Abstract
(E, E, E)-Geranylgeraniol (GGOH) is a valuable starting material for perfumes and pharmaceutical products. In the yeast Saccharomyces cerevisiae, GGOH is synthesized from the end products of the mevalonate pathway through the sequential reactions of farnesyl diphosphate synthetase (encoded by the ERG20 gene), geranylgeranyl diphosphate synthase (the BTS1 gene), and some endogenous phosphatases. We demonstrated that overexpression of the diacylglycerol diphosphate phosphatase (DPP1) gene could promote GGOH production. We also found that overexpression of a BTS1-DPP1 fusion gene was more efficient for producing GGOH than coexpression of these genes separately. Overexpression of the hydroxymethylglutaryl-coenzyme A reductase (HMG1) gene, which encodes the major rate-limiting enzyme of the mevalonate pathway, resulted in overproduction of squalene (191.9 mg liter−1) rather than GGOH (0.2 mg liter−1) in test tube cultures. Coexpression of the BTS1-DPP1 fusion gene along with the HMG1 gene partially redirected the metabolic flux from squalene to GGOH. Additional expression of a BTS1-ERG20 fusion gene resulted in an almost complete shift of the flux to GGOH production (228.8 mg liter−1 GGOH and 6.5 mg liter−1 squalene). Finally, we constructed a diploid prototrophic strain coexpressing the HMG1, BTS1-DPP1, and BTS1-ERG20 genes from multicopy integration vectors. This strain attained 3.31 g liter−1 GGOH production in a 10-liter jar fermentor with gradual feeding of a mixed glucose and ethanol solution. The use of bifunctional fusion genes such as the BTS1-DPP1 and ERG20-BTS1 genes that code sequential enzymes in the metabolic pathway was an effective method for metabolic engineering.
(E,E,E)-Geranylgeraniol (GGOH) can be used as an important ingredient for perfumes and as a desirable raw material for synthesizing vitamins A and E (4, 13). It is also known to induce apoptosis in various cancer and tumor cell lines (24, 36). GGOH is the dephosphorylated derivative of (E,E,E)-geranylgeranyl diphosphate (GGPP) (Fig. 1). GGPP is a significant intermediate of ubiquinone and carotenoid biosyntheses, especially in carotenoid-producing microorganisms and plant cells. It is also utilized as the lipid anchor of geranylgeranylated proteins. In the yeast Saccharomyces cerevisiae, GGPP is synthesized by GGPP synthase (GGPS), encoded by the BTS1 gene, which catalyzes the condensation of farnesyl diphosphate (FPP) and isopentenyl diphosphate (IPP) rather than the successive addition of IPP molecules to dimethylallyl diphosphate, geranyl diphosphate, and FPP that is detected in mammalian tissues (14). Biologically synthesized GGOH comprises only (E,E,E)-geometric isomers, and only the (E,E,E)-isomers have significant biological activities (23). The chemically synthesized form is usually obtained as mixtures of (E)- and (Z)-isomers and thus has lower potency. Therefore, there is a greater possibility of attaining efficient production of (E,E,E)-GGOH through fermentative production.
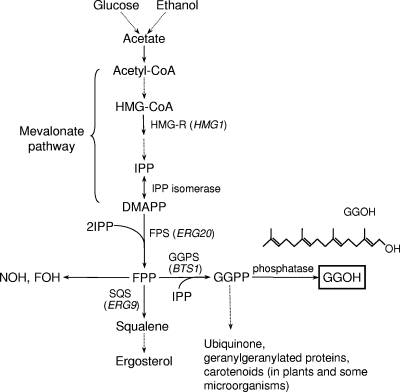
FIG. 1.
Biosynthetic pathway for GGOH in S. cerevisiae. The solid arrows indicate the one-step conversions in the biosynthesis, and the dashed arrows indicate the several steps. Intermediates: HMG-CoA, 3-hydroxy-3-methylflutaryl coenzyme A; DMAPP, dimethylallyl diphosphate. Enzymes: HMG-R, HMG-coenzyme A reductase (encoded by the HMG1 gene); FPS, FPP synthase (ERG20).
