Abstract
The white rot fungus Phanerochaete chrysosporium extensively degraded the endocrine disruptor chemical nonylphenol (NP; 100% of 100 ppm) in both nutrient-limited cultures and nutrient-sufficient cultures. The P450 enzyme inhibitor piperonyl butoxide caused significant inhibition (∼75%) of the degradation activity in nutrient-rich malt extract (ME) cultures but no inhibition in defined low-nitrogen (LN) cultures, indicating an essential role of P450 monooxygenase(s) in NP degradation under nutrient-rich conditions. A genome-wide analysis using our custom-designed P450 microarray revealed significant induction of multiple P450 monooxygenase genes by NP: 18 genes were induced (2- to 195-fold) under nutrient-rich conditions, 17 genes were induced (2- to 6-fold) in LN cultures, and 3 were induced under both nutrient-rich and LN conditions. The P450 genes Pff 311b (corresponding to protein identification number [ID] 5852) and Pff 4a (protein ID 5001) showed extraordinarily high levels of induction (195- and 167-fold, respectively) in ME cultures. The P450 oxidoreductase (POR), glutathione S-transferase (gst), and cellulose metabolism genes were also induced in ME cultures. In contrast, certain metabolic genes, such as five of the peroxidase genes, showed partial downregulation by NP. This study provides the first evidence for the involvement of P450 enzymes in NP degradation by a white rot fungus and the first genome-wide identification of specific P450 genes responsive to an environmentally significant toxicant.
Endocrine-disrupting chemicals (EDCs) are widely distributed environmental contaminants. Surfactants are among the most commonly found environmental EDCs because of their extensive applications, which range from use as industrial chemicals to inclusion in common consumer products. In the United States, Japan, and Western Europe, surfactants are employed most frequently as detergents and as agents in textile, fiber, cosmetics, and pharmaceutical manufacturing. Nearly as common are uses in mining and flotation and in petroleum, paint, lacquer, plastic manufacturing, food, pulp and paper, agrochemical, and leather and fur industries (21).
Alkylphenol ethoxylates are an important class of surfactants. Biodegradation of alkylphenol ethoxylates results in shortening of the ethoxylate chains, ultimately leading to the generation of alkylphenols, particularly nonyl- and octylphenols. Nonylphenol (NP) is the commercially predominant alkylphenol, representing nearly 85% of the total alkylphenol market. NP is a hydrophobic compound used primarily in the chemical manufacturing industry and exists as multiple congeners (11). Several congeners are relatively resistant to biodegradation and are therefore frequently detected in wastewater treatment plant effluents and rivers (15, 20, 25, 31, 48). In previous studies, 4-n-NP has been used as a test compound in risk assessment and biodegradation analyses (16, 23, 39). However, industrially generated technical-grade NP, consisting of more than 30 different isomers, is less biodegradable (14). This characteristic is due to the fact that more than 85% of these isomers possess a quaternary carbon atom in the branched alkyl chain, making them chemical contaminants of high environmental significance (28, 38, 40). Investigation of the susceptibility of technical-grade NP to biodegradation and assessment of health risks from this agent in in vitro and in vivo biological model systems are therefore warranted.
NP is known to bind to the estrogen receptor, thereby mimicking the effects of endogenous hormones, and has been shown to induce synthesis of vitellogenin and inhibit testicular growth in rainbow trout (18, 37, 41). This observation has led to increased interest in the biodegradation and elimination of this class of xenobiotic surfactants from the environment.
Certain microorganisms belonging to the bacterial and yeast groups have the ability to degrade NP (4, 5, 38, 39). Recent studies have shown the abilities of selected fungi, including white rot fungi, to degrade this chemical, albeit to various extents (33). Extracellular oxidases (laccases) have been implicated in the fungal oxidation of NP (2, 19).
The model white rot fungus Phanerochaete chrysosporium is known for its ability to oxidize a wide variety of environmental toxicants. This unique characteristic has been attributed largely to its extracellular peroxidase system. Past studies have provided ample evidence, however, that environmental toxicants can be oxidized or biodegraded even in the absence of peroxidases under nutrient-sufficient (nonligninolytic) conditions (26, 44, 46), suggesting a primary role for other oxidative enzyme systems such as P450 monooxygenases.
P. chrysosporium has recently been shown to possess an extensive P450 enzyme system, with ∼150 P450 monooxygenase genes in its genome (8, 30). Although there have been isolated reports indicating the involvement of P450 monooxygenation in the oxidation of xenobiotic chemicals in this organism, limited information on the identification of specific P450 genes/enzymes and related phase I and II metabolic genes important in such oxidations is available.
It is well known that in other biological systems, inducers of P450 monooxygenases can also be substrates for oxidation by these enzymes (1). These considerations led us to study P450 genes inducible by NP, with the aim of identifying the putative P450 catalyst(s) involved in NP degradation. The results led to the first direct evidence for the involvement of fungal P450 enzymes in the degradation of the EDC NP and functional genomic identification of specific P450 monooxygenases responsive to an environmentally significant contaminant.
MATERIALS AND METHODS
Strain and culture conditions.
The P. chrysosporium strain used in this study, BKM-F-1767 (ATCC 24725), was maintained on malt extract (ME) agar. Unless otherwise stated, the fungus was grown at 37°C in ME broth, defined low-nitrogen (LN) medium (2.4 mM N as ammonium tartrate, 100 g/liter glucose), or defined high-nitrogen (HN) medium (24 mM N as ammonium tartrate, 100 g/liter glucose) as described elsewhere (6).
Inoculum preparation.
The fungal inoculum was prepared as described previously (43). Briefly, an aqueous suspension of P. chrysosporium conidia from 5-day-old cultures on ME agar plates incubated at 37°C was prepared and adjusted to an optical density at 600 nm of 15 (equivalent to ∼108 spores/ml). Fifty milliliters of the respective sterile growth medium (without Tween 80) in a wide-mouth 2.8-liter Fernbach flask was inoculated with 1 ml of the conidial suspension (final optical density at 600 nm of 0.3), and the flask was incubated at 37°C for 48 h under stationary conditions to allow the formation of a mycelial mat. The final inoculum was obtained by blending the mycelial mat aseptically into an equivalent volume (50 ml) of the respective sterile medium by using a handheld blender (Ultra-Turrax; Tekmar Co.) for 5 min (10 intermittent pulses of 30 s each) on ice. A uniform inoculum size (10%, vol/vol) was used for all cultures.
Biodegradation experiments.
P. chrysosporium was grown in 50-ml cultures in LN, HN, or ME medium with shaking (180 rpm) at 37°C in rubber-stoppered 125-ml conical flasks. After 24 h of incubation, NP (technical grade [catalog no. 29085-8; Sigma-Aldrich Corp.]) was added to the cultures to a final concentration of 100 ppm and the incubation was continued for an additional 72 h. A parallel set of identical cultures was supplemented simultaneously with the P450 enzyme inhibitor piperonyl butoxide (PB; in methanol) at various final concentrations (100, 500, and 1,000 μM). Each treatment was conducted in triplicate. The cultures were regularly flushed with oxygen for 1 min at 24-h intervals. Two types of controls with the same amounts of NP used in the experimental cultures were prepared: (i) an uninoculated control for the estimation of the initial level of NP and the degree of any abiotic degradation was prepared using the same medium (without an inoculum) used for the experimental cultures, and (ii) a chemically killed control for the estimation of the amount of added NP adsorbed to mycelia was prepared using cultures pregrown under conditions identical to those for the experimental cultures and then treated with 10 mM sodium azide for 2 h. Following incubation, the triplicate fungal cultures/controls for each treatment were separately extracted with methylene chloride (3×) and the extracts were dried on sodium sulfate and resuspended in acetonitrile by standard methods as described previously (43). The samples were filtered through 0.45-μm glass fiber filters and analyzed using a Prostar 210/215 high-performance liquid chromatography (HPLC) system (Varian, Inc.) equipped with a C18 reverse-phase column and a UV detector. HPLC separation was achieved using a 20-min linear gradient of 50% acetonitrile in water to 100% acetonitrile as the mobile phase at a flow rate of 1 ml/min. NP was detected at 277 nm and quantified based on a standard curve generated using known concentrations of this compound.
Gene induction conditions.
