Abstract
The occurrence and spread of antibiotic-resistant bacteria (ARB) are pressing public health problems worldwide, and aquatic ecosystems are a recognized reservoir for ARB. We used culture-dependent methods and quantitative molecular techniques to detect and quantify ARB and antibiotic resistance genes (ARGs) in source waters, drinking water treatment plants, and tap water from several cities in Michigan and Ohio. We found ARGs and heterotrophic ARB in all finished water and tap water tested, although the amounts were small. The quantities of most ARGs were greater in tap water than in finished water and source water. In general, the levels of bacteria were higher in source water than in tap water, and the levels of ARB were higher in tap water than in finished water, indicating that there was regrowth of bacteria in drinking water distribution systems. Elevated resistance to some antibiotics was observed during water treatment and in tap water. Water treatment might increase the antibiotic resistance of surviving bacteria, and water distribution systems may serve as an important reservoir for the spread of antibiotic resistance to opportunistic pathogens.
The occurrence and spread of antibiotic-resistant bacteria (ARB) are pressing public health problems worldwide, and aquatic ecosystems are a recognized reservoir for ARB and antibiotic resistance genes (ARGs) (4, 6, 8, 11, 12, 15, 39). Naturally occurring ARB and ARGs in the aquatic environment are selected for and enriched for by antibiotics found in sewage and agricultural runoff, which result from the widespread and increased use of antibiotics (4, 11, 12, 15, 38). Historically, concerns about the microbial quality of drinking water have focused on the occurrence of pathogens in drinking water distribution systems (5, 34). However, the presence of trace levels of antibiotics and ARB in source water and finished drinking water may also greatly affect public health and is an emerging issue for the general public and the drinking water industry (3, 30). Although several studies have detected ARB in drinking water systems (2, 3, 20, 30, 38), most previous studies focused on cultivable bacteria and/or indicator organisms. Little is known about the fate of ARGs in drinking water systems, and it was recently proposed that ARGs are emerging contaminants (24).
We used culture-dependent methods and molecular techniques to investigate the prevalence and dynamics of heterotrophic ARB and ARGs in a drinking water source (source RW-P) and treated drinking water (source DW-P) (see Materials and Methods in the supplemental material). We tested water from a drinking water plant located in Michigan and tap water from several small cities located in Michigan and Ohio (sources TW-1, TW-2, TW-3, and TW-4). Two independent samples were collected each time at each collection site at three different times, and we used four replicates from each sample for tests. We tested bacterial resistance to the following antibiotics: amoxicillin (amoxicilline), chloramphenicol, ciprofloxacin, gentamicin, rifampin (rifampicin), sulfisoxazole, and tetracycline. We also examined the presence of eight ARGs, including beta-lactam resistance genes (blaTEM and blaSHV), chloramphenicol resistance genes (cat and cmr), sulfonamide resistance genes (sulI and sulII), and tetracycline resistance genes (tetO and tetW).