Some yeast strains accumulate ergosterol up to 4.6% dry mass (1). Thus, yeasts have the potential to produce large amounts of GGOH if it is possible to enhance and redirect the metabolic flux to GGOH synthesis. The enzyme 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMG-R), encoded by the HMG1 gene has been shown to be the major rate-limiting enzyme in the mevalonate pathway in S. cerevisiae (12). Overproduction of the catalytic domain of HMG-R in an S. cerevisiae strain resulted in squalene accumulation of up to 1% (27) and 2% (8) dry mass but did not cause any difference in the contents of isoprenoid alcohols such as farnesol (FOH) and geraniol (27). These results suggest that squalene is preferably accumulated rather than GGOH when the mevalonate pathway is enhanced by overexpression of the HMG1 gene. Squalene is synthesized through the condensation of two molecules of FPP catalyzed by squalene synthase (SQS) encoded by the ERG9 gene in S. cerevisiae (Fig. 1). The addition of an SQS inhibitor to cultures of S. cerevisiae strains resulted in the production of considerable amounts of FOH (∼77.5 mg liter−1) and relatively small amounts of GGOH (∼2.2 mg liter−1) (20). It has also been reported that SQS-deficient (Δerg9) S. cerevisiae strains, which are sterol auxotrophic, accumulated FPP in their cells (35) and excreted 1.3 mg liter−1 of FOH into the culture medium (5). Therefore, inactivation of SQS seems to enhance FOH rather than GGOH production. This is probably because of the low GGPS activity in S. cerevisiae. Indeed, a carotenoid-producing Rhodotorula yeast strain showed higher GGOH (24.4 mg liter−1) than FOH (4.4 mg liter−1) production on cultivation with an SQS inhibitor (20). Our group previously found that GGOH production could be enhanced by overexpression of the BTS1 gene in S. cerevisiae without SQS inhibition. In addition, coexpression of a fusion of the BTS1 and farnesyl diphosphate synthetase (ERG20) genes along with the HMG1 gene resulted in the production of a substantial amount of GGOH with only a small amount of FOH (C. Ohto, M. Muramatsu, E. Sakuradani, S. Shimizu, and S. Obata, submitted for publication).
These results suggest that GGOH can be produced from GGPP through some endogenous phosphatase activities when GGPP synthesis is enhanced. We therefore hypothesized that enhancement of the phosphatase activity could increase the productivity of GGOH. However, it is not clear what kind of phosphatase enhances the GGOH production. It has been reported that the products of the diacylglycerol diphosphate phosphatase (DPP1) gene and lipid phosphate phosphatase (LPP1) gene account for most of the FPP and GGPP phosphatase activities in a particulate (membrane associated) fraction of S. cerevisiae (9). In this study, we found that GGOH production could be enhanced by overexpression of these phosphatase genes. We also demonstrated that overexpression of the BTS1-DPP1 and BTS1-ERG20 fusion genes along with the HMG1 gene further increased GGOH production. Finally, we constructed a high-level GGOH-producing yeast available for industrial processes involving multicopy integration vectors. The productivity of GGOH was evaluated in test tube cultures and 10-liter jar fermentors.
MATERIALS AND METHODS
Chemicals.
[1-3H]GGPP was obtained from the Japan Radioisotope Association (Tokyo, Japan). Restriction enzymes and other modifying enzymes were purchased from Takara Bio (Shiga, Japan).
Strains and media.
