Abstract
Pseudomonas sp. strain ADP utilizes the human-made s-triazine herbicide atrazine as the sole nitrogen source. The results reported here demonstrate that atrazine and the atrazine degradation intermediates N-isopropylammelide and cyanuric acid are chemoattractants for strain ADP. In addition, the nonmetabolized s-triazine ametryn was also an attractant. The chemotactic response to these s-triazines was not specifically induced during growth with atrazine, and atrazine metabolism was not required for the chemotactic response. A cured variant of strain ADP (ADP M13-2) was attracted to s-triazines, indicating that the atrazine catabolic plasmid pADP-1 is not necessary for the chemotactic response and that atrazine degradation and chemotaxis are not genetically linked. These results indicate that atrazine and related s-triazines are detected by one or more chromosomally encoded chemoreceptors in Pseudomonas sp. strain ADP. We demonstrated that Escherichia coli is attracted to the s-triazine compounds N-isopropylammelide and cyanuric acid, and an E. coli mutant lacking Tap (the pyrimidine chemoreceptor) was unable to respond to s-triazines. These data indicate that pyrimidines and triazines are detected by the same chemoreceptor (Tap) in E. coli. We showed that Pseudomonas sp. strain ADP is attracted to pyrimidines, which are the naturally occurring structures closest to triazines, and propose that chemotaxis toward s-triazines may be due to fortuitous recognition by a pyrimidine chemoreceptor in Pseudomonas sp. strain ADP. In competition assays, the presence of atrazine inhibited chemotaxis of Pseudomonas sp. strain ADP to cytosine, and cytosine inhibited chemotaxis to atrazine, suggesting that pyrimidines and s-triazines are detected by the same chemoreceptor.
Atrazine [2-chloro-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine] is a human-made herbicide that is used worldwide to control broadleaf and grassy weeds. As one of the most heavily used herbicides in the United States, atrazine can be present in parts per million in agricultural runoffs (3), which exceeds the U.S. Environmental Protection Agency's maximum allowable contaminant level of 3 ppb in ground and surface waters (13). Atrazine is persistent in soil (34) and was once considered nontoxic to animals. However, recent studies have shown that atrazine causes sexual abnormalities in frogs (21, 22, 50), reduced testosterone production in rats (53), and elevated levels of prostate cancer in workers at an atrazine-manufacturing factory (45). These studies suggest that there is cause for concern about atrazine residues in soil, groundwater, and surface waters.
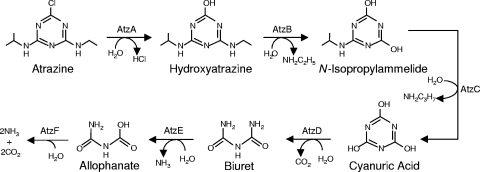
Several bacterial strains capable of mineralizing atrazine have been isolated (4, 27, 41, 49, 51, 52, 58). The best-studied atrazine-degrading strain, Pseudomonas sp. strain ADP (atrazine degrading pseudomonad), was isolated from an atrazine spill site in Minnesota (27). Strain ADP utilizes atrazine as a sole nitrogen source and mineralizes it in the process (27). The pathway of atrazine degradation in strain ADP has been characterized in detail (Fig. 1), and the genes encoding the six enzymes required for atrazine degradation have been cloned and sequenced (5, 7, 9, 10, 29, 42). The six genes are located in four distant locations on the atrazine catabolic plasmid (pADP-1) present in strain ADP (10, 29). atzA, atzB, and atzC, which encode the first three enzymes of the pathway, are constitutively expressed and highly conserved in atrazine-degrading bacteria isolated from geographically distinct locations (8, 11). Products of the atzDEF gene cluster catalyze the last three steps of atrazine degradation. This operon is divergently transcribed from atzR, the product of which has high homology to LysR-type regulatory proteins (29). AtzR and the inducer cyanuric acid are required for the expression of the atzDEF operon (14), and the operon is also subject to nitrogen control (15).
FIG. 1.
Pathway of atrazine degradation in Pseudomonas sp. strain ADP (reviewed in reference 55).
In a study investigating the bioavailability of atrazine, Park et al. provided evidence that two atrazine-degrading strains, Pseudomonas sp. strain ADP and Agrobacterium radiobacter J14a, were chemotactically attracted to atrazine (38). Chemotaxis, the ability of motile bacteria to detect and respond to specific chemicals, can help bacteria find an optimal niche for their survival and growth and may play a role in the efficient degradation of pollutants in the environment (33, 37). Chemotaxis has been shown to enhance naphthalene biodegradation in both a heterogeneous aqueous system (30) and a non-aqueous-phase liquid system (24). In addition, a chemotactic naphthalene-degrading strain caused a higher rate of naphthalene desorption than was observed with nonchemotactic and nonmotile strains (24). Pseudomonas sp. strain ADP and recombinant strains expressing atz genes have been used to remove atrazine from soil in laboratory and field scale experiments (32, 48). If chemotaxis can enhance bioavailability in environments where the chemicals are sorbed to particles, the use of a motile chemotactic strain for bioremediation would be advantageous. Aside from the practical implications of atrazine chemotaxis, we are interested in understanding the evolution of a chemotactic response to a human-made chemical that was initially synthesized just 50 years ago (23). The results reported here indicate that Pseudomonas sp. strain ADP is chemotactically attracted to atrazine, atrazine metabolites, and the nonmetabolizable structural analog ametryn. The chemotactic response is not induced during growth with atrazine in strain ADP and does not require atrazine metabolism. We demonstrated that a single chemoreceptor (Tap) mediates chemotaxis to s-triazines and structurally similar pyrimidines in Escherichia coli. Additionally, we found that Pseudomonas sp. strain ADP is attracted to pyrimidines. In competition assays, cytosine inhibited atrazine chemotaxis, and vice versa. We therefore concluded that pyrimidines and s-triazines are detected by a single chemoreceptor in Pseudomonas sp. strain ADP.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains RP437 (wild type for chemotaxis) and RP3525 (Δtap) were provided by J. S. Parkinson (28, 39, 47). UU1250(pXL4), a derivative of RP437 with all of the chromosomal methyl-accepting chemotaxis protein (MCP) genes deleted, contains a plasmid expressing the hybrid chemoreceptor Tapsr (25). For chemotaxis assays, the strains were grown at 30°C in H1 minimal salts medium (1) containing 25 mM glycerol as the carbon source; 0.5 mM each of methionine, leucine, histidine, and threonine; and 1 μg ml−1 thiamine to satisfy auxotrophic requirements. Strain UU1250(pXL4) was grown in the presence of 100 μg ml−1 ampicillin and was induced for 4 h with 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG).