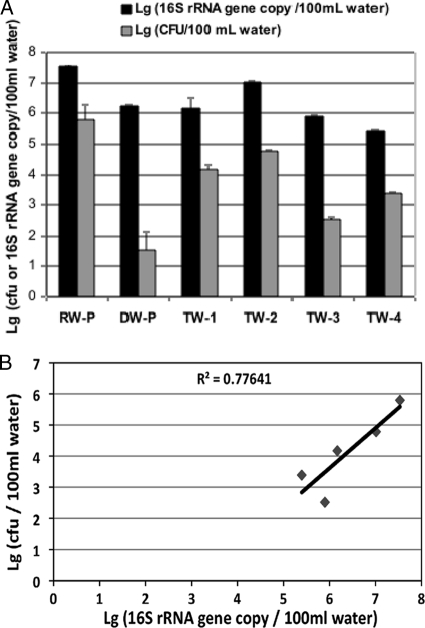
Total heterotrophic plate counts (HPC) were determined using R2A agar without added antibiotics. The water treatment process reduced the total HPC from 9.9 × 106 CFU/100 ml in source water to 68 CFU/100 ml in treated drinking water, indicating that there was efficient removal and/or deactivation of total HPC (Table 1). In contrast, the total 16S rRNA gene copy number decreased from 3.4 × 107 copies/100 ml in source water to 1.6 × 106 copies/100 ml in treated drinking water (Fig. 1). The discrepancy between the reduction in the HPC and the reduction in the total 16S rRNA gene copy number suggests that the final disinfection step effectively inactivated bacteria but most of the dead or damaged cells were still present in finished drinking water. The number of HPC in tap water ranged from 3.44 × 102 to 6.1 × 104 CFU/100 ml water, values that are lower than those for source water but significantly higher than those for treated drinking water, indicating that there is regrowth of bacteria in drinking water distribution systems. The copy numbers of total 16S rRNA genes in tap water ranged from 2.45 × 105 to 1.02 × 107 copies/100 ml water. The higher levels suggested by the 16S rRNA data are consistent with results of previous studies demonstrating that only 5 to 10% and 1% of bacteria in wastewater and soil, respectively, can be cultivated or identified by culture-based methods (9, 37). A significant correlation (P < 0.05, R2 = 0.78) was found between the 16S rRNA gene copy number and the total HPC if treated drinking water (DW-P) data were not included (Fig. 1). This suggests that cultivable bacteria in drinking water represent only a small portion of the total bacterial biomass. Including treated drinking water (DW-P) data resulted in a distorted correlation, suggesting that a large proportion of the 16S rRNA genes present came from dead and/or damaged cells. The levels of total heterotrophic bacteria were significantly higher in tap water (TW-1) than in treated drinking water (DW-P), indicating that there was bacterial regrowth in the water distribution system.
TABLE 1.
Prevalence of ARB HPC in source water, finished drinking water, and tap water from four townsa
| Sampleb | Total HPC (CFU/100 ml) | % of total HPC resistant to:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Amoxicillin | Ciprofloxacin | Chloramphenicol | Gentamicin | Rifampin | Sulfisoxazole | Tetracycline | ||
| RW-P | 1.19 × 106 | 11.67 ± 4.39 | 11.60 ± 5.92 | 4.17 ± 1.93 | 14.42 ± 5.52 | 10.85 ± 3.57 | 7.46 ± 3.87 | 1.66 ± 0.80 |
| DW-P | 68 | 39.55 ± 9.79c | 4.77 ± 4.71 | 19.45 ± 5.60c | 21.96 ± 14.43 | 47.98 ± 17.99c | 1.17 ± 1.14c | 1.50 ± 1.24 |
| TW-1 | 1.6 × 104 | 15.22 ± 2.73d | 9.99 ± 4.76 | 13.96 ± 3.70c | 13.40 ± 1.73 | 62.00 ± 8.96c | 3.34 ± 1.21 | 3.78 ± 0.93c,d |
| TW-2 | 6.04 × 104 | 3.02 ± 0.19 | 13.14 ± 0.48 | 5.49 ± 0.47 | 4.67 ± 0.21 | 28.10 ± 1.72 | 7.85 ± 0.67 | 0.08 ± 0.01 |
| TW-3 | 3.44 × 102 | 4.07 ± 0.17 | 0.18 ± 0.07 | 0.75 ± 0.39 | 2.18 ± 0.62 | 82.15 ± 1.50 | 0.33 ± 0.03 | 0.98 ± 0.38 |
| TW-4 | 2.46 × 103 | 14.33 ± 1.74 | 0.18 ± 0.05 | 2.05 ± 0.04 | 9.76 ± 0.34 | 14.23 ± 1.69 | 0.12 ± 0.001 | 0.04 ± 0.002 |
Prevalence was defined as the percentage of resistant HPC in the total HPC. The statistical analysis was done using six samples for each type and four technical replicates for each sample.