Four independent cultures (100 ml each) were grown in LN or ME medium in 250-ml flasks, as described above, with continuous shaking (180 rpm) and regular oxygenation (1 min for each flask) at 24-h intervals. Briefly, the ME cultures were grown for 24 h prior to induction by the addition of NP to a final concentration of 100 ppm, and the cultures were harvested 24 h postinduction. The LN cultures were grown for 96 h to the secondary metabolic phase (as indicated by browning), after which NP was added and the cultures were incubated for an additional 24 h before harvesting. Thus, the induction time was 24 h under either culture condition. The harvested mycelia were snap-frozen and maintained at −80°C prior to the extraction of RNA.
RNA extraction.
Total RNA was extracted from the frozen mycelia by using TRI reagent (Molecular Research Center) per the manufacturer's protocol, modified as described previously (45). The total RNA concentration was determined using an ND-1000 UV-visible spectrophotometer (NanoDrop Technologies), and the quality of RNA was checked using an Agilent 2100 bioanalyzer.
Microarray setup and data acquisition.
A custom-designed oligonucleotide microarray chip for P. chrysosporium recently developed in our laboratory was used (7, 45). Microarray slides were printed using 70-mer gene-specific oligonucleotides at the University of Cincinnati Genomics and Microarray Laboratory. The array consisted of a total of 250 genes, representing all 150 known P450 genes and 100 additional genes (including control genes and other metabolism-related genes) (7). The original gene names assigned in our earlier report (7) are retained, along with the corresponding updated names for ready reference. The additional genes included in the custom microarray comprised the following: 9 lignin peroxidase (LiP; lip) sequences (including 8 LiP isozyme-specific oligonucleotides and 1 common LiP signature oligonucleotide), 3 manganese-dependent peroxidase (MnP; mnp) genes, 2 housekeeping genes (namely, those encoding ubiquitin [ubq] and glyceraldehyde-3-phosphate dehydrogenase [gpd]), 25 mitogen-activated protein kinase genes, 9 cyclic AMP pathway genes, 4 sulfur metabolism genes, 8 nitrogen metabolism genes, 2 P450 electron transfer protein genes (those encoding cytochrome P450 oxidoreductase [POR] and cytochrome b5 reductase [cytb5r]), 3 phase I and phase II xenobiotic metabolism genes (those for microsomal epoxide hydrolase [m_eh], soluble epoxide hydrolase [s_eh], and glutathione S-transferase [gst]), and 35 other cellular metabolism-related genes. A detailed list of these genes is provided in our earlier report (7). Twenty-two of the P450 genes spotted were each represented by two separate oligonucleotides (instead of one oligonucleotide per gene) in order to verify the hybridization specificity of the chosen oligonucleotide probes. All 250 genes were spotted twice onto each microarray slide.
Differential microarray analyses of the experimental (NP-treated) cultures versus the control (untreated) cultures were performed using four biological replicates for each nutrient condition. The two nutrient conditions (those with ME and LN) were thus investigated independently. RNA samples from two of the four replicates for a given nutrient condition were labeled with a monofunctional fluorescent dye, Cy3 dye (green) for NP-treated cultures and Cy5 dye (red) for untreated controls. The remaining two replicates were labeled with the opposite dye (i.e., Cy3 for control cultures and Cy5 for experimental cultures). Following hybridization, the slides were scanned using both Cy5 (635-nm) and Cy3 (532-nm) fluorescence channels on the Axon 4000B scanner (Molecular Devices Corporation). The photomultiplier tube settings for Cy3 and Cy5 were optimized to obtain the same amounts of red and green signals, resulting in an overall pixel ratio of 1.0. The raw spot intensities (2 spots × 4 slides = 8 spots per gene) were obtained using GenePix Pro 5.4 software.
Microarray data analysis.
Data analysis was performed as described in our earlier report (7). Briefly, the data were normalized for the background and array-specific local regression was performed using the SAS statistical software package (SAS Institute Inc.). This was followed by the identification of the differentially expressed genes by fitting gene-specific mixed linear models (42). This model comes close to a three-way analysis of variance except that one of the variables (the array) is treated as a random effect. A multiple hypothesis testing adjustment of false-discovery rates (FDRs) was performed. An arbitrary cutoff of a twofold change in the transcript level was used to identify the differentially expressed genes. Expression profiles were clustered using average-linkage-based hierarchical clustering (10).
Real-time qRT-PCR analysis.
Total RNA (50 ng) was used for performing the real-time quantitative reverse transcription-PCR (qRT-PCR) analysis. Twelve genes, all of which showed differential regulation patterns in treated and untreated cultures under either or both of the nutrient conditions, were selected for this analysis. The four replicates each of NP-treated and untreated RNA samples were pooled separately, and each pool was subjected to qRT-PCR analysis in duplicate. The reaction was carried out using the Brilliant SYBR green QRT-PCR master mix kit (Stratagene) in the ABI Prism 9600 HT system (Applied Biosystems). The RT step of the reaction was carried out at 50°C for 30 min, followed by the PCR step: activation of the SureStart Taq DNA polymerase at 90°C for 10 min and then 40 cycles of amplification (denaturation at 95°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 30 s). The gene-specific primers used in this study are listed in Table 1. The level of induction was calculated as follows. The difference between the threshold cycle (CT) value for the test gene and that for the housekeeping gene gpd was calculated and used as the normalized CT value. The normalized CT value for the untreated control was then subtracted from the normalized CT value for the NP-treated sample to obtain the ΔCT value. The amount of change (n-fold) was calculated using the following formula: change = 2ΔCT.
TABLE 1.
Primers used in this study
| Gene | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|
| Pff 252a | CCGTCCACAGAACTTACCCAAGGC | GTAAGGTCGTGCGCTCAGTCCG |
| Pff 205h | GCGGGCACTGTAATTGTCGGC | CTGACCGAACCAGGGCTTCCGC |
| lpoB | CCCAAGTTCCAAGTCAAACG | GCTCGATGTCGTCGAAGATC |
| pc-2 | AGCCCGAACCCGTTCATCTTCCTC | GCAGCAGGGCACATCCACTAGG |
| glx1 | GCTCAGCAATGGCACTATG | CGAGCGTTCCAAGAAGGC |
| Pff 10a | CACACGGTCTCGCAGAGC | GGATACACTTCGTCGTCCAG |
| Pff 137a | CGAGTACTCAGACTCTCGC | GATCCACTGAAGCATGTCG |
| lipD | TCCATCGCTATCTCGCCC | ATGCGAGCGAGAACCTGA |
| agal-A | CATCACGAACAAGGCCATCATCG | GGTGTACGCCGATAGTGAC |
| xynB | CGGATCCGTCACCTACAAC | GATCGACGGCTCGTTCAC |
| gpd | GGCATTGTGCAGGGTCTCATG | GAGTAGCCCCACTCGTTGTC |
| POR | GAGCACTACCAGAACATCGTC | CAGCGTAGCTGCCTTGTCATG |
| Pff 141 | AACACGAACGGGATCAAGGAG | GTAGAACAGGTTGCTGAGGAC |
| pc-3 | CCAAGGAGGAGATAGACGAGGAC | CGAACGTTCCATGGGACAGCG |
| Pff 4a | GGACGCAACCTTGCATACA | GATTTCGGCACGCTTAGCC |
| Pff 311b | CGCTCTGGATAACACGGAGAAC | CAGAGCATCCAGGTGACC |
RESULTS AND DISCUSSION
Involvement of P450 monooxygenase(s) in NP degradation.
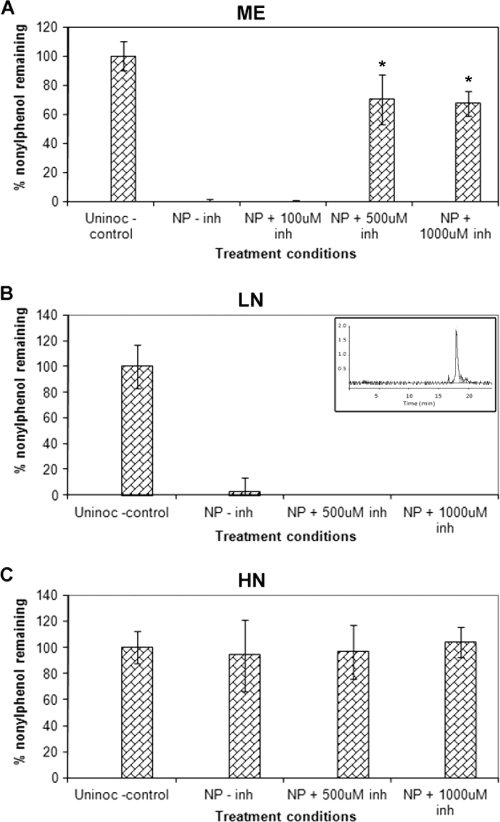
HPLC analysis showed that NP at a concentration of 100 ppm was degraded completely in ME and LN cultures (Fig. 1A and B) and not at all in HN cultures (Fig. 1C) of P. chrysosporium. Comparisons with the uninoculated controls and chemically killed controls indicated no or negligible abiotic degradation and mycelial adsorption of NP.