Pseudomonas sp. strains ADP and ADP M13-2 were kindly provided by L. P. Wackett. Pseudomonas sp. strain ADP is a wild-type isolate that can use atrazine as a sole nitrogen source (27). Strain ADP M13-2 is a derivative of strain ADP that is unable to use atrazine or cyanuric acid as a sole nitrogen source and did not contain any plasmids, based on the inability to isolate and detect plasmid DNA on gels (J. Seffernick and L. P. Wackett, personal communication). To verify that intact plasmid or deletion derivatives of pADP-1 were not present in strain ADP M13-2, we generated PCR primers based on the published sequences of atzA, atzC, and oriV (the pADP-1 origin of replication) (29) and carried out PCR with genomic DNA from strains ADP and ADP M13-2. The primers were atzA for, 5′-GCAAACGCTCAGCATCCAGC-3′; atzA rev, 5′-TCAAGGAACGCCAACTCACG-3′; atzC for, 5′-AGGCAACTATAACCTCATCC-3′; atzC rev, 5′-ATTGGGATTGTTGGTGACAG-3′; oriV for, 5′-TTCTCGGGGCCATAGAGGGC-3′; and oriV rev, 5′-CTGCGCATCACGAACTACCG-3′. These three loci are approximately equally spaced on the pADP-1 plasmid. Genomic DNA from each strain was purified using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN). The PCR conditions were as follows: 95°C for 5 min, followed by 25 cycles of 95°C for 1 min, 62.5 ± 2.5°C for 1 min, 65°C for 1.5 min, and a final extension at 65°C for 5 min. Southern hybridizations (44) with strain ADP and ADP M13-2 genomic DNA and atzA, atzC, and oriV PCR products amplified from strain ADP as probes were carried out using the DIG DNA Labeling and Detection Kit (Roche Diagnostics Corp., Indianapolis, IN).
Prior to initiating chemotaxis experiments, motile populations of strains ADP and ADP M13-2 were enriched on dilute L agar swarm plates (1, 19). For chemotaxis assays, strain ADP was grown in modified R minimal medium (12) with 11 mM glucose as the carbon source and 0.45 mM atrazine (27) and/or 1.125 mM (NH4)2SO4 (equimolar nitrogen concentrations) as the sole nitrogen source. Strain ADP M13-2 was grown in R minimal medium with 11 mM glucose and 1.125 mM (NH4)2SO4.
Chemicals.
Cytosine was obtained from Acros Organics, and thymine, uracil, and cyanuric acid were from Sigma-Aldrich; all were of the highest purity commercially available. Atrazine, N-isopropylammelide, and ametryn were kindly provided by L. P. Wackett. The purity of these compounds was verified by gas chromatography-mass spectrometry; no contaminating compounds were detected by this method (data not shown).
Chemotaxis assays.
Bacterial cells were harvested in mid-exponential phase (when the optical density at 660 nm was between 0.3 and 0.4) by centrifugation at 4,000 rpm for 5 min and washed once with chemotaxis buffer (10 mM potassium phosphate buffer [pH 7.0], 0.1 mM disodium EDTA for E. coli strains [39]; 50 mM potassium phosphate buffer [pH 7.0], 10 μM disodium EDTA, 0.05% glycerol for Pseudomonas strains [36]). The cells remained motile in chemotaxis buffer for at least 1 hour. Atrazine-grown cells were washed twice to minimize the amount of atrazine residue. Quantitative capillary assays were carried out as described previously for E. coli strains (25). High-throughput quantitative capillary assays were carried out as previously described for Pseudomonas strains (26). In competition assays (2, 31), the competing attractant was added to both the capillary solution and the cell suspension at the peak response concentration. In all experiments, negative controls (chemotaxis buffer) and positive controls (1 mM aspartate for E. coli strains; 0.1% [wt/vol] Casamino Acids for Pseudomonas strains) were included. Temporal assays were also used to confirm chemotactic responses by monitoring changes in swimming behavior in response to attractants (35, 46). Briefly, washed cells were suspended in chemotaxis buffer to an optical density at 660 nm of approximately 0.10. After the addition of an attractant, the cells were viewed directly under a microscope, and the time required for approximately 50% of the population to return to prestimulus swimming behavior was determined.
RESULTS
Atrazine, atrazine metabolites, and ametryn are attractants for strain ADP.