RW-P, source water from the drinking water treatment plant; DW-P, finished drinking water from the drinking water treatment plant; TW-1, tap water from the city where the drinking water treatment plant is located; TW-2, TW-3, and TW-4, tap water from three towns in Michigan and Ohio close to the city where the TW-1 drinking water treatment plant is located.
Significantly different from RW-P.
Significantly different from DW-P.
FIG. 1.
Heterotrophic bacteria and the 16S rRNA gene in different water samples. (A) Copy numbers of the 16S rRNA gene and numbers of heterotrophic bacteria (CFU) in 100 ml water. (B) Correlation (P < 0.05, R2 = 0.78) between the copy number of the 16S rRNA gene and the number of heterotrophic bacteria in different water samples (without the data for DW-P). RW-P, source water from the drinking water treatment plant; DW-P, finished drinking water from the drinking water treatment plant; TW-1, tap water from the city where the drinking water treatment plant is located; TW-2, TW-3, and TW-4, tap water from three towns in Michigan and Ohio close to the city where the TW-1 drinking water treatment plant is located. The statistical analysis was done using six samples for each type of water sample. Lg, log10.
The prevalence of HPC resistant to antibiotics was determined using R2A agar containing amoxicillin (4 mg/liter), chloramphenicol (16 mg/liter), ciprofloxacin (2 mg/liter), gentamicin (8 mg/liter), rifampin (2 mg/liter), sulfisoxazole (256 mg/liter), or tetracycline (8 mg/liter). Some groups of heterotrophic bacteria were resistant to all of the antibiotics at the concentrations tested in all water samples (Table 1). In the source water, 14.4% of the HPC were resistant to gentamicin and 1.7% were resistant to tetracycline. The resistance of HPC to amoxicillin, chloramphenicol, and rifampin was significantly higher (P < 0.01) in treated drinking water than in source water, while the resistance to sulfisoxazole was significantly lower (P < 0.01). Compared to treated drinking water (DW-P), the resistance of HPC to tetracycline in tap water was significantly greater and the resistance to amoxicillin was significantly lower (P < 0.01). The resistance to chloramphenicol and rifampin remained higher than the resistance in source water. The prevalence of HPC antibiotic resistance in tap water samples collected from other cities varied, but the resistance of HPC to rifampin was particularly high in all tap water samples.
A number of previous studies have reported that ARB are common in drinking water (2, 3, 19, 25, 33). We added to these studies by testing water both before and after treatment, as well as tap water. Although the bacterial concentration was effectively lower during water treatment, the prevalence of resistance to amoxicillin, rifampin, and chloramphenicol nevertheless increased significantly.
Several studies have discovered that chlorine, an agent widely used for disinfection, selects for ARB (2, 3, 9, 16, 33, 37). Armstrong et al. (2, 3) found that there was a significant increase in the proportion of multidrug-resistant (MAR) bacteria following flash mixing with chlorine. Murray et al. (16) demonstrated that the proportion of bacteria resistant to ampicillin and cephalothin (cefalotin) in sewage increased significantly following chlorination, and they observed a significant increase in the proportion of MAR strains during chlorination in laboratory experiments. Other studies demonstrated that the susceptibility of ARB to a disinfectant and the susceptibility of antibiotic-susceptible bacteria to a disinfectant are similar (7, 28), indicating that disinfection does not select ARB but instead induces the development of antibiotic resistance. Armstrong et al. (2, 3) suggested that stress-tolerant bacteria selected by chlorination might be more antibiotic resistant, and one study found that suboptimal chlorine treatment of drinking water selected for MAR Pseudomonas aeruginosa (33).
The mechanism of chlorine-induced antibiotic resistance in bacteria is unknown. It is possible that chlorine can increase expression of the multidrug efflux pumps, leading to resistance to disinfection by-products as well as antibiotics. The drinking water treatment plant that we sampled used monochloramine as a disinfectant. No previous study has reported the effects of monochlroamine disinfection on ARB, but our results suggest that monochlromaine disinfection may have an effect similar to that of chlorine disinfection.
