend bacteria in the mucosa layer. Lumen and mucosa samples were transferred into vials and stored on ice for subsequent DNA extraction.
Gene library construction.
Bacterial DNA was extracted from 180 to 220 mg of lumen or mucosa extract with a QIAmp DNA stool mini kit (Qiagen). PCR of bacterial 16S rRNA genes was performed with bacterial universal primers S-D-Bact-0008-a-S-20 and S-*-Univ-1492-a-A-19 (27). Each PCR was performed with a 50-μl reaction mixture containing 1× PCR buffer, 2 mM MgCl2, 2.5 μl dimethyl sulfoxide, 0.5 μM each primer, 200 μM deoxynucleoside triphosphates, 1.5 U Taq polymerase (Promega), and 25 ng of template DNA. Thermocycling conditions were as follows: initial denaturation at 94°C for 4 min, followed by 30 cycles of 94°C for 45 s, 56°C for 55 s, and 72°C for 1.5 min and a final elongation of 72°C for 10 min. PCR products were gel extracted with the Promega SV Gel and PCR Cleanup System. Clean PCR products were ligated into pGEM-T Easy vector (Promega) and transformed into competent Escherichia coli JM109 cells. A total of 96 recombinant clones per pig were randomly selected, and the DNA sequences were determined by the University of Washington Genomic Core with primer S-D-Bact-0008-a-S-20.
16S rRNA gene library analysis.
A total of 2,050 sequences were obtained for further analysis. All 16S rRNA gene sequences were examined with PINTAIL (2) to check for possible chimeras. By using the default settings, a total of 103 sequences were discarded. All sequences were trimmed in order to normalize their length. A further 86 sequences were discarded due to short reads (less than 610 bp) or not being classified as 16S rRNA genes. Sequences were analyzed by BLAST (1) and allocated to groups where possible. Total sequences for the treated and untreated mucus and lumen libraries were grouped by location and by treatment. To assess whether statistically significant changes in community structure were present, total and subsets of the 16S rRNA gene sequences from the lumen and mucus libraries were analyzed by ∫-LIBSHUFF (43). Sequences were first aligned in ARB (32) and used to construct a full matrix by the distance matrix ARB neighbor-joining method. The matrices were output into lower-triangular PHYLIP (11) distance matrix files and input into the ∫-LIBSHUFF program. A P value of ≤0.025 indicates a statistically supported difference in microbial libraries. For analysis in DOTUR (41), 16S rRNA gene sequences were grouped into categories, aligned in ARB, and then output into PHYLIP distance matrices as explained above for ∫-LIBSHUFF analysis. The matrices were analyzed with the furthest-neighbor algorithm, and operational taxonomic unit (OTU) data for similarity cutoffs of 5%, 3%, and 1% difference were collected. Analysis by SONS (42) yields nonparametric estimators of the ratio and richness of OTUs that were shared between libraries. The parameters Sclass and θYC generate estimates of the fraction of OTUs shared between libraries and community structure similarity. Comparisons between single pig libraries were performed with the above-mentioned alignment in ARB, DOTUR analysis, and SONS analysis. The 3% difference θYC data were collected from each pairwise comparison and used to construct a square matrix for both the lumen and mucus samples. The matrix was input into the JMP IN 5.1.2 statistical analysis program (SAS Institute, Cary, NC) and used to construct dendrograms based on Ward clustering.
Nucleotide sequence accession numbers.
The sequence data are available from the GenBank database under accession numbers FJ161980 to FJ162998 (mucus) and FJ162999 to FJ164030 (lumen).