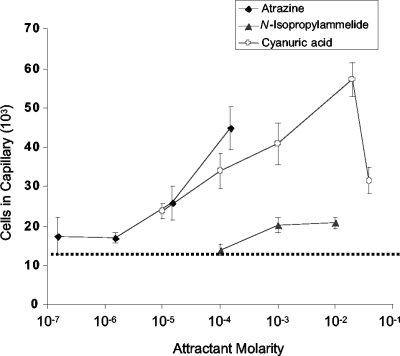
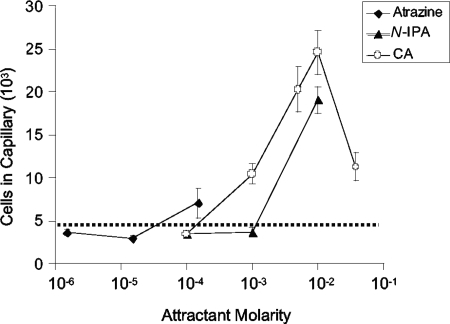
The chemotactic response of Pseudomonas sp. strain ADP to atrazine and other s-triazines was measured with the high-throughput quantitative capillary assay by comparing the numbers of cells that accumulated in a chemoattractant-containing capillary versus a buffer-containing capillary. Because of the low solubilities of s-triazines, all of the compounds were tested to the limit of solubility. Atrazine elicited a positive chemotactic response by wild-type Pseudomonas sp. strain ADP grown in R medium with glucose and atrazine, with the peak response concentration at saturation (approximately 150 μM) (Fig. 2). The average number of cells that accumulated in capillaries containing saturated atrazine was 4.48 × 104 per capillary. This represents approximately four times more cells than a capillary containing chemotaxis buffer (1.20 × 104 ± 670 per capillary). Atrazine-grown ADP was also attracted to the atrazine metabolites N-isopropylammelide and cyanuric acid (Fig. 2), which are generated in the second and third steps of atrazine degradation by hydrolytic removal of the isopropylamino and ethylamino groups, respectively, from hydroxyatrazine (Fig. 1). N-Isopropylammelide elicited a weak chemotactic response (2.08 × 104 ± 1,335 cells per capillary at the peak response concentration of 10 mM). Nevertheless, this response was significant based on Student's t test (P < 0.001). Cyanuric acid elicited a good response, with a peak response concentration of 20 mM (Fig. 2). In addition, the atrazine structural analog ametryn [2-methylthiol-4-(N-ethylamino)-6-(N-isopropylamino)-1,3,5-s-triazine; solubility, approximately 0.81 mM], which is not metabolized by strain ADP, served as a chemoattractant for atrazine-grown strain ADP (4.3 × 104 ± 5,702 per capillary; 3.6-fold over background).
FIG. 2.
Concentration response curves for chemotaxis to s-triazines by Pseudomonas sp. strain ADP. The cells were grown in minimal medium with glucose and atrazine, and suspended in chemotaxis buffer as described in Materials and Methods. High-throughput assays were performed at room temperature with various concentrations of each s-triazine up to its limit of solubility. Capillaries containing chemotaxis buffer served as the negative control; those with Casamino Acids (0.1% in chemotaxis buffer) served as the positive control. The results are the averages of at least 15 capillaries from at least two independent experiments; the error bars indicate standard errors. The average background accumulation in capillaries containing buffer only is indicated by the dotted line (approximately 1.2 × 104 ± 670 cells). The number of cells that accumulated in capillaries containing Casamino Acids (positive control) is not shown (approximately 1.5 × 105 ± 6,286 cells).
Chemotactic responses were verified with a temporal assay, which quantitatively measured the swimming behavior of bacterial cells. In chemotaxis buffer, the cells swam randomly and changed directions frequently (approximately once per 1.5 s). Upon addition of an attractant, the cells responded by altering their swimming behavior by changing direction much less frequently (smooth swimming). The time required for approximately 50% of the population of cells to return to prestimulus swimming behavior was recorded as the adaptation time (35, 46). Although response times were short and somewhat variable (Table 1), a distinct change in swimming behavior was observed with atrazine-grown strain ADP in response to atrazine, the atrazine metabolites N-isopropylammelide and cyanuric acid, and the atrazine analog ametryn. In contrast, the cells continued to swim randomly upon addition of chemotaxis buffer (negative control). These results are consistent with the results of the quantitative capillary assay.
TABLE 1.
Temporal responses of Pseudomonas sp. strains ADP and ADP M13-2 to s-triazines
| Attractanta | Mean smooth-swimming response timeb ±SD (no. of independent assays)
|
||
|---|---|---|---|
| ADP
|
ADP M13-2 [(NH4)2SO4] | ||
| Atrazine | (NH4)2SO4 | ||
| None | − (5) | − (5) | − (5) |
| CAA | 89 ± 16 (31) | 113 ± 29 (19) | 83 ± 22 (16) |
| Atrazine | 14 ± 8 (24) | 18 ± 5 (16) | 29 ± 7 (15) |
| N-Isopropylammelide | 24 ± 8 (21) | 45 ± 12 (16) | 44 ± 10 (15) |
| Cyanuric acid | 18 ± 4 (15) | 17 ± 3 (15) | 17 ± 3 (15) |
| Ametryn | 13 ± 7 (16) | 28 ± 6 (16) | 27 ± 5 (15) |
Attractants were supplied at the following concentrations: Casamino Acids (CAA), 0.2%; atrazine, 0.075 mM; N-isopropylammelide, 10 mM; cyanuric acid, 5 mM; ametryn, 0.4 mM. The s-triazine concentrations used were approximately half-saturating, except for that of cyanuric acid.
The amount of time in seconds for approximately 50% of the cells to adapt to the added attractant. The bacterial strains Pseudomonas sp. strains ADP (wild type) and ADP M13-2 (cured strain) were grown as described in Materials and Methods, with the nitrogen source indicated. −, no response detected.
The chemotactic response to atrazine and atrazine metabolites is not inducible in ADP.