The yeast strains used in this study are listed in Table 1. Synthetic dextrose (SD) medium was used for transformant selection and precultures of recombinant yeasts harboring episomal plasmids. SD medium was comprised of 20 g liter−1 glucose, 6.7 g liter−1 yeast nitrogen base without amino acids (Difco/BD Diagnostic Systems, MD), 40 μg ml−1 adenine, and 0.77 g liter−1 complete supplement mixture lacking specified amino acid(s) for auxotrophic marker(s) (BIO 101 Systems, CA), pH 6.0. Selective SD agar plates contained 20 g liter−1 of agar in SD medium. YM broth and YPD medium were used for fermentation experiments in test tubes. YM broth was comprised of 3 g liter−1 yeast extract, 3 g liter−1 malt extract, 5 g liter−1 peptone, 10 g liter−1 glucose, and adenine (40 μg ml−1). YPD medium was comprised of 10 g liter−1 yeast extract, 20 g liter−1 peptone, 20 g liter−1 glucose, and adenine (40 μg ml−1). In order to reduce the formation of nerolidol (NOH) under acidic conditions, the initial pH of the YM and YPD media was set to 7.0. Sporulation plates contained 10 g liter−1 potassium acetate, 1 g liter−1 yeast extract, 0.5 g liter−1 glucose, and 20 g liter−1 agar, pH 6.0. YMP medium was used for the precultures in the jar fermentation experiments. YMP medium was comprised of 5 g liter−1 yeast extract, 5 g liter−1 malt extract, 10 g liter−1 peptone, and 5 g liter−1 glucose (pH was not adjusted). The main culture medium for jar fermentations comprised 1 g liter−1 glucose, 0.85 g liter−1 KH2PO4, 1.5 g liter−1 MgSO4·7H2O, 5 g liter−1 (NH4)2SO4, 0.35 g liter−1 CaCl2·2H2O, 1.5 g liter−1 an antifoaming agent (Adekanol LG-109; ADEKA Co., Tokyo, Japan), and corn steep liquor adjusted to pH 5.5 with aqueous ammonia. The bacterial cultures were grown at 37°C in LB broth (10 g liter−1 tryptone, 5 g liter−1 yeast extract, and 5 g liter−1 sodium chloride) containing 100 μg ml−1 ampicillin when appropriate.
TABLE 1.
Plasmids, vectors, and yeast strains used in this study
| Plasmid, vector, or yeast strain | Relevant feature(s) and/or description | Reference or source |
|---|---|---|
| Plasmids and vectorsa | ||
| pRS434GAP | YEp (2μm), TRP1 marker; DDBJ accession no. 304854 | 25 |
| pRS435GAP | YEp (2μm), LEU2 marker; DDBJ accession no. 304858 | 25 |
| pRS436GAP | YEp (2μm), URA3 marker; DDBJ accession no. 304862 | 25 |
| pRS504GAP | Multicopy integration vector targeted to rDNA loci; TRP1d marker | This study |
| pRS524GAP | Multicopy integration vector targeted to rDNA loci; TRP1d marker | This study |
| pRS515GAP | Multicopy integration vector targeted to rDNA loci; LEU2d marker | This study |
| pDI626GAP | Multicopy integration vector targeted to Ty δ loci; URA3d marker | This study |
| Parent strains | ||
| YPH499 | MATalys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ura3-52 | Stratagene |
| YPH500 | MATα lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ura3-52 | Stratagene |
| Strains with episomal plasmids | ||
| LPP1/YPH499 | pRS436LPP1/YPH499 | This study |
| DPP1/YPH499 | pRS436DPP1/YPH499 | This study |
| BTS1/YPH499 | pRS435BTS1/YPH499 | This study |
| LPP1/BTS1/YPH499 | pRS436LPP1/pRS435BTS1/YPH499 | This study |
| DPP1/BTS1/YPH499 | pRS436DPP1/pRS435BTS1/YPH499 | This study |
| GGDP/YPH499 | pRS436GGDP/YPH499 | This study |
| GGF/GGDP/HMG1/YPH499(YEp) | pRS434HMG1/pRS435GGF/pRS436GGDP/YPH499 | This study |
| Strains with multicopy integration vectors | ||
| HMG1/YPH499 | pRS504HMG1/YPH499 | This study |
| GGDP/HMG1/YPH499 | pDI626GGDP/pRS504HMG1/YPH499 | This study |
| GGF/GGDP/HMG1/YPH499 | pRS515GGF/pDI626GGDP/pRS504HMG1/YPH499 | This study |
| GGF/GGDP/HMG1/YPH499-H | GGF/GGDP/HMG1/YPH499 HIS3 | This study |
| GGF/GGDP/HMG1/YPH500 | pRS515GGF/pDI626GGDP/pRS524HMG1/YPH500 | This study |
| GGF/GGDP/HMG1/YPH500-AK | GGDP/HMG1/YPH500 ADE2 LYS2 | This study |
| 5X10 | Diploid prototroph generated by mating GGF/GGDP/HMG1/YPH499-H with GGF/GGDP/HMG1/YPH500-AK | This study |
All the plasmids and vectors contained the TDH3 promoter and CYC1 terminator from S. cerevisiae for gene expression.
