Abstract
The diversity in regulatory phenotypes among a collection of 84 Lactococcus lactis strains isolated from dairy and nondairy origin was explored. The specific activities of five enzymes were assessed in cell extracts of all strains grown in two different media, a nutritionally rich broth and a relatively poor chemically defined medium. The five investigated enzymes, branched chain aminotransferase (BcaT), aminopeptidase N (PepN), X-prolyl dipeptidyl peptidase (PepX), alpha-hydroxyisocaproic acid dehydrogenase (HicDH), and esterase, are involved in nitrogen and fatty acid metabolism and catalyze key steps in the production of important dairy flavor compounds. The investigated cultures comprise 75 L. lactis subsp. lactis isolates (including 7 L. lactis subsp. lactis biovar diacetylactis isolates) and 9 L. lactis subsp. cremoris isolates. All L. lactis subsp. cremoris and 22 L. lactis subsp. lactis (including 6 L. lactis subsp. lactis biovar diacetylactis) cultures originated from a dairy environment. All other cultures originated from (fermented) plant materials and were isolated at different geographic locations. Correlation analysis of specific enzyme activities revealed significantly different regulatory phenotypes for dairy and nondairy isolates. The enzyme activities in the two investigated media were in general poorly correlated and revealed a high degree of regulatory diversity within this collection of closely related strains. To the best of our knowledge, these results represent the most extensive diversity analysis of regulatory phenotypes within a single bacterial species to date. The presented findings underline the importance of the availability of screening procedures for, e.g., industrially relevant enzyme activities in models closely mimicking application conditions. Moreover, they corroborate the notion that regulatory changes are important drivers of evolution.
Lactococcus lactis is an important component of many dairy starter cultures and has been studied extensively in the context of its function in these industrial processes. However, lactococci are also frequently isolated from (fermented) plant material, but the strains isolated have been described in much less molecular detail (22). In dairy products L. lactis cultures are responsible for various enzymatic conversions which affect the organoleptic characteristics of the fermented product. For instance, the caseinolytic activity of lactococcal proteases and peptidases has a major influence on cheese texture (13), and secondary bacterial metabolites are key contributors to cheese taste and flavor (19). Enzyme activities are strain specific, and simply by changing the starter culture composition one can alter product properties (19, 24). It is therefore attractive to screen culture collections and natural isolates for the activity of specific enzymes that contribute to specific flavor formation capacities during cheese production. Amino acids are the precursors of a large variety of flavor compounds, and, consequently, bacterial amino acid metabolism has been studied extensively (23, 26). Some lactic acid bacteria are able to degrade milk proteins with a complex proteolytic system that includes a number of endopeptidases (13). These peptidases are thought to influence the flavor of cheese especially after cell lysis and their release into the cheese matrix (19). Branched chain amiotransferase (BcaT) and aromatic aminotransferase (AraT) catalyze the conversion of amino acids to the corresponding α-keto acids, which are the precursors for a number of aroma compounds (25-27). These α-keto acids are, for example, a substrate for alpha-hydroxyisocaproic acid dehydrogenase (HicDH), which reduces them to the corresponding hydroxy acids (26). Another reaction related to aroma compounds is the conversion of carboxylic acids to (thio)esters by esterases (19).