RESULTS
The 16S rRNA gene amplicon pools from ileal lumen and mucus DNA extracts of the 12 pigs were used to generate 24 individual libraries, each with 96 clone sequences. The libraries contained 4.8% putative chimeric sequences according to Pintail analysis (see Table S1 in the supplemental material), similar to the 5% generally reported (2). After discarding putative chimeras or short sequences (<610 bp), a total of 2,050 sequences were obtained (see Table S1 in the supplemental material). Libraries from the same treatment and location were then compiled for comparative analyses. The ileal lumen and mucus communities were dominated by three groups: Lactobacillus bacteria, various members of the order Clostridiales, and members of Turicibacter, a poorly known genus (Table 1). Trends within the CTC treatment group showed three overall shifts: a decrease in Lactobacillus johnsonii, a concomitant increase in L. amylovorus, and a decrease in Turicibacter phylotypes (Table 1). Turicibacter is a gram-positive anaerobe and a member of the family Erysipelotrichaceae, but only one member, Turicibacter sanguinis, isolated from a blood culture of a febrile patient with acute appendicitis, has been formally described (3). DNA sequence analysis indicated that porcine ileal Turicibacter bacteria are distinct from T. sanguinis (data not shown). The relative abundance of Clostridiales and Enterobacteriaceae bacteria shifted only in mucus samples (Table 1). The relative abundance of Clostridiales bacteria in mucus decreased with the addition of CTC, but in the lumen, the numbers remained the same.
TABLE 1.
Numbers of 16S rRNA gene sequences in each of the individual libraries allocated by clustering with specific bacterial groups
| Site and group | No. of 16S rRNA gene sequences in indicated library
|
Total control | Total CTC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADa | ADCTC | AE | AECTC | BE1 | BDCTC | BE2 | BECTC | CD | CDCTC | CE | CECTC | |||
| Lumen | ||||||||||||||
| Turicibacter | 18 | 14 | 34 | 17 | 21 | 2 | 0 | 0 | 0 | 5 | 0 | 0 | 73 | 38 |
| L.johnsonii | 0 | 10 | 0 | 3 | 28 | 3 | 76 | 41 | 17 | 2 | 3 | 5 | 124 | 64 |
| L.amylovorus | 0 | 4 | 0 | 20 | 1 | 58 | 6 | 38 | 61 | 48 | 85 | 85 | 153 | 253 |
| Clostridiales | 63 | 51 | 52 | 43 | 18 | 20 | 4 | 1 | 0 | 28 | 0 | 1 | 137 | 144 |
| Mucus | ||||||||||||||
| Turicibacter | 13 | 15 | 45 | 5 | 54 | 1 | 9 | 0 | 3 | 2 | 8 | 0 | 132 | 23 |
| L.johnsonii | 1 | 2 | 0 | 2 | 5 | 0 | 56 | 16 | 12 | 0 | 5 | 22 | 79 | 42 |
| L.amylovorus | 0 | 6 | 0 | 0 | 4 | 73 | 1 | 49 | 35 | 54 | 46 | 12 | 86 | 194 |
| Clostridiales | 52 | 36 | 47 | 19 | 20 | 11 | 14 | 5 | 14 | 29 | 19 | 2 | 166 | 102 |
| Enterobacteriaceae | 5 | 6 | 0 | 12 | 0 | 0 | 1 | 1 | 12 | 0 | 3 | 18 | 21 | 37 |
| Ruminococcus | 10 | 2 | 0 | 21 | 4 | 2 | 0 | 4 | 0 | 0 | 2 | 3 | 16 | 32 |
Letters refer to the respective maternal (antenatal environment) and surrogate (postnatal environment) sows.
The apparent variation between libraries from pigs of the same treatment or group (Table 1) prompted interrogation of the data for possible roles played by antenatal or postnatal environment. Only the offspring of sows A and C were used for this purpose because three instead of two offspring of sow B were erroneously placed with the same surrogate mother. A ∫-LIBSHUFF analysis of the libraries from the lumen of piglets AD and AE (both from sow A; raised on surrogate sows D and E, respectively; and not fed CTC) versus CD and CE library pairs showed a significant difference between the library pairs (Table 2), although the piglets had been exposed to different postnatal environments. Similarly, analysis of the mucus libraries also revealed a statistically significant difference. The community composition of both the lumen and mucus microbiota therefore appeared to have been significantly influenced by the antenatal environment. The postnatal environment did not have a significant effect on the ileal lumen bacterial community, as the AD and CD library pair was not significantly different from the AE and CE library pair (Table 2). In contrast, the postnatal environment did have an effect on the ileal mucus community. This is in agreement with a recent report where the porcine colonic and fecal microbiota was influenced primarily by the housing environment (47).
TABLE 2.