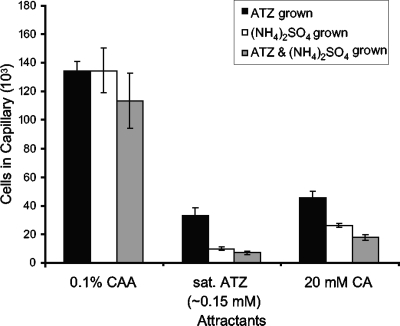
To determine whether the chemotactic response of strain ADP to atrazine and other s-triazines is induced during growth with atrazine, wild-type strain ADP cells were grown with glucose and ammonium sulfate in the presence or absence of atrazine and assayed for chemotaxis. Using the quantitative capillary assay, we showed that cells grown with ammonium sulfate only or with both ammonium sulfate and atrazine were attracted to atrazine and cyanuric acid (Fig. 3). The responses exhibited by cells grown under the above two conditions were similar but were weaker than the response exhibited by cells grown with atrazine as the sole nitrogen source (Fig. 3). The cells responded to Casamino Acids to approximately the same level after growth under all the conditions, and the background accumulations were approximately the same, as well (Fig. 3). These data suggest that atrazine does not induce the chemotactic response, but ammonium appears to repress the response. The atzDEF genes for the conversion of cyanuric acid to ammonia and CO2 (Fig. 1) are under nitrogen control (14, 15, 40), so a similar regulatory mechanism may be in place to control expression of the atrazine chemotaxis genes. We also carried out temporal assays using cells grown with ammonium as the sole nitrogen source. The adaptation times of ammonium-grown cells to all of the tested attractants were approximately the same or slightly longer than for atrazine-grown cells (Table 1). These results also indicate that chemotaxis to s-triazine compounds is not dependent on the presence of atrazine in the growth medium.
FIG. 3.
Chemotactic responses of Pseudomonas sp. strain ADP to atrazine and cyanuric acid after growth under different conditions using the high-throughput capillary assay. The cells were grown in minimal medium with glucose as a carbon source and with atrazine (ATZ), ammonium sulfate, or both as nitrogen source(s). The assays were carried out as described in the legend to Fig. 2. Atrazine was provided as a saturated solution in chemotaxis buffer (approximate concentration based on its solubility in water at 25°C, 150 μM). Cyanuric acid (CA) was provided at 20 mM in chemotaxis buffer. Casamino Acids (0.1% wt/vol) (CAA) served as the positive control. The data were corrected for background accumulation in capillaries containing buffer only for each growth condition (approximately 1.2 × 104 ± 670, 1.1 × 104 ± 605, and 9.0 × 103 ± 773 cells for atrazine-grown cells, ammonium sulfate-grown cells, and cells grown with both nitrogen sources, respectively). The results are the averages of at least 15 capillaries from at least two independent experiments; the error bars indicate standard errors.
Atrazine metabolism is not required for the chemotactic response.
To determine whether the atrazine chemotactic response is dependent on the presence of the atrazine catabolic plasmid pADP-1, a cured strain (ADP M13-2) that does not contain the atrazine catabolic plasmid was tested for chemotaxis to atrazine and atrazine metabolites. The absence of the plasmid in strain ADP M13-2 was verified by PCR and Southern blotting. PCR products of the appropriate sizes (approximately 1 to 1.4 kb) were amplified from the wild-type genomic DNA, but not from genomic DNA purified from strain ADP M13-2, with primers specific for atzA, atzC, and oriV (data not shown). Labeled atzA, atzC, and oriV PCR products amplified from strain ADP hybridized with genomic DNA from strain ADP, but not ADP M13-2 (data not shown).
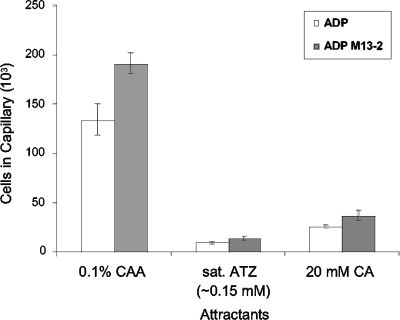
The results of both the quantitative capillary assays and temporal assays demonstrated that s-triazine compounds were all effective chemoattractants for strain ADP M13-2, and each compound elicited similar responses in both the wild-type and the cured strains (Fig. 4 and Table 1). These results indicate that specific genes required for atrazine chemotaxis are not carried on the atrazine catabolic plasmid. The results also demonstrate that atrazine metabolism is not required for the chemotactic response, because strain ADP M13-2 is unable to degrade atrazine.
FIG. 4.
Chemotactic responses of Pseudomonas sp. strains ADP and ADP M13-2 (cured strain) to s-triazines (ATZ, atrazine; CA, cyanuric acid) using the high-throughput capillary assay. The cells were grown in minimal medium with glucose as the carbon source and ammonium sulfate as the sole nitrogen source. The assays were carried out as described in the legend to Fig. 2. Casamino Acids (0.1% wt/vol) (CAA) served as the positive control. The data are corrected for background accumulation in capillaries containing buffer only for each strain (approximately 1.1 × 104 ± 605 and 1.4 × 104 ± 1,025 cells for ADP and ADP M13-2, respectively). The results are the averages of at least 15 capillaries from at least two independent experiments; the error bars indicate standard errors.
Wild-type E. coli is chemotactic toward s-triazines.
Because atrazine utilization and chemotaxis did not appear to be linked in Pseudomonas sp. strain ADP, we speculated that the detection of s-triazines might be a more general response mediated by a receptor for structurally related chemicals, such as pyrimidines. We had previously reported that pyrimidines are attractants for E. coli (25), so we tested chemotaxis to atrazine, N-isopropylammelide, and cyanuric acid in E. coli, which cannot use these s-triazines as growth substrates. E. coli RP437, which is wild type for chemotaxis, responded to N-isopropylammelide and cyanuric acid, but not atrazine (Fig. 5). Both N-isopropylammelide and cyanuric acid had the same peak response concentration (10 mM), but cyanuric acid appeared to be a better attractant based on the lower threshold concentration and greater accumulation of cells in the capillary (Fig. 5).
FIG. 5.
Concentration response curves for chemotaxis to s-triazines by E. coli RP437 (wild type). The cells were grown at 30°C in H1 minimal salts medium containing 25 mM glycerol, the required amino acids, and thiamine. Traditional quantitative capillary assays were performed at 30°C with each compound up to its limit of solubility. The results are the averages of at least 10 capillaries from at least three independent experiments; the error bars indicate standard errors. The background accumulation in capillaries containing buffer only is indicated by the dotted line (approximately 5.0 × 103 ± 624 cells). N-IPA, N-isopropylammelide; CA, cyanuric acid.