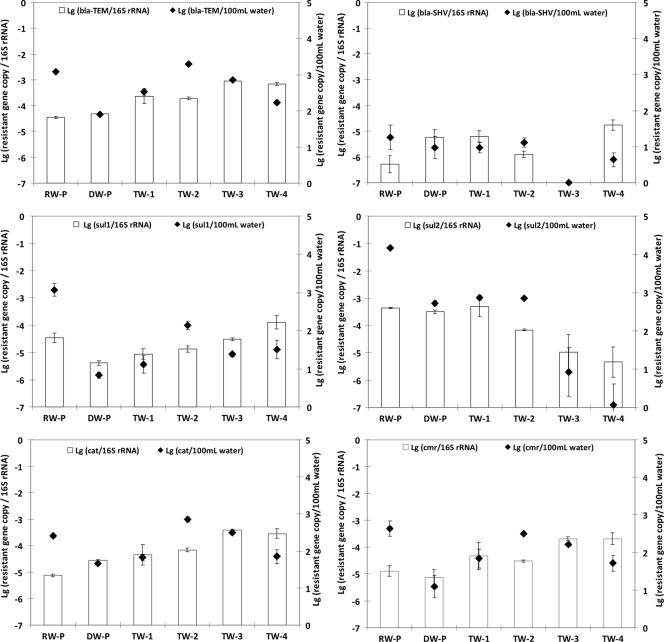
Real-time PCR was used to quantify ARGs (including cat, cmr, blaTEM, blaSHV, sulI, sulII, tetW, and tetO) in collected water samples. All ARGs tested were detected in all water samples, except for the tetO and tetW genes, which were detected only in source water (Fig. 2). The copy number of each ARG in 100 ml water was calculated and normalized to the copy number of the total 16S rRNA genes to determine the relative abundance of each ARG in the water samples. Compared to the copy number in finished water, the copy number of ARGs in tap water was significantly greater (P < 0.001), except for the blaSHV gene, whose copy number was not significantly different (P = 0.124); the tetO and tetW genes were not detected in the drinking water sample after treatment. In terms of the relative abundance of ARGs in bacterial populations, all ARG/16S rRNA gene ratios were less than −3 log. Compared to source water, treated drinking water had a higher abundance of the cat and blaSHV genes (P < 0.001) but a lower abundance of the sulI gene (P < 0.001) (Fig. 2). No significant difference in any other ARG was found. After distribution, no significant change was observed in any ARG, except that the abundance of the blaTEM gene was significantly increased (P < 0.01) compared with the abundance in treated drinking water (DW-P) or in tap water (TW-1) (Fig. 2). The ARGs were also present in tap water samples collected from other cities. The similarity of the abundance of ARGs in the different tap water samples is quite remarkable (Fig. 2). The relative abundance of all ARGs was similar to that in the TW-1 tap water sample, except that the relative abundance of sulII and blaSHV was lower in the TW-2 and TW-3 tap water samples (Fig. 2).
FIG. 2.
Quantities of ARGs in different water samples. The bars indicate the copy numbers of the resistance genes normalized to the 16S rRNA gene copy number, and the symbols indicate the absolute copy numbers of ARGs in 100 ml water. RW-P, source water from the drinking water treatment plant; DW-P, finished drinking water from the drinking water treatment plant; TW-1, tap water from the city where the drinking water treatment plant is located; TW-2, TW-3, and TW-4, tap water from three towns in Michigan and Ohio close to the city where the TW-1 drinking water treatment plant is located. The statistical analysis was done using six samples for each type of water sample. Lg, log10.
The quantities of individual ARGs were not significantly correlated with either HPC counts or 16S rRNA genes (data not shown), indicating that the ARGs tested were not evenly distributed among the bacterial populations in the water samples. However, the overall trends in quantity were similar for some ARGs and ARB. For example, in source water, treated drinking water, and tap water (TW-1), the number of heterotrophic bacteria resistant to amoxicillin, chloramphenicol, and sulfisoxazole corresponded to the proportion of genes coding for resistance to these antibiotics (blaSHV, cat, and sulI, respectively).