Bacterial gene expression is often tightly regulated, and for several genes we have detailed knowledge about the regulatory mechanisms involved. However, this detailed knowledge of regulatory characteristics of specific enzymes has been unraveled for a relatively small number of strains. An example of a well-characterized regulatory system is the pleiotropic regulator CodY, which regulates mainly the expression of genes involved in nitrogen metabolism such as amino acid biosynthesis and transport and genes of the proteolytic system (7, 12). The codY binding site is reported to be well conserved upstream of genes involved in branched-chain amino acid metabolism throughout bacterial species belonging to the Firmicutes (12). Nevertheless, the relationship of the conservation of regulatory elements like CodY and sequence conservation at the protein level to the actual enzyme activities in individual strains of a specific species or subspecies remains to be established. Previously, we have performed an extensive genotypic and phenotypic analysis of 102 lactococcus isolates from either the dairy environment or plant material (18). For detailed analysis of phylogenetic relationships, the partial sequences of six genes were determined for 89 strains, and they revealed 363 polymorphic sites on a total DNA length of 1,970 bases. The ratio of synonymous to nonsynonymous sites was <0.08 for the partial sequences of pepN, bcaT, and pepX. This clearly indicates that, at the protein sequence level, these strains are very closely related to each other. In the present study we selected 84 L. lactis strains from the previously described collection (18) and determined the specific activities of five different enzymes that are well-known to have an impact on flavor formation (aminopeptidase N [PepN], X-prolyl-dipeptidyl aminopeptidase [PepXP], BcaT, HicDH, and esterase). Of the 84 investigated strains, the majority (75 strains) belonged to the subspecies L. lactis, while 7 of these belonged to the L. lactis subsp. lactis biovar diacetylactis, and 9 strains belonged to L. lactis subsp. cremoris. All strains of L. lactis subsp. cremoris, 6 strains of L. lactis subsp. lactis biovar diacetylactis, and 16 strains of L. lactis subsp. lactis were isolated from the dairy environment. All other strains were isolated from (fermented) plant materials originating from different geographic locations. To assess the extent to which the regulatory mechanisms are conserved between these closely related strains, the enzyme activities of cells grown in different media were compared. We define the environment-dependent, strain-specific variations of enzyme activities as the regulatory phenotype. Since specific enzyme activities were compared throughout this paper, it should be noted that we have used the term regulation in the broad sense, as a cumulative effect of important mechanisms like transcription and translation or any other cellular factor that would influence specific enzyme activities of the bacterial culture. The results demonstrate distinct differences between dairy and nondairy isolates and remarkably little correlation between specific enzyme activities measured in the different media.
MATERIALS AND METHODS
Bacterial isolates and media.
A total of 84 L. lactis strains, which form a subset of an earlier described diversity study (18), were used for the experiments presented here. The subset was obtained from the NIZO culture collection (NIZO food research, Ede, The Netherlands) (see neighbor-joining tree in Fig. 4). Throughout this paper we use the taxonomic classification used by Rademaker et al. (18). The strains were grown at 30°C either in M17 broth (Oxoid Ltd., Basingstoke, Hampshire, England) supplemented with 0.5% glucose (wt/vol) (GM17) or in chemically defined medium (CDM) (17) supplemented with 0.5% glucose.
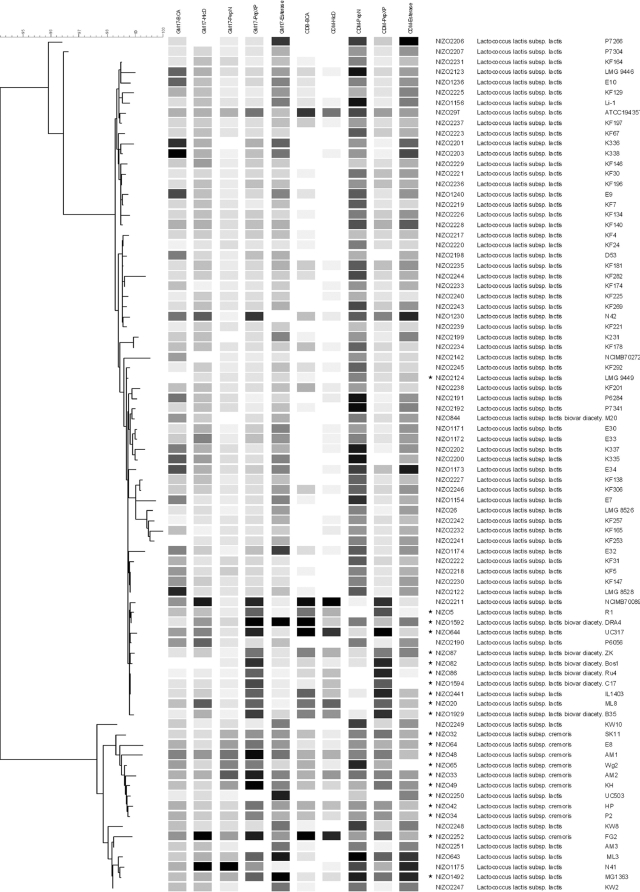
FIG. 4.
Specific enzyme activities of BcaT, HicDH, PepN, PepXP, and esterase (Est) measured in GM17 medium and CDM (as indicated at the top) are shown in grayscale in the middle. The lowest enzyme activity within each condition is shown in white; the highest enzyme activity is shown in black. Neighbor-joining cluster analysis based on five-locus multilocus sequencing from sequences determined by Rademaker et al. (18) is displayed to the left. Specific enzyme activity profiles show relatively little conservation between phylogenetically closely related strains or different screening environments. Dairy isolates are indicated (*). To the right the strain names as given in the NIZO culture collection, the taxonomic position, and the commonly used strain names are given (left to right).