∫-LIBSHUFF analysis of 16S rRNA gene libraries of ileal lumen and mucosa of pigs
| Comparison | Lumen
|
Mucosa
|
||||
|---|---|---|---|---|---|---|
| XY P valuea | YX P value | Interpretation | XY P value | YX P value | Interpretation | |
| Effect of antenatal environment (AD + AE vs CD + CE) | <0.0001 | <0.0001 | Significant | <0.0001 | 0.0002 | Significant |
| Effect of postnatal environment (AD + CD vs AE + CE) | 0.0770 | 0.4311 | Not significant | <0.0001 | 0.9599 | Significant |
| Entire control vs CTC community | 0.0026 | 0.0334 | Significant | <0.0001 | 0.2357 | Significant |
| Clostridiales | 0.0111 | 0.1066 | Significant | 0.0001 | 0.6004 | Significant |
| Lactobacillus | 0.0002 | 0.0005 | Significant | 0.0043 | 0.9998 | Significant |
| AD vs ADCTC | 0.8115 | 0.0311 | Not significant | <0.0001 | 0.3778 | Significant |
| AE vs AECTC | <0.0001 | 0.5459 | Significant | <0.0001 | 0.2594 | Significant |
| BE1 vs BDCTC | 0.0013 | <0.0001 | Significant | <0.0001 | <0.0001 | Significant |
| BE2 vs BECTC | 0.0012 | 0.0006 | Significant | 0.0171 | 0.0142 | Significant |
| CD vs CDCTC | <0.0001 | 0.2333 | Significant | 0.7006 | 0.1152 | Not significant |
| CE vs CECTC | 0.0033 | <0.0001 | Significant | <0.0001 | 0.7668 | Significant |
The consensus for ∫-LIBSHUFF analyses is that if either XY or YX has a P value of <0.025, the two communities are significantly different (43).
The apparent overall community shifts associated with CTC were tested by ∫-LIBSHUFF analysis to compare compiled data on CTC-treated and control libraries from the lumen and mucus. The analysis indicated a significant difference associated with addition of CTC to the diet (Table 2). In order to test whether CTC was associated with a shift in pigs from the same ante- and postnatal environments, pairwise comparisons between single pigs were performed by ∫-LIBSHUFF analysis. The results showed that CTC was associated with a significant shift in the community in each case, except for the lumen of pigs AD and the mucus of pigs CD (Table 2). As Clostridiales and Lactobacillus bacteria made up a major portion of the total libraries, the species and subspecies taxonomic richness became a point of interest. ∫-LIBSHUFF analysis of the Clostridiales sequences yielded a significant P value for the lumen and mucus libraries (Table 2), indicating a shift in species or subspecies makeup between the CTC-treated and control groups (Table 2). The Lactobacillus mucus and lumen P values were also significant, again indicating shifts between the treated and control pigs.
To further investigate the overall trends among the libraries, DOTUR was used to generate rarefaction curves at 5, 3, and 1% differences (see Fig. S1 in the supplemental material), the Shannon diversity indices (at 3% difference), and the number of OTUs (at a 3% difference) (Table 3). A difference of 5% between 16S rRNA gene sequences approximates the genus level, and differences of 3 and 1% approximate the species and strain levels, respectively (41). Rarefaction curves showed that both the lumen and mucus libraries appeared to approach saturation at the genus and species levels, but not at the strain level, where further richness was indicated. Shannon indices for both the lumen and mucus total libraries indicated a high level of biodiversity at the species level (Table 3) and especially at the strain level (see Table S3 in the supplemental material). The CTC-treated group was not associated with an upward or downward trend in the level of diversity. The Clostridiales group also showed a high level of taxonomic richness. The Lactobacillus community was much less diverse in the mucus than in the lumen. The number of OTUs in either the control or the treated group from the lumen and mucus was always lower than that in the sum of the control and CTC groups (Table 3). This indicated that several OTUs were unique to either the control or the CTC-treated group. The minimal overlap at the species level was not due to underrepresentation in the library because rarefaction curves at 3% difference indicated that the dominant ileal community members were represented (see Fig. S1 in the supplemental material). This low degree of overlap in OTU composition between the treated and control groups provided additional support to the ∫-LIBSHUFF data.
TABLE 3.