Bacteria may inherit resistance to some antibiotics or can develop resistance via spontaneous mutation or the acquisition of resistant genes (35). The acquisition of a resistant gene via horizontal gene transfer is the most common and easiest way for bacteria to develop antibiotic resistance both in the environment and in a host (26, 29). Many bacteria transmit ARGs, and these ARGs were recently proposed to be emerging contaminants because of their widespread occurrence in aquatic ecosystems (13, 21, 22, 24). Plasmid-mediated blaTEM and blaSHV are the most common genes coding beta-lactamases and “extended-spectrum” beta-lactamases, a major cause of resistance to beta-lactams, and they are increasingly being found in different settings worldwide (14, 23). The enzymes encoded by these genes confer unequivocal resistance to ampicillin, amoxicillin, ticarillin, and carbenicillin (32, 36). We detected blaTEM and blaSHV genes in all but one water sample, which is evidence that these genes are distributed widely in drinking water systems. The selective increases in the levels of both genes in tap water due to either water treatment or regrowth within drinking water distribution systems suggest that the spread of at least some beta-lactam-resistant determinants may occur through drinking water distribution systems.
Both tetO and tetW are tetracycline resistance genes encoding ribosomal protection proteins. Both of these genes are common in intestinal and rumen environments (1, 31); thus, their presence may indicate fecal contamination (22). If the tetO and tetW genes truly represent the level of fecal contamination, our results show that drinking water treatment was effective for eliminating and controlling fecal contamination.
The most frequent cause of bacterial resistance to chloramphenicol is enzymatic inactivation by acetylation of the drug via different types of chloramphenicol acetyltransferases encoded by cat genes (17), but other mechanisms, such as efflux systems, may also contribute to chloramphenicol resistance (18). The proportion of cat genes increased significantly following water treatment, suggesting that the drinking water treatment did not effectively remove or inactivate the chloramphenicol-resistant bacterial population. On the other hand, the cmr gene, an efflux pump gene related to chloramphenicol resistance, showed little variation in different water sources.
Sulfonamides act as competitive inhibitors of the enzyme dihydropteroate synthase in the folic acid pathway of bacterial and some eukaryotic cells. sulI and sulII encode alternative sulfonamide-resistant dihydropteroate synthases in gram-negative clinical bacteria, and both genes commonly occur (often at roughly the same frequency) in sulfisoxazole-resistant gram-negative clinical isolates (10). The drinking water treatment process significantly decreased the abundance of the sulI gene but had no significant influence on the sulII gene.
In summary, we found heterotrophic ARB and ARGs in all finished water and tap water tested, although the amounts were small. The size of the general population of bacteria followed the order source water > tap water > finished water, indicating that there was regrowth of bacteria in drinking water distribution systems; elevated resistance to some antibiotics was observed during water treatment and in tap water. We show that the quantities of most ARGs are greater in tap water than in finished water and source water. The increased levels of ARGs and specialized groups of ARB in tap water compared to finished water and source water suggest that water treatment could increase the antibiotic resistance of surviving bacteria and/or induce transfer of ARGs among certain bacterial populations. Water distribution systems could serve as an incubator for growth of certain ARB populations and as an important reservoir for the spread of antibiotic resistance to opportunistic pathogens. Drinking water treatment processes and distribution systems can impact the spread of antibiotic resistance. Rusin et al. (27) estimated that the risk of infection by bacteria in drinking water was as low as 7.3 per billion people for exposure to low levels of Aeromonas and as high as 98 per 100 patients receiving antibiotic treatment exposed to high levels of Pseudomonas (27). Whether exposure to ARB results in an increased risk to the general public, particularly individuals with compromised immune systems, the very young, the very old, or individuals with chronic conditions, is not known and deserves further study. Future research should identify factors accounting for the selective increase in antibiotic resistance and develop new methods and approaches to reduce accumulation of such resistance.