Construction of yeast episomal plasmids and a BTS1-DPP1 fusion gene.
Standard techniques for nucleic acid manipulation were performed as described by Sambrook et al. (32). The PCR primers used in this study are listed in Table 2. The plasmids pRS434GAP, pRS435GAP, and pRS436GAP (25) were used as 2μm-based yeast episomal plasmids. These plasmids contained multicloning sites flanked by the glyceraldehyde-3-phosphate dehydrogenase 3 (TDH3) gene promoter and cytochrome c (CYC1) gene terminator (CYC1t) from S. cerevisiae, and thereby the genes cloned into the multicloning sites were constitutively expressed under the control of the TDH3 promoter. These plasmids have almost the same sequences except for the selection markers (TRP1, LEU2, and URA3, respectively). The genes encoding diacylglycerol diphosphate phosphatase (DPP1), lipid phosphate phosphatase (LPP1), and GGPP synthase (BTS1) were amplified from YPH499 genomic DNA by PCR. The PCR fragments of the DPP1 and LPP1 genes were cloned into the multicloning site of pRS436GAP, and the resulting plasmids were named pRS436DPP1 and pRS436LPP1, respectively. The PCR fragment of the BTS1 gene was cloned into the multicloning site of pRS435GAP, and the resulting plasmid was named pRS435BTS1.
TABLE 2.
PCR primers used in this study
| Primer name | Sequence (5′-3′)a | Restriction site or linker |
|---|---|---|
| S-BTS1-1 | ACGCCGCGGACAATG GAGGCCAAGATAGATGAG | SacII |
| BTS1-2 | TCTGTTCATAGAACCACCACCCAATTCGGATAAGTGGTCTAT | GGGS linker |
| DPP1-3 | GGTGGTGGTTCTATGAACAGAGTTTCGTTTAT | GGGS linker |
| X-DPP1-4 | CTTTTCTCGAGTTACATACCTTCATCGGA | XhoI |
| R4 | ATGAGAGTAGCAAACGTAAGTCTAA | |
| K-R7 | TGACTGGTACCTTTCCTCTAATCAGGTTCCACC | KpnI |
| AatTRP1-50F | TTTCCGACGTCCACGTGAGTATACGTGATTAAG | Aat II |
| TRP1d-R | AGGCAAGTGCACAAACAATACTT | |
| KpnTRP1-50dR | TGACTGGTACCAGGCAAGTGCACAAACAATACTT | KpnI |
| S-R4 | TATTGAGCTCATGAGAGTAGCAAACGTAAGTCTAA | SacI |
| BB-R5 | CAGGTGCGCGCGGTAACCCAGTTCCTCACTA | Bst PI |
| AB-R6 | GTGAGGACGTCGGTTACCCGGGGCACCTGTCAC | Bst PI |
| A-R7 | CACTCGACGTCTTTCCTCTAATCAGGTTCCACC | Aat II |
| Pma-LEU2d-F | CACGTGTCGACTACGTCGTAAGGCCGTTT | Pma CI |
| Kpn-LEU2d-R | GGTACCAAGGATATACCATTCTAATGTCTGCCCC | KpnI |
| T7-F | GCGTAATACGACTCACTATAGGG | |
| 2μm-R | CCTGATGCGGTATTTTCTCCTTAC | |
| Aat-TY1F | GACGTCTGTTGGAATAAAAATCCACTATCG | Aat II |
| Sse-TY2R | CCTGCAGGATTCCGTTTTATATGTTTATATTCATTG | Sse 8387I |
| SacI-TY3F | GAGCTCGAGGAATAATCGTAATATTAGTATGTA | SacI |
| Bss-TY4R | GCGCGCTGAGAAATTTGTGGGTAATTAGATAAT | Bss HII |
| HIS3-F | ATGACAGAGCAGAAAGCCCTAG | |
| HIS3-R | CTACATAAGAACACCTTTGGTGG | |
| LYS2-F | ATGACTAACGAAAAGGTCTGGATAG | |
| LYS2-R | TTAAGCTGCTGCGGAGCTTCCAC | |
| ADE2-F | ATGGATTCTAGAACAGTTGGTATATTAG | |
| ADE2-R | TTACTTGTTTTCTAGATAAGCTTCGTAAC |
The start codon (ATG) is shown in boldface. Restriction sites and linkers are underlined.