Enzyme activity measurements.
For enzyme activity assays cells were grown overnight in 2 ml of GM17 or CDM in 96-well plates in quadruplicate. Each plate contained a sample of strain ML3 (NIZO643) as reference, and noninoculated medium was used as a negative control. Cultures were centrifuged and washed with 2 ml of 50 mM sodium phosphate buffer, pH 7.2. Subsequently, cells were disrupted using a Mini-beadbeater 96 Cell Disrupter (Merlin Diagnostic Systems, Breda, The Netherlands) with 300 μl of 0.1-mm zirconium beads (Merlin Diagnostic Systems) and 1 ml of 50 mM sodium phosphate buffer, pH 7.2. Crude extracts were prepared by four cycles of 30 s of bead beating interspaced by 2-min cooling periods on ice. The resulting lysate was centrifuged (for 10 min at 830 × g at 4°C) to remove cell debris, and the cell extract obtained was used to determine enzyme activities as an average of four independent cultures. The standard deviations in these experiments were on average 10% for PepN, 10% for PepXP, 18% for esterase, 46% for BcaT, and 49% for HicDH. Protein concentrations were determined with a bicinchoninic protein assay (Pierce, Rockford, IL). In all cases specific activities were calculated and expressed as nanomoles of substrate converted per milligram of protein per minute. The reference strain ML3 was present in all 96-well plates, and its enzyme activities showed variations from 11% to 26% in the separate experiments. All activities were normalized to strain ML3 to minimize the influence of experimental variation before activities were subjected to statistical analysis.
Esterase activities were determined by online monitoring of the release of p-nitrophenol from the substrate p-nitrophenyl butyrate in reaction mixtures consisting of 100 μl of cell extract and 145 μl of 50 mM sodium phosphate buffer (pH 7.5). The assay was started by adding 5 μl of substrate solution (10 μl of p-nitrophenol-butyrate in 478 μl of dimethyl sulfoxide). The initial rates of release of p-nitrophenyl from the ester substrate at 30°C were quantified by measuring the increase in absorbance at 410 nm, and specific activities were calculated.
Analogously, PepN activities were determined in a 96-well format by online monitoring of the release of p-nitroanilide from the substrate Lys-p-nitroanilide dihydrobromide (FA Bachem, Bubendorf, Switzerland) at 30°C by measuring absorbance at 410 nm, as described previously (8).
PepXP activities were determined by online monitoring of the release of p-nitroanilide from the substrate H-Ala-Pro-p-nitroanilide by measuring the increase in absorbance at 410 nm. Reaction mixtures consisted of 250 μl of a solution of 0.256 mg ml−1 of H-Ala-Pro-p-nitroanilide in 50 mM sodium phosphate buffer, pH 7.2. Assays were carried out at 30°C, started by adding 50 μl of cell extract, and specific activities were calculated.
HicDH and BcaT activities were determined as described earlier (3). It is important to note that BcaT activity was measured with leucine as a substrate. Out of 84 BcaT measurements in CDM, 24 showed no detectable activity. These data were omitted from the analysis.
Data analysis.
The average values of four measurements were calculated for each condition and used to compute Spearman rank correlation coefficient (R) values. Classical decision trees were generated using the “ctree” function (21) as implemented in the R program (R Development Core Team; www.r-project.org).
RESULTS
Enzyme phenotype screening.
The analysis of 84 lactococcus isolates revealed that the highest specific activities observed for each of the five analyzed enzymes are comparable with previous reports for L. lactis strains (1, 9, 20, 25). The variation of enzyme activities between the different isolates was considerable, with the largest variation observed for HicDH (249-fold) and relatively small variation for esterase (Table 1). Strain-to-strain variations were similar if measured in either GM17 medium or CDM with the exception of PepN. PepN activities varied approximately 120-fold between strains grown in GM17 medium and only approximately 5-fold for strains grown in CDM. In order to analyze the regulatory control of enzyme activities, correlation coefficients comparing the specific activities after growth in the different media were calculated for the total data set and for subsets defined on the basis of strain origin or phylogenetic clustering (Fig. 1 and 2).
TABLE 1.