Supplementary Material
Acknowledgments
This work was supported by the Graham Environmental Sustainability Institute at the University of Michigan.
Footnotes
Published ahead of print on 6 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. L., J. J. Calomiris, and R. J. Seidler. 1982. Selection of antibiotic-resistant standard plate count bacteria during water treatment. Appl. Environ. Microbiol. 44:308-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, J. L., D. S. Shigeno, J. J. Calomiris, and R. J. Seidler. 1981. Antibiotic-resistant bacteria in drinking water. Appl. Environ. Microbiol. 42:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baquero, F., J. L. Martinez, and R. Canton. 2008. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19:260-265. [DOI] [PubMed] [Google Scholar]
- 5.Berry, D., C. Xi, and L. Raskin. 2006. Microbial ecology of drinking water distribution systems. Curr. Opin. Biotechnol. 17:297-302. [DOI] [PubMed] [Google Scholar]
- 6.Cooke, M. D. 1975. Antibiotic resistance in coliform and fecal coliform bacteria from natural wasters and effluents. N. Z. J. Mar. Freshwater Res. 10:391-397. [Google Scholar]
- 7.Fraise, A. P. 2002. Susceptibility of antibiotic-resistant cocci to biocides. J. Appl. Microbiol. 92(Suppl.):158S-162S. [PubMed] [Google Scholar]
- 8.Gonzal, S. M., C. P. Gerba, and J. L. Melnick. 1979. Transferable drug resistance in bacteria of coastal canal water and sediment. Water Res. 13:349-356. [Google Scholar]
- 9.Hiraishi, A. 1998. Respiratory quinone profiles as tools for identifying different bacterial population in activated sludge. J. Gen. Appl. Microbiol. 34:39-56. [Google Scholar]
- 10.Huovinen, P., L. Sundstrom, G. Swedberg, and O. Skold. 1995. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 39:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klare, I., H. Heier, H. Claus, G. Bohme, S. Marin, G. Seltmann, R. Hakenbeck, V. Antanassova, and W. Witte. 1995. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb. Drug Resist. 1:265-272. [DOI] [PubMed] [Google Scholar]
- 12.Kummerer, K. 2004. Resistance in the environment. J. Antimicrob. Chemother. 54:311-320. [DOI] [PubMed] [Google Scholar]
- 13.Lachmayr, K. L., L. J. Kerkhof, A. G. Dirienzo, C. M. Cavanaugh, and T. E. Ford. 2009. Quantifying nonspecific TEM β-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 75:203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez, J. L. 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321:365-367. [DOI] [PubMed] [Google Scholar]
- 16.Murray, G. E., R. S. Tobin, B. Junkins, and D. J. Kushner. 1984. Effect of chlorination on antibiotic resistance profiles of sewage-related bacteria. Appl. Environ. Microbiol. 48:73-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray, I. A., and W. V. Shaw. 1997. O-Acetyltransferases for chloramphenicol and other natural products. Antimicrob. Agents Chemother. 41:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsen, I. W., I. Bakke, A. Vader, O. Olsvik, and M. R. El-Gewely. 1996. Isolation of cmr, a novel Escherichia coli chloramphenicol resistance gene encoding a putative efflux pump. J. Bacteriol. 178:3188-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathak, S. P., and K. Gopal. 2008. Prevalence of bacterial contamination with antibiotic-resistant and enterotoxigenic fecal coliforms in treated drinking water. J. Toxicol. Environ. Health A 71:427-433. [DOI] [PubMed] [Google Scholar]
- 20.Pavlov, D., C. M. de Wet, W. O. Grabow, and M. M. Ehlers. 2004. Potentially pathogenic features of heterotrophic plate count bacteria isolated from treated and untreated drinking water. Int. J. Food Microbiol. 92:275-287. [DOI] [PubMed] [Google Scholar]