A BTS1-DPP1 fusion gene was constructed by fusing the DPP1 gene to the 3′ end of the BTS1 gene as follows. A BTS1 fragment was amplified by PCR using primers S-BTS1-1 and BTS1-2 and the pRS435BTS1 plasmid as a template. A DPP1 fragment was amplified using primers DPP1-3 and X-DPP1-4 and the pRS436DPP1 plasmid as a template. These two fragments were purified by agarose gel electrophoresis and then mixed and fused by PCR using the primer pair of S-BTS1-1 and X-DPP1-4, as described Nikawa and Kawabata (21). The BTS1-DPP1 fusion gene fragment contained SacII at the 5′ end and XhoI at the 3′ end, and the Gly-Gly-Gly-Ser (GGGS) linker between the BTS1 and DPP1 genes, as shown in Fig. 2A. The DPP1-BTS1 fusion gene was cloned into the multicloning site of pRS436GAP, and the resulting plasmid was named pRS436GGDP.
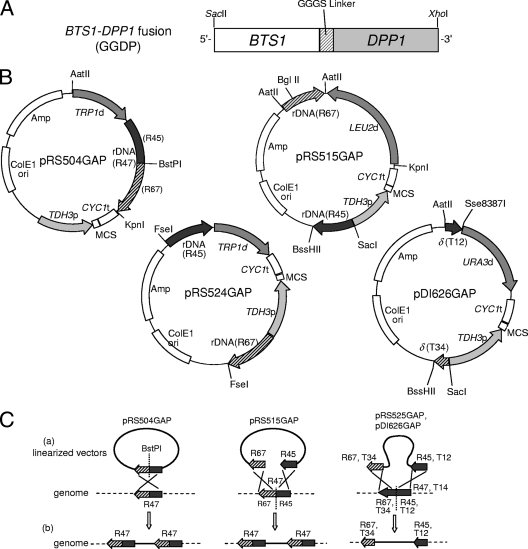
FIG. 2.
Structures of the BTS1-DPP1 fusion gene and multicopy integration vectors. (A) Structures of the diacylglycerol diphosphate phosphatase (DPP1) gene and GGPS (BTS1) fusion gene. GGGS linker, the Gly-Gly-Gly-Ser sequence additionally inserted between the genes as a linker sequence. (B) Structures of the multicopy-integration vectors. MCS, multicloning site. (C) Schematic representation of the genomic integration of the vectors. The linearized vectors before (a) and after (b) homologous recombination into the genomic target sequences are shown. In pRS504GAP, the E. coli sequences (ColE1 ori and Amp) are integrated together; in pRS515GAP, the E. coli sequences are eliminated before integration; and in pRS525GAP and pDI626GAP, the E. coli sequences are eliminated before integration. A tandem repeat of the target sequence does not occur with these vectors.
The episomal plasmid for overexpression of the HMG1 gene was constructed by inserting a gene fragment into the multicloning site of pRS434GAP, and the resulting plasmid was named pRS434HMG1. The episomal plasmid for overexpression of the BTS1-ERG20 fusion gene (pRS435GGF) was constructed previously (C. Ohto et al., submitted).
Construction of multicopy integration vectors.