FIG. 1.
Effects of the P450 inhibitor (inh) PB on the degradation of NP by P. chrysosporium under different nutrient conditions. Final concentrations of PB of 100, 500, and 1,000 μM were used. Results for ME broth cultures (A) and cultures in defined LN medium (B) and defined HN medium (C) are shown. Bars represent average percentages of nondegraded NP. Asterisks indicate statistically significant values (P ≤ 0.05). The inset in panel B shows a typical HPLC peak profile for NP (technical grade). Error bars represent the standard errors calculated for three biological replicates. Uninoc, uninoculated.
The analysis of degradation in ME cultures indicated that this activity is mediated by enzymes other than the ligninolytic peroxidases. This pattern occurs because the expression of the peroxidases is suppressed under high-nutrient conditions, such as those in ME cultures (43). The observed significant abrogation of the degradation activity (by ∼75%) by the added eukaryotic P450 enzyme inhibitor PB (Fig. 1A) suggested that a cytochrome P450 enzyme(s) plays a key oxidizing role in the NP degradation process in P. chrysosporium. The abrogation activity of PB was concentration dependent (Fig. 1A), further suggesting that P450 enzyme activity is the underlying basis of NP degradation in this organism.
Under the defined LN conditions, the results were quite different. Unlike nutrient-rich ME cultures, LN cultures showed no effect of PB (at even the highest tested concentration of 1,000 μM) on the overall degradation of NP (Fig. 1B). This finding suggested either that the P450 enzymes are not involved in NP oxidation under LN conditions or that the P450 enzymes are readily replaceable by the predominant coexpressed peroxidases, which are the primary oxidizing enzymes involved in various degradation reactions under nutrient-limited conditions. Alternatively, PB may be labile in the LN cultures due to peroxidase and/or other ligninolytic enzyme activities and thus not available as a P450 inhibitor under these conditions. Furthermore, the possibility that other oxidative enzymes are involved under these conditions cannot be excluded.
The fact that we observed no detectable degradation of NP under the defined HN conditions (Fig. 1C), which are known to completely suppress peroxidase expression, lends credence to the hypothesis that extracellular peroxidases are involved in NP degradation in LN cultures.
It was interesting that there were major differences in the NP degradation patterns in HN and ME media, despite the fact that both these media may be considered to be nutrient rich. One possible explanation may be the compositional differences between the two media. HN medium is a synthetic medium containing defined amounts of the nutrients, especially inorganic nitrogen (24 mM) and glucose (1%). ME medium is a complex nutrient medium containing peptone and ME as the organic sources of nitrogen and carbon (maltose) and other nutrients in addition to the added carbon (2% glucose). It can therefore be speculated that the different nitrogen levels (24 mM in HN medium compared to 2.4 mM in LN medium and an estimated 8 mM in ME medium), along with the different types of nitrogen and carbon sources used in these nutrient media, may provide an explanation for the observed degradation patterns, possibly involving the differential P450 expression patterns. In contrast to the P450 genes, the peroxidase genes are known to be distinctly regulated by the nitrogen and/or carbon level (22, 32).
NP-inducible P450 monooxygenase genes and related phase I and phase II metabolic genes.
Our recent studies had shown that P450 genes in P. chrysosporium are differentially regulated under the two biodegradation-relevant nutrient conditions (those with limited and sufficient nutrients) and in response to several xenobiotic chemicals (6, 7, 9, 36, 45, 47). However, genome-wide analysis to identify all P450 monooxygenases responsive to a given xenobiotic toxicant in this system has not been reported yet.
PB exerted differential effects on NP degradation under different nutrient (ME versus LN) conditions, suggesting that the induction of NP-oxidizing P450 enzymes is tightly regulated by the nutrient status. We therefore used the microarray approach to identify the specific P450 genes involved in the NP oxidation process under the two nutrient conditions, one (the presence of ME) under which the P450 enzymes seemed to play a clear role and the other (LN) under which the P450 enzymes seemed to be unimportant or dispensable (replaceable by peroxidases) in the NP oxidation process.
All genes that showed differential patterns of regulation by NP under either nutrient condition (in LN or ME medium) are listed in Table 2 with their corresponding Joint Genome Institute (JGI) gene models (version 1 and version 2), as well as the CYP nomenclature in the case of P450 monooxygenase genes. Twelve genes were selected for confirmation using qRT-PCR. Except for pc-2, these genes were either induced or repressed by NP under only one of the two nutrient conditions. Overall, the induction patterns in the microarray analysis corroborated those observed in the qRT-PCR analysis (Table 3). Nevertheless, qRT-PCR yielded relatively higher change (n-fold) values, especially for the highly inducible genes, possibly because of the spot saturation effect during microarray hybridization.
TABLE 2.
New and old gene model assignments for NP-induced genes from P. chrysosporium and corresponding nutritional regulation information
| Genea | JGI gene ID
|
JGI protein ID (version 2) | P450 familyc | Nutritional regulation under LN vs HN conditionsd | |
|---|---|---|---|---|---|
| Version 1b | Version 2 | ||||
| P450 genes induced by NP in ME cultures | |||||
| PC-2 | ug.20.35.1 | e_gwh2.2.241.1 | 129114 | CYP63A2 | U(HN) |
| Pff 10a | pc.21.108.1 | e_gww2.9.198.1 | 132113 | CYP512H1 | U(HN) |
| Pff 129 | pc.81.21.1 | fgenesh1_pg.C_scaffold_4000627 | 3368 | CYP5141D1 | NR |
| Pff 141 | pc.1.6.1 | e_gww2.8.145.1 | 138737 | CYP5035A5 | NR |
| Pff 164a | ug.1.19.1 (pc.1.261.1) | e_gww2.5.195.1 | 130996 | CYP53C2 | U(HN) |
| Pff 164c | ug.1.25.1 | e_gwh2.5.180.1 | 121157 | CYP5037B2 | NR |
| Pff 169f | pc.83.7.1 | fgenesh1_pg.C_scaffold_1000770 | 770 | CYP5144A9 | U(HN) |
| Pff 173 | genewise2nd.66.11.1 (genscan.66.16.1) | fgenesh1_pg.C_scaffold_2000954 | 1976 | CYP5150B1 | U(HN) |
| Pff 205h | genewise2nd.24.9.1 (pc.24.11.1) | e_gww2.1.401.1 | 133311 | CYP5144A7 | U(HN) |