Minimum and maximum enzyme activities of 84 strains measured after growth in either in GM17 or CDM
| Enzyme | Enzyme activity (nmol/min/mg of protein) with the indicated mediuma
|
Prediction errorb
|
||||||
|---|---|---|---|---|---|---|---|---|
| Min
|
Max
|
Diversity (max/min)
|
Above the 90th percentile | Below the 10th percentile | ||||
| GM17 | CDM | GM17 | CDM | GM17 | CDM | |||
| BcaT | 58.7 | ND | 1033.5 | 436.8 | 17.6 | ND | 100 | 92 |
| HicD | 39.5 | 39.6 | 9826.1 | 4648 | 249 | 117.4 | 55.6 | 92 |
| PepN | 2.4 | 42.2 | 289.7 | 202.4 | 121.1 | 4.8 | 77.8 | 92 |
| PepXP | 20.7 | 15.8 | 694.5 | 250.5 | 33.5 | 15.9 | 66.7 | 93.3 |
| Esterase | 7 | 2.8 | 42.9 | 20 | 6.1 | 7.1 | 44.4 | 93.3 |
Diversity, an estimate of the diversity of enzyme activities; Min, minimum; Max, maximum; ND, not done (below the detection limit).
The environment-to-environment prediction error of activities.
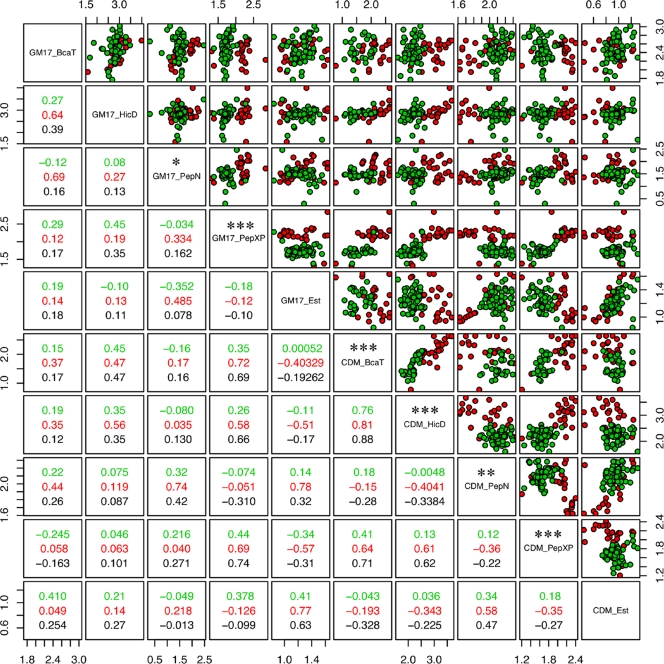
FIG. 1.
Correlation matrix and pair plots of all enzyme measurements in the two different environments. Dairy isolates are shown in red; nondairy isolates are shown in green. Spearman rank correlation coefficients (R) are given for each group in the corresponding color of the lower panels. The correlation coefficient for the total data is given in black. Each dot plot displays the activities of the horizontally and vertically projected pair of enzymes indicated in the diagonal. The axes pair plots have a logarithmic scale. Significance levels of a two-tailed t test comparing dairy and nondairy isolates are given above the description in the diagonal panel (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Dairy isolates can be clearly distinguished from nondairy isolates, but correlations between measurements in the different media are relatively poor.
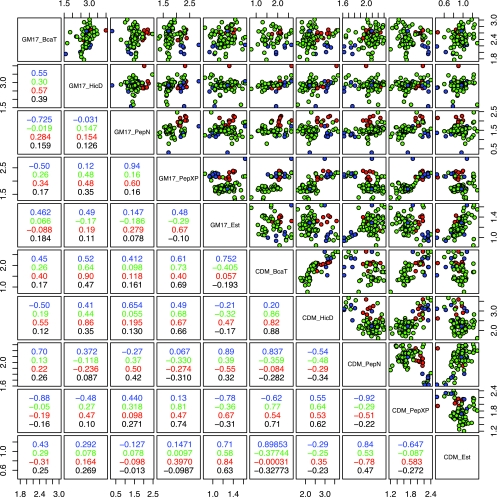
FIG. 2.
Correlation matrix and pair plots as described in the legend of Fig. 1, but strains are sorted by taxonomic position: green, L. lactis subsp. lactis; blue, L. lactis subsp. lactis biovar diacetylactis; and red, L. lactis subsp. cremoris. Analysis based on the taxonomic position of strains shows increased correlation only for some enzyme activities, but overall correlations between measurements in different environments remain poor.