Shannon diversity indices and numbers of OTUs of selected phylotypes from DOTUR done at a similarity cutoff of 3% difference
| Group | Shannon diversity index
|
No. of OTUs
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lumen
|
Mucosa
|
Lumen
|
Mucosa
|
|||||||||
| Control | CTC | Total | Control | CTC | Total | Control | CTC | Total | Control | CTC | Total | |
| Community | 2.60 | 2.16 | 2.68 | 2.41 | 2.58 | 2.64 | 47 | 45 | 77 | 39 | 55 | 72 |
| Clostridiales | 2.07 | 2.15 | 2.25 | 1.97 | 2.28 | 2.32 | 17 | 20 | 29 | 21 | 24 | 38 |
| Lactobacillus | 1.70 | 0.96 | 1.91 | 0.75 | 0.75 | 0.84 | 20 | 16 | 34 | 3 | 9 | 10 |
In order to compare the structural overlap between individual libraries, the data were analyzed by SONS. SONS analysis yields nonparametric estimators of the ratio and richness of OTUs that are shared between libraries (42). Pairwise comparisons between single lumen and mucus libraries were performed by SONS analysis at a 3% sequence difference. The θYC value is an estimate of community structure similarity. Dendrograms obtained after conversion of θYC values to distances by Ward's clustering did not reveal a clear pattern (Fig. 2). Libraries did not cluster according to one particular factor. Offspring of sow A clustered according to the antenatal environment, responding to CTC treatment, while offspring of sows B and C clustered in some cases according to the postnatal environment or CTC treatment. Yet analysis of pooled lumen or mucus libraries indicated an overall shift (Table 4). The Sclass values were all about 0.5, indicating that many OTUs were unique to either the control or treated groups. The θYC values obtained showed that the community structures of pooled libraries were different in response to CTC.
FIG. 2.
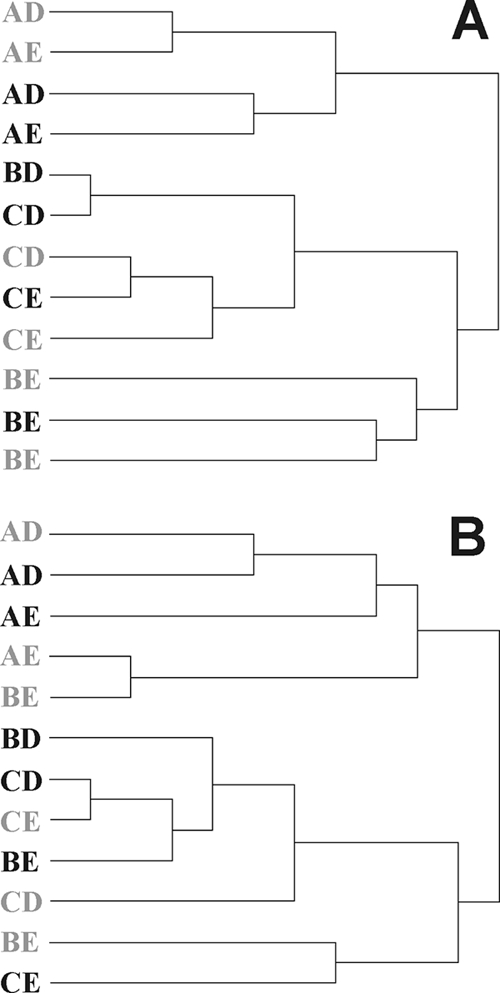
Dendrograms of the pairwise nonparametric estimates of lumen (A) and mucus (B) community structure similarity (θYC values) between various CTC-treated (gray letters) and control (black letters) pig libraries. The θYC values were calculated by SONS analysis, and the dendrograms were constructed by Ward clustering.
TABLE 4.
SONS analysis of compiled control versus CTC-treated libraries performed at a similarity cutoff of 3% difference
| Group | Sclass
|
ThetaYC
|
||
|---|---|---|---|---|
| Lumen | Mucosa | Lumen | Mucosa | |
| Community | 0.4752 | 0.4681 | 0.7685 | 0.4957 |
| Clostridiales | 0.4737 | 0.4167 | 0.9282 | 0.7276 |
| Lactobacillus | 0.5833 | 0.4615 | 0.7921 | 0.7279 |
DISCUSSION
The 16S rRNA clone library data suggested shifts in the microbial communities in the ilea of pigs fed the growth-promoting antibiotic CTC. This was supported by ∫-LIBSHUFF analysis of both the lumen and mucus libraries (Table 2). Theta (θYC) values obtained by SONS analysis showed that the community structures of pooled libraries were different in control and treated pigs (Table 4). Furthermore, the CTC-treated ileal microbiota harbored a large proportion of unique OTUs, as indicated by the low Sclass values. The surface properties of the ileal tissue of these pigs was previously characterized by lectin binding profiles and differed between the CTC-fed and control pigs, consistent with a host response to an altered bacterial community (15). Examination of both the control and CTC-treated libraries found that nearly 80 to 98.2% of the recovered sequences originated from three phylogenetic groups: Lactobacillus, Clostridiales, and Turicibacter (Table 1). Since they make up the majority of the total microbial ileal populations, they may play key roles in how changes in intestinal microbes might affect the pig.