Tap mediates chemotaxis to s-triazines in E. coli.
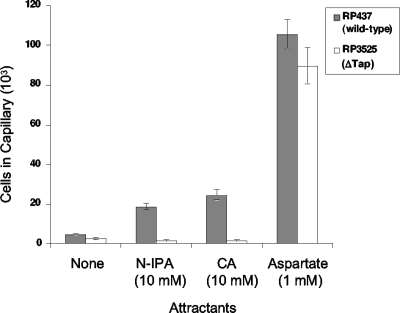
Previous studies demonstrated that Tap, the transducer for dipeptide attractants, also mediates chemotaxis to pyrimidines in E. coli (25, 28). Because pyrimidines are the closest naturally occurring structural analogs of s-triazines, we tested the hypothesis that s-triazines are recognized by the pyrimidine chemoreceptor Tap. Using quantitative capillary assays, the responses of the Δtap mutant strain RP3525 to N-isopropylammelide and cyanuric acid were determined. The peak attractant concentrations determined with wild-type RP437 (Fig. 5) were used for each s-triazine in this experiment. RP3525 (Δtap) cells did not respond to either compound, but the cells responded to the positive-control attractant aspartate (Fig. 6). These results indicate that Tap is required for s-triazine chemotaxis. In an earlier study, we constructed a hybrid chemoreceptor, Tapsr, which has the periplasmic sensing domain of Tap and the cytoplasmic signaling domain of Tsr (25). We showed that Tapsr mediates chemotaxis to thymine, uracil, and the dipeptide Pro-Leu when expressed in a strain that had all of the chromosomally encoded MCP genes deleted (25). To measure the chemotactic response of this strain [UU1250(pXL4)], which has only one chemoreceptor (Tapsr), cells were grown and induced as described in Materials and Methods and then tested using quantitative capillary assays. The number of cells that accumulated in capillaries containing 10 mM cyanuric acid was 16,400 ± 1,140, which was approximately fourfold over background (4,250 ± 450 cells per capillary). These results indicate that the hybrid chemoreceptor Tapsr mediates chemotaxis toward cyanuric acid. Based on these results and the data from our previous studies, the periplasmic domain of Tap appears to be responsible for detecting both pyrimidines and s-triazines.
FIG. 6.
Chemotactic responses of E. coli RP437 (wild type) and RP3525 (Δtap) to s-triazines. The cells were grown and assayed as described in the legend to Fig. 5. N-Isopropylammelide (N-IPA) was provided as a saturated solution in chemotaxis buffer (approximate concentration based on its solubility in water, 10 mM). The cyanuric acid (CA) was 10 mM in chemotaxis buffer. Capillaries containing chemotaxis buffer served as the negative control; those with 1 mM aspartate served as the positive control. The results are the averages of at least 10 capillaries from at least three independent experiments; the error bars indicate standard errors.
Strain ADP is chemotactic toward pyrimidines.
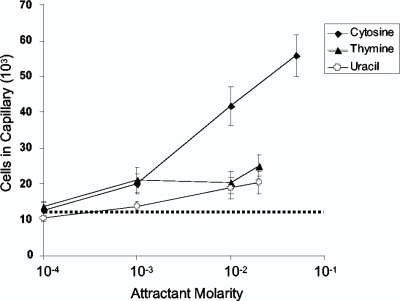
Since s-triazines and pyrimidines are both chemoattractants for E. coli, we tested whether pyrimidines are also chemoattractants for strain ADP. Wild-type ADP cells grown in R medium with glucose and atrazine responded to cytosine at concentrations from 1 to 50 mM, with the peak response at 50 mM (Fig. 7). The average number of cells that accumulated in capillaries containing 50 mM cytosine was 5.59 × 104 ± 5,793 per capillary, which was approximately fivefold higher than background. ADP responded very weakly (about twofold over background) to relatively high concentrations of thymine and uracil (Fig. 7). These results indicate that strain ADP is also chemotactic toward pyrimidines.
FIG. 7.
Concentration response curves for chemotaxis to pyrimidines by Pseudomonas sp. strain ADP. The cells were grown and assayed as described in the legend to Fig. 2. The average background accumulation of cells in capillaries containing buffer only is indicated by the dotted line (approximately 1.2 × 104 ± 670 cells). The results are the averages of at least 15 capillaries from at least two independent experiments; the error bars indicate standard errors.
Cytosine, atrazine, and cyanuric acid compete for detection by the same receptor.
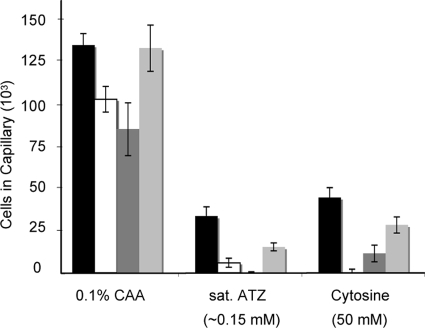
Competition chemotaxis assays were carried out by including a competing attractant at its peak response concentration in the cell suspension and capillary in order to evaluate whether the same or different chemoreceptors are used to detect pyrimidines and s-triazines. The chemotactic response of Pseudomonas sp. strain ADP to atrazine was reduced by over 80% when 50 mM cytosine was present in the cell suspension and was reduced by approximately 50% when 20 mM cyanuric acid was present (Fig. 8). Similar reductions in the response to cytosine were observed when either atrazine or cyanuric acid was present in the cell suspension and capillary (Fig. 8). The presence of saturating atrazine also reduced the response to 20 mM cyanuric acid by approximately 75% (data not shown). In control experiments, the presence of atrazine in the cell suspension completely eliminated the response to atrazine, and the presence of cytosine abolished the response to cytosine (Fig. 8). These results suggest that atrazine, cyanuric acid, and cytosine are detected by the same chemoreceptor.