| Pff 252d | genewise2nd.24.17.1 (pc.24.19.1) | e_gww2.1.429.1 | 132914 | CYP5144D4 | U(LN) |
| Pff 311b | pc.142.11.1 | fgenesh1_pg.C_scaffold_9000456 | 5852 | CYP5136A2 | NR |
| Pff 311c | pc.142.5.1 | e_gww2.9.179.1 | 131921 | CYP5144C7 | NR |
| Pff 388a | pc.181.9.1 | Not assigned | CYP5141A1 | NR | |
| Pff 4a | pc.16.161.1 (genscan.16.7.1) | fgenesh1_pg.C_scaffold_8000176 | 5001 | CYP5136A3 | U(HN) |
| Pff 5 | ug.11.23.1 | fgenesh1_pg.C_scaffold_11000011 | 6326 | CYP5145A3 | NR |
| Pff 7 | genewise2nd.38.22.1 | e_gwh2.2.246.1 | 129675 | CYP5139A1 | NR |
| Pff 78 | ug.128.46.1 | e_gww2.3.561.1 | 137301 | CYP5036A3 | NR |
| Pff 82b | ug.43.44.1 | fgenesh1_pg.C_scaffold_22000065 | 9198 | CYP5035C1 | NR |
| P450 genes induced by NP under LN conditions | |||||
| foxy1 | ug.73.16.1 (pc.73.11.1) | e_gww2.3.23.1 | 137601 | CYP505D4 | U(HN) |
| pc-2 | ug.20.35.1 | e_gwh2.2.241.1 | 129114 | CYP63A2 | U(HN) |
| Pff 137a | pc.30.118.1 | fgenesh1_pg.C_scaffold_18000091 | 8720 | CYP512B5 | NR |
| Pff 15c | ug.50.27.1 | fgenesh1_pg.C_scaffold_5000216 | 3654 | CYP5147A1 | NR |
| Pff 169f | pc.83.7.1 | fgenesh1_pg.C_scaffold_1000770 | 770 | CYP5144A9 | U(HN) |
| Pff 205d | genewise2nd.24.5.1 (pc.24.8.1) | e_gww2.1.391.1 | 133327 | CYP5144A3 | NR |
| Pff 242b | ug.73.16.1 | e_gww2.3.23.1 | 137601 | CYP505D4 | U(HN) |
| Pff 242c | ug.73.15.1 (pc.73.10.1) | e_gww2.3.57.1 | 137435 | CYP505D3 | U(HN) |
| Pff 249a | ug.53.54.1 (genscan.53.52.1) | fgenesh1_pg.C_scaffold_20000051 | 8961 | CYP5035A3 | U(LN) |
| Pff 252a | pc.24.27.1 | gwh2.1.540.1 | 28946 | CYP5142A3 | U(HN) |
| Pff 361a | pc.73.4.1 | gww2.3.2.1 | 34980 | CYP505D2 | U(LN) |
| Pff 361b | ug.73.17.1 (pc.73.3.1) | Not assigned | CYP505D1 | U(HN) | |
| Pff 51 | pc.14.210.1 | fgenesh1_pg.C_scaffold_10000101 | 6018 | CYP5136A5 | NR |
| Pff 57 | pc.15.127.1 | gwh2.15.175.1 | 33411 | CYP5036A1 | U(HN) |
| Pff 7 | genewise2nd.38.22.1 | e_gwh2.2.246.1 | 129675 | CYP5139A1 | NR |
| Pff 86f | ug.20.43.1 (pc.20.56.1) | e_gwh2.2.406.1 | 129578 | CYP5142C1 | U(HN) |
| Pff 9 | ug.97.52.1 | fgenesh1_pg.C_scaffold_13000094 | 7306 | CYP5035A2 | NR |
| Peroxidase genes differentially regulated by NP under LN conditions | |||||
| lipA (M22720) | pc.85.52.1 | fgenesh1_pm.C_scaffold_19000014 | 10957 | ||
| lipC (M36815) | pc.85.38.1 | e_gww2.19.87.1 | 131738 | ||
| lipD (M18743) | pc.19.174.1 | fgenesh1_pg.C_scaffold_11000496 | 6811 | ||
| lipF (M80213) | pc.21.40.1 | e_gwh2.9.293.1 | 122202 | ||
| lipH (M24082) | pc.85.15.1 | e_gwh2.19.97.1 | 121806 | ||
| lipJ (AF140062) | genewise2nd.85.6.1 | e_gww2.19.111.1 | 131709 | ||
| lpoB (M37701) | pc.85.50.1 | e_gwh2.19.98.1 | 121822 | ||
| mnp1 (M60672) | pc.15.23.1 | e_gww2.15.98.1 | 140708 | ||
| mnp3 (U10306) | pc.27.12.1 | fgenesh1_pg.C_scaffold_1000878 | 878 | ||
| Transcription factor genes and signal transduction genes/gene homologs differentially regulated by NP in ME or LN cultures | |||||
| agal-A (AF246262) | pc.11.202.1 | e_gwh2.11.209.1 | 125033 | ||
| xynB-A (AF301904) | pc.120.11.1 | e_gww2.6.463.1 | 133788 | ||
| POR (AAG31349.1) | pc.51.97.1 | fgenesh1_kg.C_scaffold_12000002 | 11093 | ||
| cbhI-2 (S40817) | ug.139.1.1 | e_gww2.14.106.1 | 137042 | ||
| CDH (U46081) | pc.5.157.1 | fgenesh1_kg.C_scaffold_12000007 | 11098 | ||
| YAP2 (S68847) | genscan.183.4.1 | fgenesh1_pg.C_scaffold_5000600 | 4038 | ||
| TEC1 (M32797) | pc.100.54.1 | e_gwh2.13.284.1 | 126075 | ||
| Cellulose binding protein gene ac2 (AJ003224) | pc.47.15.1 | e_gwh2.2.646.1 | 129325 | ||
| gstA (AF425746) | pc.3.88.1 | fgenesh1_pg.C_scaffold_1000503 | 503 | ||
| OSM1 (Af184980) | pc.31.2.1 | e_gww2.13.222.1 | 135546 | ||
| pbs2 (NP_012407) | pc.200.8.1 | e_gww2.15.132.1 | 140816 | ||
| creA (AJ272151) | genewise2nd.23.8.1 | fgenesh1_pg.C_scaffold_8000320 | 5145 | ||
| uap (AB083397) | pc.93.60.1 | e_gwh2.22.25.1 | 121582 | ||
| glx1 (L47286) | pc.52.9.1 | fgenesh1_kg.C_scaffold_6000005 | 11068 | ||
| CBHI.1 (Z22528) | pc.139.26.1 | e_gwh2.14.89.1 | 127029 | ||
| STE12 (X16112) | pc.56.6.1 | gwh2.9.410.1 | 33036 | ||
NCBI accession numbers are shown in parentheses. Pff gene numbering is the same as that used in reference 7; the Pff numbering corresponds to the original scaffold numbers used by David Nelson on his cytochrome P450 webpage (http://drnelson.utmem.edu/whiterot.fasta.html).
Gene ID assigned in JGI genome annotation version 1. Numbers in parentheses represent the original (prerevision) JGI gene models of the draft genome as used by Doddapaneni and Yadav (7); these gene models were subsequently replaced by the revised JGI version 1 gene models.
Designations were assigned by the International Nomenclature Committee.
Nutritional regulation information was derived from Doddapaneni and Yadav (7). U(HN) indicates gene induction under HN conditions, U(LN) indicates gene induction under LN conditions, and NR means nonregulated.
TABLE 3.
Verification of the microarray-based induction patterns of selected genes by qRT-PCR analysisa
| Gene name | Change (n-fold) in ME cultures as determined by:
|
Change (n-fold) under LN conditions as determined by:
|
||
|---|---|---|---|---|
| Microarray | qRT-PCR | Microarray | qRT-PCR | |
| Pff 4a | 166.96 ± 0.091* | 537.36 ± 153.38 | 1.1 | 2.05 ± 0.64 |
| xynB-A | 22.55 ± 0.098* | 10.62 ± 3.47 | 1.41 ± 0.20 | 0.15 ± 2.02 |
| gpd | −1.51 ± 0.051* | 1.01 ± 0.01 | −1.08 ± 0.26 | −1.96 ± 0.15 |
| pc-2 | 8.49 ± 0.065* | 8.27 ± 3.42 | 3.93 ± 0.19* | 16.77 ± 3.18 |
| Pff 311b | 194.94 ± 0.055* | 506.95 ± 22.32 | −1.5 ± 0.22 | 2.82 ± 0.80 |
| Pff 141 | 5.12 ± 0.092* | 17.57 ± 0.77 | 1.52 ± 0.16 | 3.08 ± 0.33 |
| agal-A | 2.58 ± 0.081* | 2.33 ± 1.50 | 1.4 ± 0.20 | 3.41 ± 0.33 |
| pc-3 | 1.97 ± 0.081* | 4.59 ± 1.07 | −1.31 ± 0.19 | 1.76 ± 0.81 |
| POR | 3.82 ± 0.073* | 3.60 ± 0.24 | −1.1 ± 0.20 | −1.27 ± 0.03 |
| Pff 10a | 2.64 ± 0.135* | 1.19 ± 0.14 | 1.87 ± 0.18* | 5.62 ± 1.52 |
| Pff 205h | 3.04 ± 0.208* | 1.64 ± 0.52 | 6.6 | 10.02 ± 3.51 |
| Pff 252a | −2.39 ± 0.138* | −1.46 ± 0.41 | 5.11 ± 0.3* | 4.26 ± 1.88 |
| glx | −1.21 ± 0.119 | 0.03 ± 2.61 | −10.44 ± 0.48* | −5.43 ± 0.31 |
| lipD | 1.33 ± 0.23 | −0.08 ± 1.58 | −25.68 ± 0.49* | −6.09 ± 0.47 |
| lpoB | −1.16 ± 0.076 | −0.03 ± 1.67 | −7.2 ± 0.20* | −14.88 ± 0.80 |
| Pff 137a | −13.36 ± 0.136* | −3.28 ± 1.81 | 6.12 ± 0.31* | 9.10 ± 0.53 |
The microarray values are averages ± standard errors obtained by evaluating eight independent spots for each gene. The qRT-PCR values are averages ± standard errors obtained from duplicate samples. Asterisks indicate values that are statistically significant (P ≤ 0.05) and correspond to an FDR of <0.1.