Multicopy integration vector pRS504GAP was constructed as follows. A part of the ribosomal DNA (rDNA) sequence of S. cerevisiae was amplified by PCR using primers R4 and K-R7 with YPH499 genomic DNA as a template, yielding R47 rDNA sequence [rDNA(R47)]. The TRP1d marker, which was truncated in the promoter region of the TRP1 marker gene, was amplified by PCR using primers AatTRP1d-50F and TRP1d-R with pRS434GAP as a template. The rDNA(R47) and TRP1d fragments were fused by PCR, followed by replacement with the AatII-KpnI site of pRS434GAP, which contained the TRP1 and 2μm ori, and the resulting vector was named pRS504GAP (Fig. 2B). The HMG1 gene was amplified by PCR, followed by insertion into the multicloning site of pRS504GAP. The resulting vector was named pRS504HMG1. pRS504HMG1 was linearized by BstPI digestion before use for yeast transformation.
Multicopy integration vector pRS515GAP was constructed as follows. In the pRS515GAP vector, the rDNA(R47) sequence used in pRS504GAP was divided into two parts, as shown in Fig. 2C. The rDNA sequences of the upstream (R45) and downstream (R67) recombination sites were amplified by PCR using the primer pair S-R4 and BB-R5 and the pair AB-R6 and A-R7 and the plasmid pRS504GAP as a template. The TRP1d marker was amplified by PCR using primers AatTRP1d-50F and KpnTRP1-50dR and the plasmid pRS434GAP as a template. The R45, R67, and TRP1d fragments were ligated into the SacI-BssHII, AatII, and AatII-KpnI sites of pRS434GAP, respectively, and the resulting vector was named pRS514GAP. The LEU2d marker, which was truncated in the promoter region of the LEU2 marker gene, was amplified by PCR using primers Pma-LEU2dF and Kpn-LEU2dR and the plasmid pRS435GAP as a template, followed by subcloning into the pCR-Blunt-II TOPO vector (Invitrogen, CA) to yield TOPO-LEU2d. The TOPO-LEU2d vector was digested with PmaCI and XhoI, and the resulting LEU2d fragment was blunt ended with T4 DNA polymerase. The pRS514GAP vector was digested with PmaCI and KpnI, blunt ended with KOD Plus DNA polymerase (Toyobo, Tokyo, Japan), and ligated with the LEU2d fragment to yield pRS515GAP (Fig. 2B). The SacII-MluI fragment of pRS435GGF, which contains the BTS1-ERG20 fusion gene, was ligated into the same site of pRS515GAP to yield pRS515GGF. pRS515GGF was linearized by BssHII-BglII digestion before use for yeast transformation.
Vectors pRS504GAP and pRS515GAP were designed to be introduced in a gene insertion manner with and without introduction of the Escherichia coli sequences, respectively, as shown in Fig. 2C.
Multicopy integration vector pRS524GAP was constructed as follows. The R45 and R67 sequences in pRS514GAP were converted into each other with insertion of FseI sites by PCR, as shown in Fig. 2B. The HMG1 gene was amplified by PCR and inserted into the multicloning site of pRS524GAP, and the resulting vector was named pRS524HMG1. pRS524HMG1 was linearized by FseI digestion before use for yeast transformation.
Multicopy integration vector pDI626GAP was constructed as follows. The 2μm ori of pRS436GAP was eliminated by PCR using phosphorylated primers T7-F and 2μm-R, followed by self-ligation of the PCR product to yield pRS436-2 μm. The Ty δ sequences for upstream and downstream recombination sites (named T12 and T34, respectively) were amplified by PCR using the primer pair Aat-TY1F and Sse-TY2R and the pair SacI-TY3F and Bss-TY4R, respectively. The genomic DNA of YPH499 was used as a template. The T12 and T34 fragments were ligated into the AatII-Sse8387I and SacI-BssHII sites of pRS436-2μm, respectively, to yield pDI626GAP (Fig. 2B). The promoter region of the URA3 marker gene was truncated in pDI626GAP. The SacII-XhoI fragment of pRS434GGDP was ligated into the same site of pDI626GAP to yield pDI626GGDP. pDI626GGDP was linearized by AatII-BssHII digestion before use for yeast transformation.
Vectors pRS524GAP and pDI626GAP were designed to be introduced in a gene replacement manner without introduction of the E. coli sequences, as shown in Fig. 2C. This type of integration vector can be more stably maintained in the genome than gene insertion type vectors such as pRS504GAP and pRS515GAP because the tandem repeat of the target sequence is not present.