FIG. 8.
Competition chemotaxis assays. Pseudomonas sp. strain ADP cells were grown as described in the legend to Fig. 2. The responses to saturating atrazine and 50 mM cytosine (in the capillaries) were tested in the absence of competing attractant (black bars) or when 50 mM cytosine (white bars), saturating (sat.) atrazine (dark-gray bars), or 20 mM cyanuric acid (light-gray bars) was added to the cell suspension and capillaries. Capillaries containing Casamino Acids (0.1% in chemotaxis buffer) served as the positive control. The data were corrected for background accumulation in capillaries containing buffer and the corresponding competing compound for each condition (approximately 1.2 × 104 ± 670 cells for experiments without competing attractant and 1.5 × 104 ± 2,100, 4.8 × 103 ± 640, and 7.2 × 103 ± 780 cells for experiments with cytosine, atrazine, or cyanuric acid, respectively, as the competing attractant in the cell suspension and the capillary). The results are the averages of at least eight capillaries; the error bars indicate standard errors.
DISCUSSION
Bacterial chemotaxis to a wide variety of environmentally relevant chemicals has been demonstrated (33, 37). In several cases, genes required for chemotaxis are colocalized and often coordinately regulated with genes for bacterial degradation. For example, Pseudomonas putida G7 is chemotactically attracted to naphthalene, and the response is inducible during growth with naphthalene or the naphthalene degradation intermediate salicylate (16). The nahY gene, which encodes the methyl-accepting chemotaxis protein required for the response to naphthalene, is located in an operon with genes for naphthalene degradation (17). Toluene chemotaxis in P. putida F1 is also an inducible response, requiring the same two-component regulatory system for the expression of the toluene pathway structural genes and toluene chemotaxis (36). The chemotactic response of Ralstonia eutropha JMP134 to the human-made herbicide 2,4-dichlorophenoxyacetate (2,4-D) is induced during growth with 2,4-D. The tfdK gene, which is colocalized with genes for 2,4-D degradation, encodes a major facilitator superfamily transporter that is required for chemotaxis to 2,4-D (20). Another major facilitator superfamily transporter, PcaK, is involved in the chemotactic response to 4-hydroxybenzoate in P. putida PRS2000. The pcaK gene is in an inducible operon with genes for 4-hydroxybenzoate metabolism (18). Biodegradation pathways are often encoded on large catabolic plasmids; this is the case with the naphthalene and 2,4-D degradation pathways. Genes encoding MCPs or other chemotaxis proteins are present on catabolic plasmids in several strains (6, 16, 17, 20, 43, 54). The results presented here, however, demonstrate that atrazine degradation and chemotaxis are not genetically linked in strain ADP, as the presence of the atrazine catabolic plasmid pADP-1 is not necessary for the chemotactic response, and the genes for atrazine chemotaxis and degradation are not coordinately regulated. In addition, no obvious chemotaxis genes were identified on the completely sequenced pADP-1 plasmid (29). These results indicate that the cells must have one or more chromosomally encoded chemoreceptor(s) for the detection of s-triazines.
An earlier study reported chemotaxis to atrazine by Pseudomonas sp. strain ADP (38). Our data confirm and extend these results to include the atrazine degradation intermediates cyanuric acid and N-isopropylammelide. We also provide evidence that atrazine metabolism is not required, indicating that the response is not due to energy taxis but represents the direct detection of atrazine. This result is consistent with the demonstrated ability of strain ADP to detect the nonmetabolizable atrazine analog ametryn.
In general, the relative chemotactic responses (the ratio of the accumulation of cells in attractant-containing capillaries to that of control capillaries) of strain ADP to s-triazines were low but significant (two- to fivefold over background, depending on the compound, strain, and growth conditions). However, the ability to detect these alternative human-made nitrogen sources in the environment is expected to confer a physiological benefit on the relatively rare bacteria that are capable of atrazine degradation. In addition, the chemotactic response could potentially enhance the biodegradation of s-triazines in the environment, especially since strain ADP was shown to be capable of detecting environmentally relevant concentrations of atrazine. We believe that these findings are also important because strain ADP demonstrates a specific chemotactic response to a series of chemicals that has been present in the environment for only approximately 50 years. These results bring up the question of what type of chemoreceptor might be capable of detecting synthetic s-triazine compounds.
Pyrimidines are structurally similar to s-triazines, and we previously showed that cytosine is an attractant for P. putida strains (26). Pseudomonads are known to utilize pyrimidines as nitrogen sources, or in some cases carbon and nitrogen sources (56, 57), so these organisms could benefit from the presence of a chemotaxis system that allows the detection of pyrimidines. In addition, several of the enzymes involved in the degradation of atrazine appear to have been recruited from pyrimidine pathways, as the products of atzA, atzB, and atzC are all members of a large family of amidohydrolases that includes, for example, cytosine deaminase (42). It therefore seems reasonable that the ability to sense atrazine could be fortuitous and due to the presence of a broad-specificity pyrimidine chemoreceptor that can also detect s-triazines. If so, other bacteria that are chemotactic to pyrimidines may have the ability to sense s-triazines. We therefore used E. coli as a model organism to test this hypothesis. We previously demonstrated that pyrimidines are attractants for E. coli and that the response is mediated by the methyl-accepting chemotaxis protein Tap (25). Here, we showed that wild-type E. coli is attracted to the s-triazine compounds N-isopropylammelide and cyanuric acid, which do not provide any growth benefit to E. coli, and we demonstrated that s-triazines are not detected by the Tap mutant. These results indicate that chemotactic responses to structurally related pyrimidines and s-triazines are mediated by a single chemoreceptor in E. coli. As might be expected for fortuitously detected structurally related compounds, the responses of E. coli to s-triazines were significantly weaker (Fig. 5 and 6) than those to pyrimidines (11- and 19-fold responses to uracil and thymine, respectively [25]). Although we have not yet identified the chemoreceptor for atrazine in Pseudomonas sp. strain ADP, we propose that chemotaxis to s-triazines and pyrimidines may be mediated by a single chemoreceptor in this organism. In addition to cyanuric acid and N-isopropylammelide, strain ADP is able to respond to atrazine, which is not detected by E. coli. The ability to detect atrazine might be a newly evolved ability of the chemoreceptor(s) that detects pyrimidines and other s-triazine compounds in Pseudomonas sp. strain ADP. The demonstration that the presence of cytosine reduces the response to atrazine and that atrazine reduces the response to cytosine in competition chemotaxis assays (Fig. 8) strongly suggests that the two compounds are detected by the same chemoreceptor. Previous competition assays with E. coli demonstrated a wide range of inhibition, from slight effects to complete abolishment of responses; these differences were suggested to depend on the affinity of the receptor for the attractants being studied (2, 31). We are currently working to identify the chemoreceptor(s) required for pyrimidine and s-triazine chemotaxis in strain ADP; this information will contribute to our understanding of the mechanisms used by bacteria to sense and respond to human-made chemicals.