Our results supported previous findings from mouse and human GI tract analyses (21, 51) and from pigs (47) emphasizing that each individual carries a unique intestinal bacterial community, determined primarily by its genotype and secondarily by its environment. Any putative effect of a treatment would therefore need to be determined against this background variation between individual pigs. This was underpinned by clustering of the θYC values of individual libraries (Fig. 2). The reasons underlying the establishment of unique GI microbiota in each individual pig are not clear. Piglets are exposed to a diverse array of bacteria after birth, but only some of these become established. The intestinal bacterial community develops, adapting to changes in diet, such as after weaning (25, 28). The composition of the microbiota also becomes increasingly diverse with progression through the pig GI tract (24, 39, 45). Although the experiment was not designed to address the role of the ante- and postnatal environments, ∫-LIBSHUFF results indicated that the antenatal environment plays a role in the establishment of the ileal microbiota (Table 2). The postnatal environment and housing conditions appeared to be a lesser determining factor in the ileum, contrasting the findings for porcine feces of a much larger group of animals (47). Human twins have more similar intestinal communities than do married couples or unrelated individuals, giving some of the first evidence that genotype is a major contributor to microbial selection in the intestine (51). A later study with mice provides further support for the role genetics plays in microbial diversity (21). Conceivably, the ileum community is under different constraints, due in part to the greater surface-to-volume ratio, the lower bacterial density, and the active nature of the ileal tissue (for example, through excretion of mucus). The community-determining role of the porcine ileal physiology should therefore be investigated in more detail.
The ileal communities were dominated by lactic acid bacteria, as has been reported for recently weaned pigs fed various diets. Overall, a shift from L. johnsonii to L. amylovorus associated with CTC treatment was observed. This raises the question of whether they may possibly play a role in the growth-promoting effect. L. johnsonii, the predominant lactic acid bacterium in control pigs, is autochthonous to the porcine GI tract (27). Lactobacillus bacteria are known to produce an array of bacteriocins, and many isolates have shown promise as probiotics (22, 26, 37). Various strains of L. johnsonii and L. amylovorus display probiotic activity and also produce antibacterial peptides (33). The shift from an L. johnsonii-dominated to an L. amylovorus-dominated microbiota may be due both to suppression of the latter by L. johnsonii in control pigs and to the greater CTC susceptibility of L. johnsonii, leading to a bloom of L. amylovorus. Phylogenetic data cannot be used to extrapolate functional properties from those of well-characterized type strains. Therefore, the growth-promoting effect associated with a shift from a series of strains related to L. johnsonii to L. amylovorus cannot be interpreted unambiguously, but a number of general traits may be considered. L. johnsonii is not known to be pathogenic to pigs. Several isolates of L. johnsonii degrade bile (10) and may thereby influence the absorption of lipid components of the diet. L. amylovorus is amylolytic and able to contribute to saccharification of the high proportion of starch in the diet (see Table S2 in the supplemental material). A recent Dutch study using Hypor × Pietrain piglets fed a diet rich in maize starch found that L. reuteri and L. amylovorus dominate the ileum (24).
Clostridiales is a group of bacteria that have been consistently found in high numbers in the pig intestine (6, 18, 27). The noted decrease in the mucosal CTC library is not particularly surprising because tetracycline is used to treat some Clostridium infections (4). On the other hand, an increase in Clostridiales bacteria in antibiotic-treated animals has been noted in chickens (31). Yet studies have not explored either the possibilities of benefits from intestinal colonization by nonpathogenic Clostridiales bacteria or the distribution of resistance genes among them, so their persistence in the ecosystem and their effect on the growth of pigs cannot yet be explained. The taxonomic richness of the Clostridiales bacteria was detected at the species level, with little overlap between the ileal control and CTC-treated communities. CTC may allow an increase in the presence of a group of Clostridiales bacteria, which are usually present in low abundance due to suppression by other microbes.