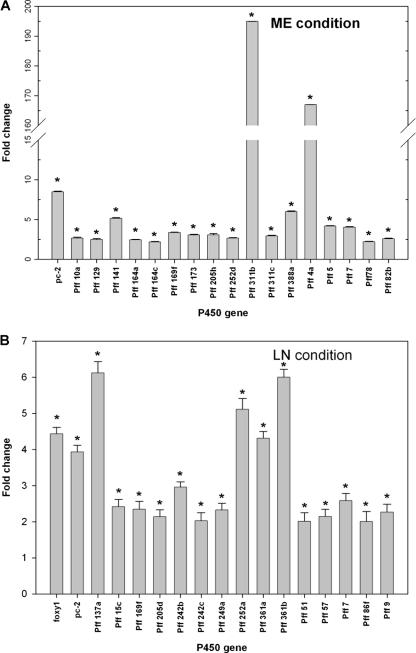
Interestingly, the microarray data showed that NP differentially induced several P450 genes (cutoff = twofold) under the two nutrient conditions: 18 genes in ME cultures and 17 genes in LN cultures (Fig. 2). The level of induction in ME cultures varied, ranging from a mean ± standard error of 2.20- ± 0.07-fold (P = 0.0001; FDR = 0.0011) for the Pff 78 gene (corresponding to protein identification number [ID] 137301) to 194.94- ± 0.092-fold (P = 0.0000; FDR = 0.0000) for the Pff 311b gene (protein ID 5852). On the other hand, the level of induction under LN conditions was much lower than that in the ME cultures, ranging from 2.01- ± 0.24-fold (P = 0.016; FDR = 0.07) for Pff 51 (protein ID 6018) to 6.12- ± 0.31-fold (P = 0.00002; FDR = 0.0009) for Pff 137a (protein ID 8720).
FIG. 2.
Induction of P450 genes in response to NP in P. chrysosporium cultures grown under ME (A) and LN (B) conditions, as determined by the custom P450 microarray analysis. Bars represent average levels of induction (n-fold) of individual P450 genes. Asterisks indicate the statistically significant values (P ≤ 0.05; FDR < 0.1). Four biological replicates for each nutrient condition were used. Error bars represent the standard errors calculated based on change (n-fold) values for eight spots (those on four slides with two spots per slide) for each gene.
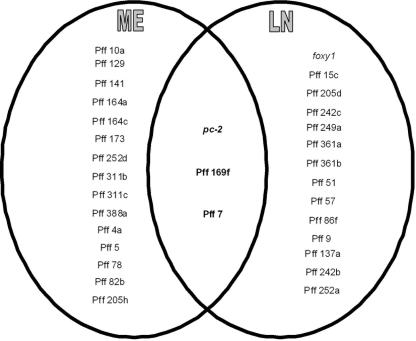
Of the NP-induced P450 genes, 15 genes were specifically induced (2.16- to 194.94-fold) in ME cultures whereas 14 genes were specifically induced, albeit to a much lower extent (2.01- to 6.12-fold), in LN cultures (Fig. 3). This finding suggests that P450 genes in P. chrysosporium are induced by NP in a nutrient-specific manner. Nevertheless, three of the NP-induced genes were common to the two culture conditions, although their induction levels differed significantly between the two conditions (Fig. 3). The common induction of these genes (pc-2 [protein ID 129114], Pff 169f [protein ID 770], and Pff 7 [protein ID 129675]) implies that their induction is compound (NP) specific and that they may be involved in NP degradation under either nutrient condition. Of the three commonly induced genes, pc-2, which belongs to the CYP63 family, has been shown to be induced by alkanes, polycyclic aromatic hydrocarbons, and aromatics with alkyl substitutions (6, 36, 47). No such transcriptional induction information is yet available for the other two genes (Pff 169f and Pff 7). However, considering that the P450 enzyme inhibitor did not abrogate the degradation activity in LN cultures, their involvement under LN conditions is not essential for the degradation process.
FIG. 3.
Comparative profiling of P. chrysosporium P450 genes inducible by NP under different nutrient conditions. Only data for those P450 genes that showed greater than twofold induction under either nutrient condition (in LN or ME medium) are included. Genes represented in the overlapping center region are those that showed induction under both the nutrient conditions.
An important aspect of this comparative profiling is that two genes induced in ME cultures only, namely, Pff 311b (protein ID 5852) and Pff 4a (protein ID 5001), showed extraordinarily high levels of induction (∼195- and 167-fold, respectively). Both of these genes belong to the CYP617/CYP547 clan of the P. chrysosporium P450ome (8). In order to delineate the functional and evolutionary relationships of the P450 enzymes encoded by these two genes, we analyzed the protein sequences against those in the NCBI protein database by using BLAST. These proteins showed relatedness to the members of the CYP4F family of cytochrome P450 proteins, which are known to catalyze the hydroxylation of fatty acids in higher eukaryotic systems (3, 34). This observation suggested further that these P450s might be involved primarily in the degradation of NP, which has a chemical substructure (a long alkyl side chain) that resembles the aliphatic backbones of the fatty acids and other related aliphatic compounds. Interestingly, the dramatic differences between the levels of these two P450s in ME versus LN cultures are consistent with the observed differential effects of the P450 enzyme inhibitor on the NP degradation activities in ME and LN cultures (Fig. 1).
Our previous analysis had shown that several cytochrome P450 genes are linked tandem as clusters in the P. chrysosporium genome (7). The P450 genes that belong to the tandem gene clusters in the genome showed independent (nonassortative) regulation in ME cultures (7). In contrast, two of the gene clusters, each containing two genes (genes Pff 361a [protein ID 34980] and Pff 361b [protein ID not assigned] and genes Pff 242b [protein ID 137601] and Pff 242c [protein ID 137435]), showed coordinated regulation under LN conditions (Table 2). Interestingly, members of these two gene clusters belong to the CYP505 family of cytochrome P450 genes. The observed divergence in the induction patterns of the majority of the NP-induced P450 genes (except Pff 361a, Pff 361b, Pff 242b, and Pff 242c) under the two nutrient conditions tested, despite the genes' being linked tandem and structurally related (encoding proteins belonging to the same family), indicates the possible evolutionary diversification that these genes have undergone in order to acquire diverse functions.
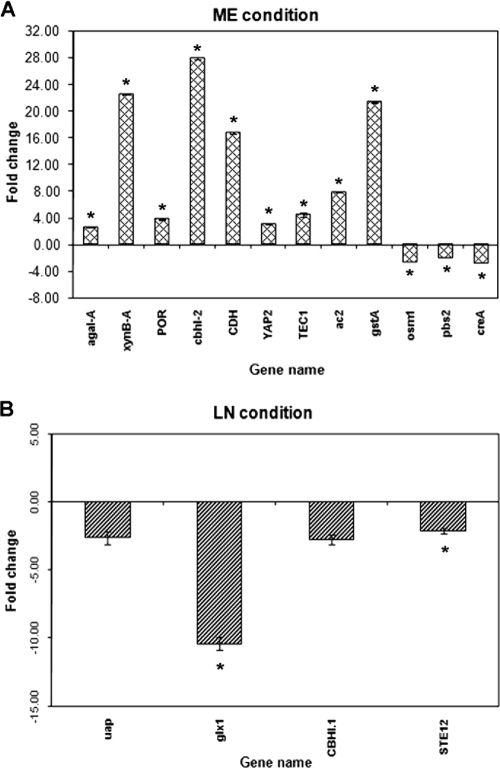
Evidence for the role of P450 enzymes in the degradation of NP in ME cultures was further substantiated by the following observation: the P450 oxidoreductase gene (POR; protein ID 11093) encoding one of the two P450 electron transfer proteins (POR and cytochrome b5 reductase [protein ID 128228]) and the phase II metabolic enzyme glutathione S-transferase gene (gst; protein ID 503) were coinduced (3.82- ± 0.073-fold [P = 0.00001; FDR = 0.0015] and 21.33- ± 0.092-fold [P < 0.00000; FDR < 0.00000], respectively) in the presence of NP in ME cultures (see Fig. 5A). POR is required for the transfer of electrons from NADPH to cytochrome P450 monooxygenases during the P450 catalytic cycle, leading to substrate hydroxylation. In contrast, glutathione S-transferase is a phase II xenobiotic metabolizing enzyme involved in the further modification of the hydroxylated products generated by the P450 monooxygenase activity. Unlike that in ME cultures, the change in the expression of gst (protein ID 503) in LN cultures in response to NP treatment did not exceed the cutoff (twofold).