Comparison dairy versus nondairy isolates.
The comparison of dairy and nondairy isolates revealed that the screening environment is of great importance for the identification of correlations between the investigated groups. For instance, BcaT activities of cells grown in GM17 were not correlated with niche origin, but a highly significant difference was measured when the same analysis was performed in CDM (Fig. 1). Furthermore, the average PepN activity levels measured in nondairy isolates were significantly lower than in dairy isolates when the strains were grown in GM17 medium (ca. 35 versus 63 nmol/min/mg protein; P = 0.015), while they were significantly higher when the same strains were grown in CDM (ca. 132 versus 97 nmol/min/mg protein; P = 0.002). In contrast, HicDH and BcaT activities of nondairy isolates were significantly lower than those of dairy isolates (P < 0.001) only when the strains were grown in CDM. Furthermore, nondairy isolates appeared to have a lower PepXP activity level than dairy isolates (P < 0.001), which was the measurement found to be the most independent of the growth medium. A decision tree analysis showed that, based on the PepXP measurements, dairy and nondairy isolates could be predicted with a prediction error of 5.9%. This demonstrates that clustering of the two groups based on specific enzyme activities is possible, but the choice of the investigated enzyme activity and the screening environment are crucial for this successful identification.
Comparison based on taxonomic position.
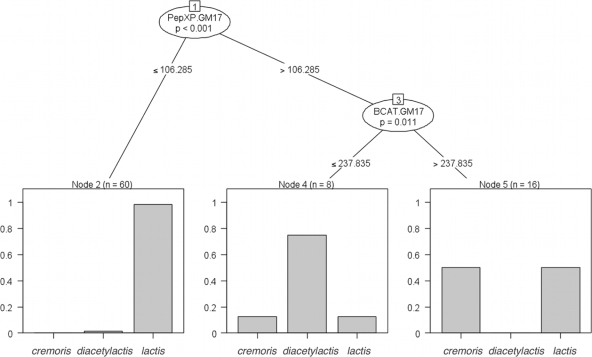
The correlation coefficients between specific activities for all 84 strains grown in the two different media were 0.17 for BcaT, 0.35 for HicDH, 0.42 for PepN, 0.74 for PepXP, and 0.63 for esterase. Some correlation coefficients increased when only a particular subspecies/biovar was analyzed. For example, HicDH activities in both media displayed a clearly correlated activity level only within L. lactis subsp. cremoris while PepXP measurements displayed a distinctly higher correlation within L. lactis subsp. lactis. Moreover, the PepN activities of GM17 medium-grown L. lactis subsp. cremoris cultures were significantly higher than activities in cultures of L. lactis subsp. lactis (ca. 112 versus 37 nmol/min/mg protein; P < 0.0001) (Fig. 2). This difference was not observed in CDM-grown cultures. A classical decision tree allowed the identification of L. lactis subsp. lactis based on PepXP activities (prediction error, 13.2%) and further showed that L. lactis subsp. lactis biovar diacetylactis could be identified by having high PepXP and low BcaT activities in GM17 (prediction error, 14.3%) (Fig. 3). The identification of L. lactis subsp. lactis biovar diacetylactis as shown in Fig. 3 is only possible if the BcaT activities are measured in GM17 medium. Error rates of decision trees based on other enzyme activities drastically increased, reflecting the relatively poor correlations between taxonomic positions and most enzyme activities measured. Overall, phylogenetically very closely related strains appear to display highly diverse enzyme activities (Fig. 4).
FIG. 3.
Decision tree to predict phylogenetic classification based on enzyme activities. Low PepXP activity is a predictor for L. lactis subsp. lactis. High PepXP activity with low BcaT activity (if measured in GM17 medium) is a predictor for L. lactis subsp. lactis biovar diacetylactis. cremoris, L. lactis subsp. cremoris; diacetylactis, L. lactis subsp. lactis biovar diacetylactis; lactis, L. lactis subsp. lactis.
Regulatory diversity.