Acknowledgments
This work was supported by a grant from the University of California Toxic Substances Research and Teaching Program.
We thank Sandy Parkinson for providing E. coli strains, Jennifer Seffernick and Larry Wackett for providing Pseudomonas sp. strains ADP and ADP M13-2 and s-triazine compounds and for helpful discussions, and Carrie Harwood and Jack Meeks for helpful suggestions.
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. [DOI] [PubMed] [Google Scholar]
- 2.Adler, J., G. L. Hazelbauer, and M. M. Dahl. 1973. Chemotaxis toward sugars in Escherichia coli. J. Bacteriol. 115:824-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbash, J. E., G. P. Thelin, D. W. Kolpin, and R. J. Gilliom. 2001. Major herbicides in ground water: results from the National Water-Quality Assessment. J. Environ. Qual. 30:831-845. [DOI] [PubMed] [Google Scholar]
- 4.Behki, E. R., E. Topp, W. Dick, and P. Germon. 1993. Metabolism of the herbicide atrazine by Rhodococcus strains. Appl. Environ. Microbiol. 59:1955-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boundy-Mills, K. L., M. L. de Souza, R. T. Mandelbaum, L. P. Wackett, and M. J. Sadowsky. 1997. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl. Environ. Microbiol. 63:916-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis, J. J., and G. J. Zylstra. 2004. Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 341:753-768. [DOI] [PubMed] [Google Scholar]
- 7.de Souza, M. L., M. J. Sadowsky, and L. P. Wackett. 1996. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J. Bacteriol. 178:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Souza, M. L., J. Seffernick, B. Martinez, M. J. Sadowsky, and L. P. Wackett. 1998. The atrazine catabolism genes atzABC are widespread and highly conserved. J. Bacteriol. 180:1951-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Souza, M. L., L. P. Wackett, K. L. Boundy-Mills, R. T. Mandelbaum, and M. J. Sadowsky. 1995. Cloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine. Appl. Environ. Microbiol. 61:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Souza, M. L., L. P. Wackett, and M. J. Sadowsky. 1998. The atzABC genes encoding atrazine catabolism are located on a self-transmissible plasmid in Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 64:2323-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devers, M., G. Soulas, and F. Martin-Laurent. 2004. Real-time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil. J. Microbiol. Methods 56:3-15. [DOI] [PubMed] [Google Scholar]
- 12.Eaton, R. W., and D. W. Ribbons. 1982. Metabolism of dibutylphthalate and phthalate by Micrococcus sp. strain 12B. J. Bacteriol. 151:48-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Environmental Protection Agency. 2009. National primary drinking water regulations. Environmental Protection Agency, Washington, DC.
- 14.Garcia-Gonzalez, V., F. Govantes, O. Porrua, and E. Santero. 2005. Regulation of the Pseudomonas sp. strain ADP cyanuric acid degradation operon. J. Bacteriol. 187:155-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Gonzalez, V., F. Govantes, L. J. Shaw, R. G. Burns, and E. Santero. 2003. Nitrogen control of atrazine utilization in Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 69:6987-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm, A. C., and C. S. Harwood. 1997. Chemotaxis of Pseudomonas putida to the polyaromatic hydrocarbon naphthalene. Appl. Environ. Microbiol. 63:4111-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm, A. C., and C. S. Harwood. 1999. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 181:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwood, C. S., N. N. Nichols, M.-K. Kim, J. L. Ditty, and R. E. Parales. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harwood, C. S., M. Rivelli, and L. N. Ornston. 1984. Aromatic acids are chemoattractants for Pseudomonas putida. J. Bacteriol. 160:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawkins, A. C., and C. S. Harwood. 2002. Chemotaxis of Ralstonia eutropha JMP134(pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Appl. Environ. Microbiol. 68:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes, T. B., A. Collins, M. Lee, M. Mendoza, N. Noriega, A. A. Stuart, and A. Vonk. 2002. Hermaphroditic, demasculinized frogs after exposure to the herbicide atrazine at low ecologically relevant doses. Science 99:5476-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes, T. B., K. Haston, M. Tsui, A. Hoang, C. Haeffele, and A. Vonk. 2003. Atrazine-induced hermaphroditism at 0.1 ppb in American leopard frogs (Rana pipiens): laboratory and field evidence. Environ. Health Perspect. 111:568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroschwitz, J. I., and M. Howe-Grant. 1995. Kirk-Othmer encyclopedia of chemical technology, 4th ed. John Wiley and Sons, New York, NY.