FIG. 5.
Changes in the expression of transcription factor genes and other signal transduction genes responsive to NP in P. chrysosporium grown under different nutrient conditions as revealed by microarray analysis. Bars represent the average changes (n-fold) in expression of only those genes that showed changes greater than the cutoff of ±2.0-fold. Asterisks indicate the statistically significant values (P ≤ 0.05; FDR < 0.1). Four biological replicates were used for each nutrient condition. Error bars represent the standard errors calculated based on change (n-fold) values for eight spots (those on four slides with two spots per slide) for each gene.
In contrast to those upregulated, 8 P450 genes were downregulated (2.15- to 13.36-fold) in ME cultures and 10 P450 genes were downregulated (2.02- to 4.15-fold) in LN cultures in response to NP (data not shown). The observed downregulation may be due partly to the general toxicity of NP to the fungus, as reported earlier by Kollmann et al. (24), or to the rerouting of the fungal protein synthesis machinery for de novo synthesis of the induced P450s.
Table 2 lists P450 genes that showed differential induction patterns in the presence of NP under the tested nutrient-rich (ME) versus nutrient-limited (LN) conditions (this study), along with the respective information on nitrogen regulation under normal conditions (in HN and LN media without NP treatment) from our previous study (7). Seven of the 18 P450 genes that showed induction in NP-treated ME cultures in this study also showed induction in untreated HN cultures in our previous study (7) (Table 2). Two of the 17 genes that were induced in NP-treated LN cultures in this study likewise showed induction in the untreated LN cultures in our previous study (7) (Table 2). In contrast, 1 of the 18 genes that showed induction in NP-treated ME cultures in this study showed induction in untreated LN cultures in the earlier study and 9 of the 17 genes induced in NP-treated LN cultures (this study) were induced in untreated HN cultures (7) (Table 2). These observations collectively indicate that there is a direct regulatory (inducing/repressing) effect of the treatment chemical (NP) on the genes, in addition to the effect of the nutrient conditions. The data from these two studies (present and previous) also point to the fact that even though both HN and ME media may be considered to provide high-nutrient conditions, these media support different P450 induction patterns, possibly due to the differences in their carbon and nitrogen levels and chemical natures (organic versus inorganic). This result is consistent with the pattern differences observed for the NP degradation activity in the two media.
Effects of NP on peroxidase genes.
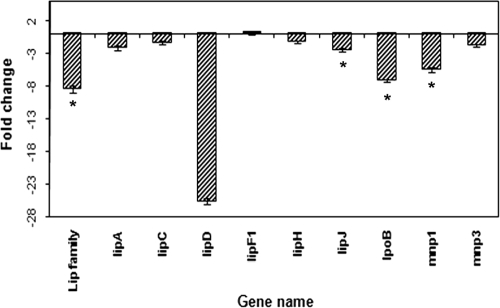
Hybridization signals from LiP genes in NP-treated LN cultures varied from those in the untreated cultures (Fig. 4). Four LiP genes (lipA [protein ID 10957], lipD [protein ID 6811], lipJ [protein ID 131709], and lpoB [protein ID 121822]) were downregulated in the presence of NP, with statistical significance being found in three cases (those of lipD [downregulated 25.68- ± 0.49-fold; P = 0.00006; FDR = 0.0016], lipJ [downregulated 2.47- ± 0.24-fold; P = 0.0043; FDR = 0.033], and lpoB [downregulated 7.20- ± 0.20-fold; P = 0.000003; FDR = 0.0004]). The LiP-specific common oligonucleotide spot also showed a signal intensity lowered by 7.20- ± 0.20-fold (P = 0.005; FDR = 0.03), indicating overall downregulation of the LiP gene pool in the presence of NP. The expression changes in other LiP genes, lipC (protein ID 131738), lipF (protein ID 122202), and lipH (protein ID 121806), did not exceed the cutoff of twofold. In NP-treated ME cultures, the LiP genes showed no or negligible hybridization signals.
FIG. 4.
Regulation of the peroxidase genes in P. chrysosporium in response to NP under LN culture conditions as revealed by microarray analysis. Bars represent average changes (n-fold) in gene expression; those with an asterisk indicate the statistically significant values (P ≤ 0.05; FDR < 0.1). Four biological replicates were used for each nutrient condition. Error bars represent the standard errors calculated based on change (n-fold) values for eight spots (those on four slides with two spots per slide) for each gene.
Among the MnP genes (Fig. 4), mnp1 (protein ID 140708) and mnp3 (protein ID 878) showed considerable hybridization signals in LN cultures. mnp3 showed no significant change in expression, whereas mnp1 was downregulated (5.51- ± 0.31-fold [P = 0.0004; FDR = 0.0064]) by NP. On the other hand, none of the MnP genes spotted showed considerable hybridization signals in ME cultures; mnp3 showed a low hybridization signal representing a change of less than twofold (data not shown).
As expected, the expression of the peroxidases (LiPs and MnPs) was detected only in LN cultures. Four of the eight LiP genes and one of the three MnP genes (mnp1) were expressed at higher levels in untreated than in NP-treated LN cultures. This finding indicated that treatment with NP at 100 ppm directly or indirectly inhibits the expression of some of the peroxidase genes. Furthermore, the expression of another ligninolytic condition-specific gene, that for glyoxal oxidase (glx1; protein ID 11038), was downregulated 10.44- ± 0.48-fold (P = 0.0006; FDR = 0.0083) in the NP-treated LN cultures, consistent with the repression of peroxidase genes, the major genes specific to the ligninolytic conditions. Overall, lower levels of expression of peroxidase genes and other genes specific to ligninolytic conditions in NP-treated than in untreated LN cultures may be due to the suppressing effect of NP on the common transcription network for these genes. Nevertheless, the peroxidases in the NP-treated LN cultures seemed to be enough to catalyze NP oxidation without the need for the involvement of P450 monooxygenases. However, the involvement of other oxidative enzymes in NP oxidation in LN cultures cannot be ruled out. Despite a detectable inhibitory effect on some components of the ligninolytic metabolic machinery, NP addition did not seem to affect the overall NP-degrading potential of the fungus, as indicated by comparable percentages of NP degradation in LN and ME cultures. This finding reinforces the idea of a contributory but replaceable/dispensable role for P450s under LN conditions.
Effects of NP on other metabolic and regulatory genes.
The expression of the two tested housekeeping genes gpd (protein ID 132198) and ubq (protein ID 10733) remained unchanged (differences in transcript levels were within the cutoff range of ±2.00-fold) under the ME and LN degradation conditions (Table 3).
(i) Effects in ME cultures.
Several of the genes catalyzing the breakdown of polysaccharides and other sugars, including those involved in the hydrolysis of xylan (endo-1,4-β-xylanase B [xynB-A; protein ID 133788]), cellulose (cellulose binding protein gene ac2; protein ID 129325), cellobiose (cbhI-2 [protein ID 137042] and CDH [protein ID 11098]), and galactose (agal-A; protein ID 125033), were significantly upregulated in ME cultures (Fig. 5A), indicating coregulation of the alternate carbon source-utilizing enzymes during NP degradation. This observation is consistent with the statistically significant reduction (2.90- ± 0.000-fold; P = 0.00019; FDR = 0.0014) in the expression of the catabolite repressor protein encoded by creA (protein ID 5145) in the presence of NP under these growth conditions; creA is a known suppressor of xylanases and cellulases in the cellulolytic fungus Trichoderma reesei (17, 29). While the physiological relevance of the induction of these hydrolytic enzymes is not clear, it may be assumed that NP exposure prepares the fungal system for assimilating alternate carbon sources to sustain the nutrient environment required for P450-mediated biodegradation.