CTC decreased the relative abundance of Turicibacter bacteria in the porcine ileum. Turicibacter is a relatively unknown group of bacteria. Recent reports of 16S rRNA gene and ribosomal intergenic spacer analysis data indicate the presence of Turicibacter bacteria in the pig (12, 23) and rat (30) GI tracts. Three sequences from porcine GI tracts that were communicated prior to the description of the species are highly homologous to Turicibacter (27). Turicibacter has also been reported from insect hindguts, dairy wastewaters, and raw milk (8, 9, 34). However, as only one species has been cultured (3), the physiological diversity of the members of this genus is unknown. As the isolated strain is a putative pathogen, it is possible that the Turicibacter bacteria present in the pig may cause subclinical infection or have some other deleterious effect on the GI tract. If so, Turicibacter phylotypes present in the pig ileum may have deleterious effects and a CTC-associated decrease may help explain the growth-promoting effect. A number of porcine Turicibacter bacteria must be isolated in order to study the effect these uncharacterized bacteria may exert on the dynamics of the GI tract, especially the ileal compartment.
Differences between lumen- and mucosa-associated bacteria have been noted in the past (39, 40, 45), and data from this study also support variation between the two locations. This differs from a previous report in which the mucosa bacterial microbiota was viewed as a subset of the lumen microbiota (27). Interestingly, several of the variances noted in our study can only be seen when approaching the information from different levels of sequence similarity criteria. A greater taxonomic richness was observed in mucosa when viewed at the genus level (5% difference), but at the species and subspecies levels (3 and 1% differences) there was a higher level of taxonomic richness in the lumen.
The composition of the microbiota varied considerably between individual pigs, as revealed by θYC values. While the observed variation between untreated pigs obscures the possible effect of CTC, an overall comparison by SONS analysis did show that, overall, both treated and control pigs had a series of unique OTUs in both the lumen and mucus. Overall shifts associated with CTC included decreases in the abundance of L. johnsonii, Clostridiales, and Turicibacter and increases in L. amylovorus.
Supplementary Material
Acknowledgments
We thank Yun Luo and Laura Weyrich for technical assistance and Stephanus Venter for helpful comments on the manuscript. The anonymous reviewers are thanked for useful comments to improve analysis of the data.
This work was supported by a grant from the Center for Infectious Disease Research and Vaccinology/2010 initiative of the State of South Dakota and by the South Dakota Agricultural Experiment Station. We acknowledge the use of the SDSU-FGCF, supported in part by NSF/EPSCoR grant 0091948 and by the State of South Dakota.
Footnotes
Published ahead of print on 17 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
Journal series publication 3627 from the South Dakota Agricultural Experiment Station.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosshard, P. P., R. Zbinden, and M. Altwegg. 2002. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 52:1263-1266. [DOI] [PubMed] [Google Scholar]
- 4.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier, C. T., M. R. Smiricky-Tjardes, D. M. Albin, J. E. Wubben, V. M. Gabert, B. Deplancke, D. Bane, D. B. Anderson, and H. R. Gaskins. 2003. Molecular ecological analysis of porcine ileal microbiota responses to antimicrobial growth promoters. J. Anim. Sci. 81:3035-3045. [DOI] [PubMed] [Google Scholar]
- 6.Cotta, M. A., T. R. Whitehead, and R. L. Zeltwanger. 2003. Isolation, characterization and comparison of bacteria from swine faeces and manure storage pits. Environ. Microbiol. 5:737-745. [DOI] [PubMed] [Google Scholar]
- 7.Cromwell, G. L. 2002. Why and how antibiotics are used in swine production. Anim. Biotechnol. 13:7-27. [DOI] [PubMed] [Google Scholar]
- 8.Delbès, C., L. Ali-Mandjee, and M. C. Montel. 2007. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 73:1882-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egert, M., U. Stingl, L. D. Bruun, B. Pommerenke, A. Brune, and M. W. Friedrich. 2005. Structure and topology of microbial communities in the major gut compartments of Melolontha melolontha larvae (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 71:4556-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins, C. A., S. A. Moser, and D. C. Savage. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147:3403-3412. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—phylogeny inference package. Cladistics 5:164-166. [Google Scholar]
- 12.Gagnon, N., G. Talbot, P. Ward, D. Roy, M. Dupuis, E. Farnworth, T. A. Tompkins, and M. Lessard. 2007. Evaluation of bacterial diversity in the gut of piglets supplemented with probiotics using ribosomal intergenic spacer analysis. Can. J. Anim. Sci. 87:207-219. [Google Scholar]
- 13.Gaskins, H. R. 2001. Intestinal bacteria and their influence on swine growth, p. 585-608. In A. J. Lewis and L. L. Southern (ed.), Swine nutrition. CRC Press, Inc., Boca Raton, FL.