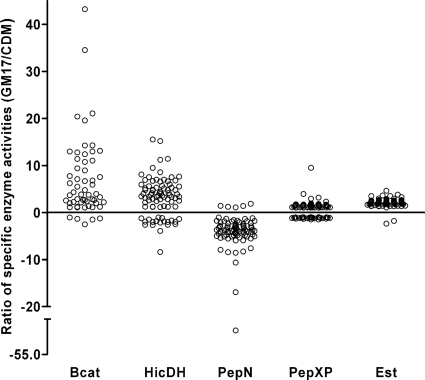
To quantify the level of regulatory diversity, the ratio of the specific enzyme activities in the two media employed was calculated and revealed considerable medium-dependent differences. The specific activities of BcaT, HicDH, PepN, PepXP, and esterase differed up to 43-, 16-, 52-, 10-, and 5-fold between the measurements in the two media, respectively (Fig. 5). To assess the extent to which the measurements from one environment can predict the outcome in a different environment, we projected the data for the measurements after growth in GM17 medium onto results obtained after cells were grown in CDM. This comparison showed that the environment-to-environment prediction of strains with specific enzyme activities below the 10th or above the 90th percentile resulted in error rates ranging from 44 to 100% (Table 1). Six out of 10 predictions have an error rate close to 90%, which is what one would expect if the values were randomly distributed. We therefore consider the predictive value of enzyme measurements obtained from one environment of very limited (or even no) use for different environments.
FIG. 5.
Regulatory diversity as expressed by the ratio of specific enzyme activities if measured either in GM17 medium or in CDM. Clouds represent the different enzyme measurements indicated on the x axis. Each circle represents an individual strain. The differential regulation in the two investigated environments shows large strain-specific variations especially for the activities of BcaT, HicDH, and PepN.
DISCUSSION
The enzymatic conversion of biomolecules by means of fermentation is an important process in the food industry. To improve such conversions, there is a continuous process of strain improvement, with the focus on acquiring and/or selecting microorganisms that have the desired phenotype. Ongoing developments in the field of high-throughput screening open new avenues for the discovery of industrially relevant phenotypes. An important approach is the mining of biodiversity by screening natural bacterial isolates for required enzymatic activities. However, it is often difficult or impossible to perform screening procedures under the same environmental conditions envisioned for the application of the strains. It is commonly accepted that the screening conditions are likely to influence the expression level of bacterial enzymes. Consequently, the relatively poor predictive value of laboratory-based screening results may be anticipated when these are extrapolated to the strain's performance in the application environment. To increase our insight into this diversity at a regulatory level, we determined five enzyme activities of 84 closely related L. lactis strains in two different growth media. The activities observed with dairy isolates of the L. lactis subsp. lactis and L. lactis subsp. subsp. cremoris are in agreement with a more limited, previously performed analysis by Crow et al. (5), which also included some of the strains used in the present study. In this study it was shown that PepN activity levels were almost twice as high in isolates belonging to L. lactis subsp. cremoris as in isolates of L. lactis subsp. lactis. Our measurements in GM17 medium confirm this finding, but we also show that this difference is not observed if cells are grown in CDM (Fig. 2). It is important to realize that Crow et al. grew the strains in reconstituted skim milk, which, for certain strains, was supplemented with yeast extract and/or glucose. Various peptidases of dairy strains are known to be important for the bacterial utilization of milk proteins (13), which is in agreement with their high number of amino acid auxotrophies (2, 6, 10). Consequently, the finding that PepXP activity levels in dairy isolates are high compared to nondairy isolates is probably a reflection of the adaptation to the dairy environment. Nevertheless, the dairy-related regulatory adaptation in terms of control of specific enzyme levels appears to follow different paths for PepN activity. PepN is significantly increased in dairy isolates only for strains grown in CDM while an opposite conclusion may be reached when the same activity is measured in dairy isolates grown in GM17 medium. Both PepN and BcaT are regulated by the pleiotropic regulator CodY, which strongly represses responsive genes in the presence of high peptide concentrations. Consistent with this mechanism, higher PepN activities were found in cells grown in peptide-free CDM. In contrast, the BcaT measurements display the opposite response (lower activities in CDM), for which we lack an appropriate explanation to date. Nevertheless, the observation that dairy isolates display a significantly increased BcaT activity in CDM supports that some form of regulation underlies this observation. Although the codY binding site is well conserved in Firmicutes (12), our data indicate that its regulation differs clearly between dairy and nondairy isolates. Another significant difference found is the increased activity of HicDH in dairy isolates for strains grown in CDM. HicDH belongs to a family of hydroxy acid dehydrogenases, which play an important role in NAD+ regeneration in the cell. This finding together with the upregulation of a L. lactis lactate dehydrogenase (ldh) during the ripening of cheese (Bachmann et al., unpublished data) suggests a dairy-specific redox response at the level of either adaptation or regulation.