Two transcription factor genes, namely, TEC1 and YAP2, were upregulated severalfold by NP in ME cultures (Fig. 5A). TEC1 (protein ID 126075), a homolog of the gene involved in Ty1 and Ty1-mediated gene expression, is involved in pseudohyphal development in Saccharomyces cerevisiae, and YAP2 (protein ID 126075), a homolog of the AP-1-like stress-induced transcriptional activator gene, is involved in oxidative-stress responses (12, 35). The NP induction of TEC1 and YAP2 genes suggests a role in the survival of the fungus under the NP biodegradation conditions through the enhancement of hyphal length, thereby allowing more access to the nutrients and combating the increased oxidative stress in the presence of NP. The stress-responsive genes osm1 (protein ID 135546) and pbs2 (protein ID 140816) were downregulated during NP degradation under these growth conditions (Fig. 5A).
(ii) Effects in LN cultures.
The P. chrysosporium homolog of the fungal stress-responsive gene for uric acid xanthine permease (uap; protein ID 121582) showed significantly lower expression in the presence of NP than in the absence of NP under LN conditions (Fig. 5B). This finding is in contrast to the observed upregulation of uap in Aspergillus nidulans and P. chrysosporium in response to other forms of chemical stress (13, 27). This result may be a consequence of the general toxicity/chemical stress or an inhibition effect of NP on the metabolism of the fungus and/or may be due to metabolic rerouting of the fungal enzyme synthesis machinery to attain the induced levels of P450 enzymes and other inducible proteins. Of the two cellobiose hydrolysis genes, CBHI.1 (protein ID 127029) showed 2.76- ± 0.35-fold downregulation (P = 0.1140; FDR = 0.185) (Fig. 5B). In contrast, the cellulose hydrolysis genes did not seem to be significantly regulated under these culture conditions.
An important aspect of this study was the use of technical-grade NP, a formulation containing multiple congeners, as opposed to the model compound 4-n-NP, which consists of a single congener. Use of the technical grade was prompted by the fact that such a mixture of congeners is more difficult to degrade than a single congener, thereby making technical-grade NP a more environmentally representative form of this chemical. In this context, it can be assumed that individual congeners of NP would have induction potentials different from those of the mixture of congeners used in this study. Consequently, the observed profile of P450 gene induction by technical-grade NP is a net result of the inductions by the individual NP congeners present therein and is more representative of the natural scenario occurring in the environment.
In conclusion, this study has demonstrated a role for P450 monooxygenases in the biodegradation of NP by P. chrysosporium and has led to the identification of multiple P450 monooxygenase genes specifically induced under NP biodegradation conditions. The NP-inducible P450 monooxygenases of P. chrysosporium are differentially regulated by nutrient conditions, however. The identification of the specific NP-responsive genes in this study would allow their heterologous expression and use as tools to elucidate the catalytic activities and substrate specificities of the P450 biocatalysts for NP degradation. This study is also the first to report genome-wide induction of the P450 monooxygenase genes in response to a specific xenobiotic toxicant class in white rot fungi. Our future work will include transcriptional profiling for the identification of P450 genes that are responsive to the individual congeners of NP. Such xenobiotic class/congener-specific information will help in engineering versions of the intact fungus with increased activities for the degradation of specific environmental pollutants.
Acknowledgments
This work was supported by the NIH/NIEHS grants ES10210 and ES015543 to J.S.Y., a University of Cincinnati research incentive award (to J.S.Y.), and NIH/NIEHS Center for Environmental Genetics (CEG) grant P30-ES06096.
We thank Mary Beth Genter and Stuart Baxter for critical reading and editing of the manuscript. We acknowledge Mario Medvedovic for his help in microarray data analysis.
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Anzenbacher, P., and E. Anzenbacherová. 2001. Cytochrome P450 and metabolism of xenobiotics. Cell. Mol. Life Sci. 58:737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabana, H., J. L. Jiwan, R. Rozenberg, V. Elisashvili, M. Penninckx, S. N. Agathos, and J. P. Jones. 2007. Elimination of endocrine disrupting chemicals nonylphenol and bisphenol A and personal care product ingredient triclosan using enzyme preparation from the white rot fungus Coriolopsis polyzona. Chemosphere 67:770-778. [DOI] [PubMed] [Google Scholar]
- 3.Christmas, P., K. Tolentino, V. Primo, K. Z. Berry, R. C. Murphy, M. Chen, D. M. Lee, and R. J. Soberman. 2006. Cytochrome P-450 4F18 is the leukotriene B4 omega-1/omega-2 hydroxylase in mouse polymorphonuclear leukocytes: identification as the functional orthologue of human polymorphonuclear leukocyte CYP4F3A in the down-regulation of responses to LTB4. J. Biol. Chem. 281:7189-7196. [DOI] [PubMed] [Google Scholar]
- 4.Corvini, P. F., R. J. Meesters, A. Schaffer, H. F. Schroder, R. Vinken, and J. Hollender. 2004. Degradation of a nonylphenol single isomer by Sphingomonas sp. strain TTNP3 leads to a hydroxylation-induced migration product. Appl. Environ. Microbiol. 70:6897-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries, Y. P., Y. Takahara, Y. Ikunaga, Y. Ushiba, M. Hasegawa, Y. Kasahara, H. Shimomura, S. Hayashi, Y. Hirai, and H. Ohta. 2001. Organic nutrient-dependent degradation of branched nonylphenol by Sphingomonas sp. YT isolated from a river sediment sample. Microbes Environ. 16:240-249. [Google Scholar]
- 6.Doddapaneni, H., and J. S. Yadav. 2004. Differential regulation and xenobiotic induction of tandem P450 monooxygenase genes pc-1 (CYP63A1) and pc-2 (CYP63A2) in the white-rot fungus Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 65:559-565. [DOI] [PubMed] [Google Scholar]
- 7.Doddapaneni, H., and J. S. Yadav. 2005. Microarray-based global differential expression profiling of P450 monooxygenases and regulatory proteins for signal transduction pathways in the white rot fungus Phanerochaete chrysosporium. Mol. Genet. Genomics 274:454-466. [DOI] [PubMed] [Google Scholar]
- 8.Doddapaneni, H., R. Chakraborty, and J. S. Yadav. 2005. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: evidence for gene duplications and extensive gene clustering. BMC Genomics 6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doddapaneni, H., V. Subramanian, and J. S. Yadav. 2005. Physiological regulation, xenobiotic induction, and heterologous expression of P450 monooxygenase gene pc-3 (CYP63A3), a new member of the CYP63 gene cluster in the white-rot fungus Phanerochaete chrysosporium. Curr. Microbiol. 50:292-298. [DOI] [PubMed] [Google Scholar]
- 10.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Commission. 2002. European Union risk assessment report. Office for Official Publication of the European Communities, Luxembourg.
- 12.Gavrias, V., A. Andrianopoulos, C. J. Gimeno, and W. W. Timberlake. 1996. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 19:1255-1263. [DOI] [PubMed] [Google Scholar]
- 13.Gorfinkiel, L., G. Diallinas, and C. Scazzocchio. 1993. Sequence and regulation of the uapA gene encoding a uric acid-xanthine permease in the fungus Aspergillus nidulans. J. Biol. Chem. 268:23376-23381. [PubMed] [Google Scholar]
- 14.He, Y., and H. K. Lee. 1996. Separation of structural homologues of alkylphenols and isomers of 4-nonylphenol by cyclodextrin-modified micellar electrokinetic chromatography. J. Chromatogr. A 749:227-236. [Google Scholar]
- 15.Heemken, O. P., H. Reincke, B. Stachel, and N. Theobald. 2001. The occurrence of xenoestrogens in the Elbe River and the North Sea. Chemosphere 45:245-259. [DOI] [PubMed] [Google Scholar]
- 16.Hu, J. Y., G. H. Xie, and T. Aizawa. 2002. Products of aqueous chlorination of 4-nonylphenol and their estrogenic activity. Environ. Toxicol. Chem. 21:2034-2039. [PubMed] [Google Scholar]
- 17.Ilmen, M., M. L. Onnela, S. Klemsdal, S. Keranen, and M. Penttila. 1996. Functional analysis of the cellobiohydrolase I promoter of the filamentous fungus Trichoderma reesei. Mol. Gen. Genet. 253:303-314. [DOI] [PubMed] [Google Scholar]
- 18.Jobling, S., D. Sheahan, J. A. Osborne, P. Matthiessen, and P. Sumpter. 1996. Inhibition of testicular growth in rainbow trout (Oncorhynchus mykiss) exposed to estrogenic alkylphenolic chemicals. Environ. Toxicol. Chem. 15:194-202. [Google Scholar]
- 19.Junghanns, C., M. Moeder, G. Krauss, C. Martin, and D. Schlosser. 2005. Degradation of the xenoestrogen nonylphenol by aquatic fungi and their laccases. Microbiology 151:45-57. [DOI] [PubMed] [Google Scholar]
- 20.Kannan, K., T. L. Keith, C. G. Naylor, C. A. Staples, S. A. Snyder, and J. P. Giesy. 2003. Nonylphenol and nonylphenol ethoxylates in fish, sediment, and water from the Kalamazoo River, Michigan. Arch. Environ. Contam. Toxicol. 44:77-82. [DOI] [PubMed] [Google Scholar]
- 21.Karsa, D. R. 1987. Industrial applications of surfactants. Royal Society of Chemistry, London, United Kingdom.