- 14.Gaskins, H. R., C. T. Collier, and D. B. Anderson. 2002. Antibiotics as growth promotants: mode of action. Anim. Biotechnol. 13:29-42. [DOI] [PubMed] [Google Scholar]
- 15.George, S., Y. Oh, S. Lindblom, S. Vilain, A. J. Rosa, D. H. Francis, V. S. Brözel, and R. S. Kaushik. 2007. Lectin binding profile of the small intestine of five-week-old pigs in response to the use of chlortetracycline as a growth promotant and under gnotobiotic conditions. J. Anim. Sci. 85:1640-1650. [DOI] [PubMed] [Google Scholar]
- 16.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 17.Hill, E. G., and N. L. Larson. 1955. Effect of chlortetracycline supplementation on growth and feed utilization of unsuckled baby pigs obtained by hysterectomy. J. Anim. Sci. 14:1116-1121. [Google Scholar]
- 18.Hill, J. E., S. M. Hemmingsen, B. G. Goldade, T. J. Dumonceaux, J. Klassen, R. T. Zijlstra, S. H. Goh, and A. G. Van Kessel. 2005. Comparison of ileum microflora of pigs fed corn-, wheat-, or barley-based diets by chaperonin-60 sequencing and quantitative PCR. Appl. Environ. Microbiol. 71:867-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, R. L., J. F. Elam, G. W. Anderson, L. L. Gee, J. Fowler, and J. R. Couch. 1953. Further evidence as to the possible mechanism involved in the growth-promoting responses obtained from antibiotics. J. Nutr. 51:507-513. [DOI] [PubMed] [Google Scholar]
- 20.Jukes, T. H. 1955. Antimetabolites and antibiotics as tools for research on blood formation. Am. J. Clin. Nutr. 3:56-63. [DOI] [PubMed] [Google Scholar]
- 21.Jussi, V., E. Erkki, and T. Paavo. 2005. Comparison of cellular fatty acid profiles of the microbiota in different gut regions of BALB/c and C57BL/6J mice. Antonie van Leeuwenhoek 88:67-74. [DOI] [PubMed] [Google Scholar]
- 22.Kim, P. I., M. Y. Jung, Y. H. Chang, S. Kim, S. J. Kim, and Y. H. Park. 2007. Probiotic properties of Lactobacillus and Bifidobacterium strains isolated from porcine gastrointestinal tract. Appl. Microbiol. Biotechnol. 74:1103-1111. [DOI] [PubMed] [Google Scholar]
- 23.Kishimoto, A., K. Ushida, G. O. Phillips, T. Ogasawara, and Y. Sasaki. 2006. Identification of intestinal bacteria responsible for fermentation of gum arabic in pig model. Curr. Microbiol. 53:173-177. [DOI] [PubMed] [Google Scholar]
- 24.Konstantinov, S. R., A. Awati, H. Smidt, B. A. Williams, A. D. Akkermans, and W. M. de Vos. 2004. Specific response of a novel and abundant Lactobacillus amylovorus-like phylotype to dietary prebiotics in the guts of weaning piglets. Appl. Environ. Microbiol. 70:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinov, S. R., A. A. Awati, B. A. Williams, B. G. Miller, P. Jones, C. R. Stokes, A. D. Akkermans, H. Smidt, and W. M. de Vos. 2006. Post-natal development of the porcine microbiota composition and activities. Environ. Microbiol. 8:1191-1199. [DOI] [PubMed] [Google Scholar]
- 26.Lee, H. S., S. E. Gilliland, and S. Carter. 2001. Amylolytic cultures of Lactobacillus acidophilus: potential probiotics to improve dietary starch utilization. J. Food Sci. 66:338-344. [Google Scholar]
- 27.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leser, T. D., R. H. Lindecrona, T. K. Jensen, B. B. Jensen, and K. Moller. 2000. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl. Environ. Microbiol. 66:3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Libby, D. A., and P. J. Schaible. 1955. Observations on growth responses to antibiotics and arsonic acids in poultry feeds. Science 121:733-734. [DOI] [PubMed] [Google Scholar]