- 24.Law, A. M., and M. D. Aitken. 2003. Bacterial chemotaxis to naphthalene desorbing from a nonaqueous liquid. Appl. Environ. Microbiol. 69:5968-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, X., and R. E. Parales. 2008. Chemotaxis of Escherichia coli to pyrimidines: a new role for the signal transducer Tap. J. Bacteriol. 190:972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, X., P. L. Wood, J. V. Parales, and R. E. Parales. 2009. Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J. Bacteriol. 191:2909-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandelbaum, R. T., D. L. Allen, and L. P. Wackett. 1995. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 61:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manson, M. D., V. Blank, G. Brade, and C. F. Higgins. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321:253-256. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marx, R. B., and M. D. Aitken. 2000. Bacterial chemotaxis enhances naphthalene degradation in a heterogeneous aqueous system. Environ. Sci. Technol. 34:3379-3383. [Google Scholar]
- 31.Mesibov, R., and J. Adler. 1972. Chemotaxis toward amino acids in Escherichia coli. J. Bacteriol. 112:315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newcombe, D., and D. E. Crowley. 1999. Bioremediation of atrazine-contaminated soil by repeated applications of atrazine-degrading bacteria. Appl. Microbiol. Biotechnol. 51:877-882. [DOI] [PubMed] [Google Scholar]
- 33.Pandey, G., and R. K. Jain. 2002. Bacterial chemotaxis toward environmental pollutants: role in bioremediation. Appl. Environ. Microbiol. 68:5789-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papiernik, S. K., and R. F. Spalding. 1998. Atrazine, deethylatrazine, and deisopropylatrazine persistence measured in groundwater in situ under low-oxygen conditions. J. Agric. Food Chem. 46:749-754. [DOI] [PubMed] [Google Scholar]
- 35.Parales, R. E. 2004. Nitrobenzoates and aminobenzoates are chemoattractants for Pseudomonas strains. Appl. Environ. Microbiol. 70:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parales, R. E., J. L. Ditty, and C. S. Harwood. 2000. Toluene-degrading bacteria are chemotactic to the environmental pollutants benzene, toluene, and trichloroethylene. Appl. Environ. Microbiol. 66:4098-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parales, R. E., and C. S. Harwood. 2002. Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr. Opin. Microbiol. 5:266-273. [DOI] [PubMed] [Google Scholar]
- 38.Park, J.-H., Y. Feng, P. Ji, T. C. Voice, and S. A. Boyd. 2003. Assessment of bioavailability of soil-sorbed atrazine. Appl. Environ. Microbiol. 69:3288-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porrua, O., M. Garcia-Jaramillo, E. Santero, and F. Govantes. 2007. The LysR-type regulator AtzR binding site: DNA sequences involved in activation, repression and cyanuric acid-dependent repositioning. Mol. Microbiol. 66:410-427. [DOI] [PubMed] [Google Scholar]
- 41.Radosevich, M., S. J. Traina, Y.-L. Hao, and O. H. Tuovinen. 1995. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl. Environ. Microbiol. 61:297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadowsky, M. J., Z. Tong, M. L. de Souza, and L. P. Wackett. 1998. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J. Bacteriol. 180:152-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samanta, S. K., and R. K. Jain. 2000. Evidence for plasmid-mediated chemotaxis of Pseudomonas putida towards naphthalene and salicylate. Can. J. Microbiol. 46:1-6. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Sass, J., and P. W. Brandt-Rauf. 2003. Cancer incidence among triazine herbicide manufacturing workers. J. Occup. Environ. Med. 45:343-344. [DOI] [PubMed] [Google Scholar]
- 46.Shioi, J., C. V. Dang, and B. L. Taylor. 1987. Oxygen as attractant and repellent in bacterial chemotaxis. J. Bacteriol. 169:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slocum, M. K., and J. S. Parkinson. 1985. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: null phenotypes of the tar and tap genes. J. Bacteriol. 163:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strong, L. C., H. McTavish, M. J. Sadowsky, and L. P. Wackett. 2000. Field-scale remediation of atrazine-contaminated soil using recombinant Escherichia coli expressing atrazine chlorohydrolase. Environ. Microbiol. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 49.Struthers, J. K., K. Jayachandran, and T. Moorman. 1998. Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil. Appl. Environ. Microbiol. 64:3368-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan, K. B., and K. M. Spence. 2003. Effects of sublethal concentrations of atrazine and nitrate on metamorphosis of the African clawed frog. Environ. Toxicol. Chem. 22:627-635. [PubMed] [Google Scholar]
- 51.Topp, E., W. M. Mulbry, H. Zhu, S. M. Nour, and D. Cuppels. 2000. Characterization of s-triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils. Appl. Environ. Microbiol. 66:3134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Topp, E., H. Zhu, S. M. Nour, S. Houtot, M. Lewis, and D. Cuppels. 2000. Characterization of an atrazine-degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils. Appl. Environ. Microbiol. 66:2773-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trentacoste, S. V., A. S. Friedmann, R. T. Youker, C. B. Breckenridge, and B. R. Zirkin. 2001. Atrazine effects on testosterone levels and androgen-dependent reproductive organs in peripubertal male rats. J. Androl. 22:142-148. [PubMed] [Google Scholar]
- 54.van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Röthlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 55.Wackett, L. P., M. J. Sadowsky, B. Martinez, and N. Shapir. 2002. Biodegradation of atrazine and related s-triazine compounds: from enzymes to field studies. Appl. Microbiol. Biotechnol. 58:39-45. [DOI] [PubMed] [Google Scholar]
- 56.West, T. P. 1991. Pyrimidine base and ribonucleoside utilization by the Pseudomonas alcaligenes group. Antonie van Leeuwenhoek 59:263-268. [DOI] [PubMed] [Google Scholar]
- 57.West, T. P., and C. P. Chu. 1986. Utilization of pyrimidines and pyrimidine analogues by fluorescent pseudomonads. Microbios 47:149-157. [PubMed] [Google Scholar]
- 58.Yanze-Kontchou, C., and N. Gschwind. 1994. Mineralization of the herbicide atrazine as a carbon source by a Pseudomonas strain. Appl. Environ. Microbiol. 60:4297-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]