The specific activities determined in the two different environments demonstrate a clear correlation for PepXP, suggesting that it is regulated similarly in both media or, alternatively, that it is not regulated at all. When considering the correlations of all other specific enzyme activities measured in the 84 strains, we conclude these to be rather poor, indicating that these enzymes are strongly and differentially regulated by environmental conditions in a strain-specific manner. The correlation coefficients appeared to increase only for some subcollections sorted either by strain origin or by different subspecies/variants. The variation of PepN activities is 25-fold lower in strains grown on CDM than in strains grown on GM17 medium. This indication of strong regulation, combined with a very low correlation coefficient between the measurements in the two media, suggests very diverse regulatory characteristics in individual strains. Based on the fact that only strains of the species L. lactis were used in this study, the observed diversity of regulatory phenotypes was unexpectedly high in our opinion. Intriguingly, the strongest correlation observed in the presented data set was the coregulation of the BcaT and HicDH activity levels when cells were grown in CDM (R = 0.88). In contrast, only a very weak correlation between these two measurements was observed between strains when they were grown in GM17 medium, which is consistent with findings reported previously (3). Since both BcaT and HicDH are involved in branched-chain amino acid metabolism, their coregulation seems plausible. Taken together, the presented data demonstrate that strong correlations can be found in our data set. However, we only found few of these strong correlations, and we are not aware of a way to predict these, which would be essential for the design and interpretation of, e.g., screening procedures in environments other than the intended application. The strong correlations between strain origin (Fig. 1), the HicDH and BcaT activity level (in CDM, P value of <1−10), or between the PepXP measurements in the different environments (P value of <1−10) are highly significant, which demonstrates the reliability of our data and strengthens the conclusions on correlations that we did not find.
The finding that very closely related strains show a large variation in their regulatory responses is consistent with a number of experimental evolution studies that indicate that the change of regulatory mechanisms belongs to the first events during adaptive evolution (4, 15, 16). In fact, drastic changes in gene regulation can be seen after as little as 1,000 generations of experimental evolution (14) (Bachmann et al., unpublished).
Despite the extensive characterization of L. lactis throughout the last decades, the extent of the regulatory diversity was never described in such detail before. To the best of our knowledge, these results represent the most extensive diversity analysis of regulatory phenotypes within a single bacterial species to date. We report that four out of five measured activities (PepN, HicDH, BcaT, and esterase) show very diverse regulatory responses within the species L. lactis, and it may be expected that other strain-specific enzyme activity patterns may be expected under alternative growth conditions (7, 11). Although some variation on the regulatory level was to be expected, the obtained results revealed an unexpectedly high level of diversity. Moreover, our data strongly support the notion that enzyme activities measured in a particular medium or under a specific environmental condition are of limited value (or even no value) for the prediction of activities in a different environment. For this reason, it is essential that screening efforts should employ environmental conditions that are as close as possible to those encountered during the process of interest. This conclusion is likely to be valid beyond the field of lactic acid bacteria. Furthermore, our data corroborate findings described for a number of experimental evolution experiments, suggesting that regulatory changes are important drivers in evolutionary adaptation processes.
Acknowledgments
This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
Published ahead of print on 10 July 2009.