- 22.Kersten, P., and D. Cullen. 2007. Extracellular oxidative systems of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Fungal Genet. Biol. 44:77-87. [DOI] [PubMed] [Google Scholar]
- 23.Kim, Y. S., T. Katase, S. Sekine, T. Inoue, M. Makino, T. Uchiyama, Y. Fujimoto, and N. Yamashita. 2004. Variation in estrogenic activity among fractions of a commercial nonylphenol by high performance liquid chromatography. Chemosphere 54:1127-1134. [DOI] [PubMed] [Google Scholar]
- 24.Kollmann, A., A. Brault, I. Touton, J. Dubroca, V. Chaplain, and C. Mougin. 2003. Effect of nonylphenol surfactants on fungi following the application of sewage sludge on agricultural soils. J. Environ. Qual. 32:1269-1276. [DOI] [PubMed] [Google Scholar]
- 25.Kolpin, D. W., E. T. Furlong, M. T. Meyer, E. M. Thurman, S. D. Zaugg, L. B. Barber, and H. T. Buxton. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999-2000: a national reconnaissance. Environ. Sci. Technol. 36:1202-1211. [DOI] [PubMed] [Google Scholar]
- 26.Kullman, S. W., and F. Matsumura. 1996. Metabolic pathways utilized by Phanerochaete chrysosporium for degradation of the cyclodiene pesticide endosulfan. Appl. Environ. Microbiol. 62:593-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurihara, H., H. Wariishi, and H. Tanaka. 2002. Chemical stress-responsive genes from the lignin-degrading fungus Phanerochaete chrysosporium exposed to dibenzo-p-dioxin. FEMS Microbiol. Lett. 212:217-220. [DOI] [PubMed] [Google Scholar]
- 28.Lalah, J. O., K. W. Schramm, B. Henkelmann, D. Lenoir, A. Behechti, K. Gunther, and A. Kettrup. 2003. The dissipation, distribution and fate of a branched 14C-nonylphenol isomer in lake water/sediment systems. Environ. Pollut. 122:195-203. [DOI] [PubMed] [Google Scholar]
- 29.Mach, R. L., J. Strauss, S. Zeilinger, M. Schindler, and C. P. Kubicek. 1996. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 6:1273-1281. [DOI] [PubMed] [Google Scholar]
- 30.Martinez, D., L. F. Larrondo, N. Putnam, M. D. Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, P. Ramaiya, J. C. Detter, F. Larimer, P. M. Coutinho, B. Henrissat, R. Berka, D. Cullen, and D. Rokhsar. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695-700. [DOI] [PubMed] [Google Scholar]
- 31.Rice, C. P., I. Schmitz-Afonso, J. E. Loyo-Rosales, E. Link, R. Thoma, L. Fay, D. Altfater, and M. J. Camp. 2003. Alkylphenol and alkylphenol-ethoxylates in carp, water, and sediment from the Cuyahoga River, Ohio. Environ. Sci. Technol. 37:3747-3754. [DOI] [PubMed] [Google Scholar]
- 32.Singh, D., and S. Chen. 2008. The white-rot fungus Phanerochaete chrysosporium: conditions for the production of lignin-degrading enzymes. Appl. Microbiol. Biotechnol. 81:399-417. [DOI] [PubMed] [Google Scholar]
- 33.Soares, A., K. Jonasson, E. Terrazas, B. Guieysse, and B. Mattiasson. 2005. The ability of white-rot fungi to degrade the endocrine-disrupting compound nonylphenol. Appl. Microbiol. Biotechnol. 66:719-725. [DOI] [PubMed] [Google Scholar]
- 34.Stark, K., B. Wongsud, R. Burman, and E. H. Oliw. 2005. Oxygenation of polyunsaturated long chain fatty acids by recombinant CYP4F8 and CYP4F12 and catalytic importance of Tyr-125 and Gly-328 of CYP4F8. Arch. Biochem. Biophys. 441:174-181. [DOI] [PubMed] [Google Scholar]
- 35.Stephen, D. W., S. L. Rivers, and D. J. Jamieson. 1995. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol. Microbiol. 3:415-423. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian, V., and J. S. Yadav. 2008. Regulation and heterologous expression of P450 enzyme system components of the white rot fungus Phanerochaete chrysosporium. Enzyme Microb. Technol. 43:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sumpter, J. P., and S. Jobling. 1995. Vitellogenesis as a biomarker for estrogenic contamination of the aquatic environment. Environ. Health Perspect. 103:173-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanghe, T., W. Dhooge, and W. Verstraete. 1999. Isolation of a bacterial strain able to degrade branched nonylphenol. Appl. Environ. Microbiol. 65:746-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallini, G., S. Frassinetti, F. D'Andrea, G. Catelani, and M. Agnolucci. 2001. Biodegradation of 4-(1-nonyl)phenol by axenic cultures of the yeast Candida aquaetextoris: identification of microbial breakdown products and proposal of a possible metabolic pathway. Int. Biodeterior. Biodegrad. 47:133-140. [Google Scholar]
- 40.Wheeler, T. F., J. R. Heim, M. R. LaTorre, and A. B. Janes. 1997. Mass spectral characterization of p-nonylphenol isomers using high-resolution capillary GC-MS. J. Chromatogr. Sci. 35:19-30. [Google Scholar]
- 41.White, R., S. Jobling, S. A. Hoare, J. P. Sumpter, and M. G. Parker. 1994. Environmentally persistent alkylphenolic compounds are estrogenic. Endocrinology 135:175-182. [DOI] [PubMed] [Google Scholar]
- 42.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 43.Yadav, J. S., and C. A. Reddy. 1993. Degradation of benzene, toluene, ethylbenzene, and xylenes (BTEX) by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 59:756-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yadav, J. S., and C. A. Reddy. 1992. Non-involvement of lignin peroxidases and manganese peroxidases in 2,4,5-trichlorophenoxyacetic acid degradation by Phanerochaete chrysosporium. Biotech. Lett. 14:1089-1092. [Google Scholar]
- 45.Yadav, J. S., and H. Doddapanani. 2003. Genome-wide expression profiling and xenobiotic inducibility of P450 monooxygenase genes in the white rot fungus Phanerochaete chrysosporium, p. 333-340. In P. Anzenbacher and J. Hudecek (ed.), Cytochrome P450, biochemistry, biophysics and drug metabolism. Monduzzi Editore, Bologna, Italy.
- 46.Yadav, J. S., D. L. Lawrence, B. A. Nuck, T. W. Federle, and C. A. Reddy. 2001. Biotransformation of linear alkylbenzene sulfonate (LAS) by Phanerochaete chrysosporium: oxidation of alkyl side-chain. Biodegradation 12:443-453. [DOI] [PubMed] [Google Scholar]
- 47.Yadav, J. S., H. Doddapaneni, and V. Subramanian. 2006. P450ome of the white rot fungus Phanerochaete chrysosporium: structure, evolution and regulation of expression of genomic P450 clusters. Biochem. Soc. Trans. 34:1165-1169. [DOI] [PubMed] [Google Scholar]
- 48.Ying, G. G., B. Williams, and R. Kookana. 2002. Environmental fate of alkylphenols and alkylphenol ethoxylates: a review. Environ. Int. 28:215-226. [DOI] [PubMed] [Google Scholar]