- 30.Licht, T. R., B. Madsen, and A. Wilcks. 2007. Selection of bacteria originating from a human intestinal microbiota in the gut of previously germ-free rats. FEMS Microbiol. Lett. 277:205-209. [DOI] [PubMed] [Google Scholar]
- 31.Lu, J., C. L. Hofacre, and M. D. Lee. 2006. Emerging technologies in microbial ecology aid in understanding the effect of monensin in the diets of broilers in regard to the complex disease necrotic enteritis. J. Appl. Poultry Res. 15:145-153. [Google Scholar]
- 32.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makras, L., V. Triantafyllou, D. Fayol-Messaoudi, T. Adriany, G. Zoumpopoulou, E. Tsakalidou, A. Servin, and L. De Vuyst. 2006. Kinetic analysis of the antibacterial activity of probiotic lactobacilli towards Salmonella enterica serovar Typhimurium reveals a role for lactic acid and other inhibitory compounds. Res. Microbiol. 157:241-247. [DOI] [PubMed] [Google Scholar]
- 34.McGarvey, J. A., W. G. Miller, S. Sanchez, and L. Stanker. 2004. Identification of bacterial populations in dairy wastewaters by use of 16S rRNA gene sequences and other genetic markers. Appl. Environ. Microbiol. 70:4267-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, P. R., A. Evenson, T. D. Luckey, E. McCoy, C. A. Elvehjem, and E. B. Hart. 1946. Use of sulfasuxidine, streptothricin, and streptomycin in nutritional studies with the chick. J. Biol. Chem. 165:437-441. [PubMed] [Google Scholar]
- 36.Page, S. W. 2003. Mode of action, p. 2-1-2-14. In S. W. Page (ed.), The role of enteric antibiotics in livestock production. Avcare, Canberra, Australia.
- 37.Parvez, S., K. A. Malik, S. Ah Kang, and H. Y. Kim. 2006. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 100:1171-1185. [DOI] [PubMed] [Google Scholar]
- 38.Phillips, I., M. Casewell, T. Cox, B. De Groot, C. Friis, R. Jones, C. Nightingale, R. Preston, and J. Waddell. 2004. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 53:28-52. [DOI] [PubMed] [Google Scholar]
- 39.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell, E. G. 1979. Types and distribution of anaerobic bacteria in the large intestine of pigs. Appl. Environ. Microbiol. 37:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarz, S., and E. Chaslus-Dancla. 2001. Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 32:201-225. [DOI] [PubMed] [Google Scholar]
- 45.Simpson, J. M., V. J. McCracken, B. A. White, H. R. Gaskins, and R. I. Mackie. 1999. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J. Microbiol. Methods 36:167-179. [DOI] [PubMed] [Google Scholar]
- 46.Smith, D. L., A. D. Harris, J. A. Johnson, E. K. Silbergeld, and J. G. Morris, Jr. 2002. Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc. Natl. Acad. Sci. USA 99:6434-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, C. L., B. Wang, and A. J. Holmes. 2008. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. ISME J. 2:739-748. [DOI] [PubMed] [Google Scholar]
- 48.Visek, W. J. 1978. The mode of growth promotion by antibiotics. J. Anim. Sci. 46:1447-1469. [Google Scholar]
- 49.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 50.Wang, G. C., and Y. Wang. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142:1107-1114. [DOI] [PubMed] [Google Scholar]
- 51.Zoetendal, E. G., A. D. L. Akkermans, W. M. Akkermans-van Vliet, J. Arjan, G. M. de Visser, and W. M. de Vos. 2001. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb. Ecol. Health Dis. 13:129-134. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.