REFERENCES
- 1.Alting, A. C., W. Engels, S. van Schalkwijk, and F. A. Exterkate. 1995. Purification and characterization of cystathionine β-lyase from Lactococcus lactis subsp. cremoris B78 and its possible role in flavor development in cheese. Appl. Environ. Microbiol. 61:4037-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayad, E. H., A. Verheul, C. De Jong, J. T. Wouters, and G. Smit. 1999. Flavour forming abilities and amino acid requirements of Lactococcus lactis strains form artisanal and non-dairy origin. Int. Dairy J. 9:725-735. [Google Scholar]
- 3.Brandsma, J. B., E. Floris, A. R. D. Dijkstra, L. Rijnen, J. A. Wouters, and W. C. Meijer. 2008. Natural diversity of aminotransferases and dehydrogenase activity in a large collection of Lactococcus lactis strains. Int. Dairy J. 18:1103-1108. [Google Scholar]
- 4.Cooper, T. F., D. E. Rozen, and R. E. Lenski. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:1072-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crow, V. L., R. Holland, G. G. Pritchard, and T. Coolbear. 1994. The diversity of potential cheese ripening characteristics of lactic acid starter bacteria: 2. The levels and subcellular distributions of peptidase and esterase activities Int. Dairy J. 4:723-742. [Google Scholar]
- 6.Delorme, C., J. J. Godon, S. D. Ehrlich, and P. Renault. 1993. Gene inactivation in Lactococcus lactis: histidine biosynthesis. J. Bacteriol. 175:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Hengst, C. D., S. A. van Hijum, J. M. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332-34342. [DOI] [PubMed] [Google Scholar]
- 8.Exterkate, F. A. 1984. Location of peptidases outside and inside the membrane of Streptococcus cremoris. Appl. Environ. Microbiol. 47:177-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez, L., M. M. Beerthuyzen, J. Brown, R. J. Siezen, T. Coolbear, R. Holland, and O. P. Kuipers. 2000. Cloning, characterization, controlled overexpression, and inactivation of the major tributyrin esterase gene of Lactococcus lactis. Appl. Environ. Microbiol. 66:1360-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godon, J. J., C. Delorme, J. Bardowski, M. C. Chopin, S. D. Ehrlich, and P. Renault. 1993. Gene inactivation in Lactococcus lactis: branched-chain amino acid biosynthesis. J. Bacteriol. 175:4383-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guedon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guedon, E., B. Sperandio, N. Pons, S. D. Ehrlich, and P. Renault. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151:3895-3909. [DOI] [PubMed] [Google Scholar]
- 13.Kunji, E. R., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie van Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 14.Le Gac, M., M. D. Brazas, M. Bertrand, J. G. Tyerman, C. C. Spencer, R. E. Hancock, and M. Doebeli. 2008. Metabolic changes associated with adaptive diversification in Escherichia coli. Genetics 178:1049-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelosi, L., L. Kuhn, D. Guetta, J. Garin, J. Geiselmann, R. E. Lenski, and D. Schneider. 2006. Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics 173:1851-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philippe, N., E. Crozat, R. E. Lenski, and D. Schneider. 2007. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays 29:846-860. [DOI] [PubMed] [Google Scholar]
- 17.Poolman, B., and W. N. Konings. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rademaker, J. L., H. Herbet, M. J. Starrenburg, S. M. Naser, D. Gevers, W. J. Kelly, J. Hugenholtz, J. Swings, and J. E. van Hylckama Vlieg. 2007. Diversity analysis of dairy and nondairy Lactococcus lactis isolates, using a novel multilocus sequence analysis scheme and (GTG)5-PCR fingerprinting. Appl. Environ. Microbiol. 73:7128-7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smit, G., B. A. Smit, and W. J. Engels. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591-610. [DOI] [PubMed] [Google Scholar]
- 20.Tan, P. S., K. M. Pos, and W. N. Konings. 1991. Purification and characterization of an endopeptidase from Lactococcus lactis subsp. cremoris Wg2. Appl. Environ. Microbiol. 57:3593-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torsten, H., H. Kurt, and Z. Achim. 2006. Unbiased recursive partitioning. J. Comput. Graph. Stat. 15:651-674. [Google Scholar]
- 22.van Hylckama Vlieg, J. E., J. L. Rademaker, H. Bachmann, D. Molenaar, W. J. Kelly, and R. J. Siezen. 2006. Natural diversity and adaptive responses of Lactococcus lactis. Curr. Opin. Biotechnol. 17:183-190. [DOI] [PubMed] [Google Scholar]
- 23.Weimer, B., K. Seefeldt, and B. Dias. 1999. Sulfur metabolism in bacteria associated with cheese. Antonie van Leeuwenhoek 76:247-261. [PubMed] [Google Scholar]
- 24.Whetstine, M. E., M. A. Drake, J. R. Broadbent, and D. McMahon. 2006. Enhanced nutty flavor formation in cheddar cheese made with a malty Lactococcus lactis adjunct culture. J. Dairy Sci. 89:3277-3284. [DOI] [PubMed] [Google Scholar]
- 25.Yvon, M., E. Chambellon, A. Bolotin, and F. Roudot-Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11:185-201. [Google Scholar]
- 27.Yvon, M., S. Thirouin, L. Rijnen, D. Fromentier, and J. C. Gripon. 1997. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl. Environ. Microbiol. 63:414-419. [DOI] [PMC free article] [PubMed] [Google Scholar]