- 21.Pei, R., J. Cha, K. H. Carlson, and A. Pruden. 2007. Response of antibiotic resistance genes (ARG) to biological treatment in dairy lagoon water. Environ. Sci. Technol. 41:5108-5113. [DOI] [PubMed] [Google Scholar]
- 22.Pei, R., S. C. Kim, K. H. Carlson, and A. Pruden. 2006. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40:2427-2435. [DOI] [PubMed] [Google Scholar]
- 23.Poole, K. 2004. Resistance to beta-lactam antibiotics. Cell. Mol. Life Sci. 61:2200-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruden, A., R. Pei, H. Storteboom, and K. H. Carlson. 2006. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 40:7445-7450. [DOI] [PubMed] [Google Scholar]
- 25.Ramteke, P. W., A. Gaur, S. P. Pathak, and J. W. Bhattacharjee. 1990. Antibiotic resistance of coliforms in drinking water in rural areas. Indian J. Med. Res. 91:185-188. [PubMed] [Google Scholar]
- 26.Rowe-Magnus, D. A., and D. Mazel. 1999. Resistance gene capture. Curr. Opin. Microbiol. 2:483-488. [DOI] [PubMed] [Google Scholar]
- 27.Rusin, P. A., J. B. Rose, C. N. Haas, and C. P. Gerba. 1997. Risk assessment of opportunistic bacterial pathogens in drinking water. Rev. Environ. Contam. Toxicol. 152:57-83. [DOI] [PubMed] [Google Scholar]
- 28.Rutala, W. A., M. M. Stiegel, F. A. Sarubbi, and D. J. Weber. 1997. Susceptibility of antibiotic-susceptible and antibiotic-resistant hospital bacteria to disinfectants. Infect. Control Hosp. Epidemiol. 18:417-421. [DOI] [PubMed] [Google Scholar]
- 29.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz, T., W. Kohnen, B. Jansen, and U. Obst. 2003. Detection of antibiotic-resistant bacteria and their resistance genes in waste-water, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 43:325-335. [DOI] [PubMed] [Google Scholar]
- 31.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seetulsingh, P. S., L. M. Hall, and D. M. Livermore. 1991. Activity of clavulanate combinations against TEM-1 beta-lactamase-producing Escherichia coli isolates obtained in 1982 and 1989. J. Antimicrob. Chemother. 27:749-759. [DOI] [PubMed] [Google Scholar]
- 33.Shrivastava, R., R. K. Upreti, S. R. Jain, K. N. Prasad, P. K. Seth, and U. C. Chaturvedi. 2004. Suboptimal chlorine treatment of drinking water leads to selection of multidrug-resistant Pseudomonas aeruginosa. Ecotoxicol. Environ. Saf. 58:277-283. [DOI] [PubMed] [Google Scholar]
- 34.Szewzyk, U., R. Szewzyk, W. Manz, and K. H. Schleifer. 2000. Microbiological safety of drinking water. Annu. Rev. Microbiol. 54:81-127. [DOI] [PubMed] [Google Scholar]
- 35.Tenover, F. C. 2006. Mechanisms of antimicrobial resistance in bacteria. Am. J. Med. 119:S3-S10. (Discussion, 119:S62-S70.) [DOI] [PubMed] [Google Scholar]
- 36.Wu, P. J., K. Shannon, and I. Phillips. 1994. Effect of hyperproduction of TEM-1 beta-lactamase on in vitro susceptibility of Escherichia coli to beta-lactam antibiotics. Antimicrob. Agents Chemother. 38:494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zellers, L. 1996. Fatty acid patterns of microbial phospholipids and lipopolysaccharides. In F. Schinner, R. Ohlinger, and E. Kandeler (ed.), Methods in soil biology. Springer Verlag, Heidelberg, Germany.
- 38.Zhang, X. X., T. Zhang, and H. H. Fang. 2009. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 82:397-414. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y., C. F. Marrs, C. Simon, and C. Xi. 2009. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 407:3702-3706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.