Yeast transformation.
Yeasts were transformed with a Frozen-EZ Yeast Transformation II kit (Zymo Research, CA). About 1 μg of episomal plasmid or about 5 μg of linearized multicopy integration vector was used for the transformation, followed by selection on selective SD agar plates. The colonies formed on the plates were checked by PCR mapping for the correct insertion of the plasmid vectors.
Estimation of copy numbers.
The copy numbers of the HMG1, BTS1-DPP1, and BTS1-ERG20 genes introduced into the genome with multicopy integration vectors were estimated by quantitative real-time PCR using a Smart Cycler system and Ex Taq R-PCR, version 1.0, with SYBR green I (both from Takara Bio), based on the manufacturer's instructions. The yeast genomic DNA was extracted with a Dr. GenTLE (from yeast) high-recovery kit (Takara Bio). The copy numbers were estimated by comparison with an internal reference gene with a known copy number (ERG9 gene; one copy in the genome). The copy numbers of the native genes (one copy) were subtracted from each estimated copy number.
Preparation of cell extracts.
The cell extracts used in the phosphatase activity assays were prepared as follows. Cells were grown at 30°C in selective SD medium for 24 h with shaking (130 rpm). Cells were harvested by centrifugation at 10,000 rpm for 5 min, washed with distilled water, and then resuspended in extraction buffer (120 mM citrate, pH 4.3) supplemented with protease inhibitors (10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 10 μg liter−1 aprotinin, 10 μg liter−1 leupeptin, and 1 μg liter−1 pepstatin). Cells were mixed with glass beads (425 to 600 μm; acid washed; Sigma, MO) and disrupted by vortexing vigorously for 10 min at 4°C. The suspensions of broken cells were centrifuged at 500 × g for 5 min to remove the unbroken cells, and the supernatants were used for the phosphatase assays. All the extraction procedures were carried out at 4°C.
Phosphatase activity.
The phosphatase activity toward GGPP (GGPPase) was assayed as the formation of geranylgeraniol from GGPP using a modification of the method of Bansal and Vaidya (2). The assay mixture (100 μl) was comprised of citrate buffer (120 mM, pH 4.3), cell extract, and 5 μM cold GGPP spiked with [1-3H]GGPP (370 to 1,110 GBq mmol−1) and was incubated for 30 min at 37°C. The reaction was initiated by the addition of the extract and stopped with 200 μl of an alcoholic KOH solution (95% ethanol-40% KOH [1:1]). Five hundred microliters of n-hexane was added to extract the GGOH formed. The mixture was vortexed for 1 min and then centrifuged at 18,000 × g for 10 min. Three hundred microliters of the organic layer was used for radioactivity counting with an LSC-3500 liquid scintillation counter (Aloka, Tokyo, Japan). The specific GGPPase activity was calculated from a standard curve obtained using known numbers of units of acid phosphatase (Nippon Boehringer Ingelheim, Tokyo, Japan).
Production of prenyl alcohols in test tube cultures.
The production of prenyl alcohols (FOH, NOH, and GGOH) and squalene was evaluated in test tube cultures with more than three independent clones of recombinants. The precultures were grown in selective SD medium for 2 days at 30°C, followed by inoculation (1/100, vol/vol) into 2.5 ml of YM broth or 2 ml of YPD medium in test tubes equipped with stainless molten caps and cultivation at 30°C for 4 days with shaking (130 rpm). The cell optical density at 600 nm was measured after 4 days of cultivation. Prenyl alcohols were extracted from whole cells with n-pentane, and their amounts were determined by using a gas chromatography/mass spectrometry system as previously described (25). For determination of the squalene content with prenyl alcohols, cells were disrupted before extraction because squalene cannot be extracted efficiently without cell disruption. The cultures (2 ml) were transferred to 2-ml sample tubes and centrifuged at 20,000 × g for 10 min. Then, the supernatants were decanted into new test tubes, followed by extraction as described above. About 0.4 ml of acid-washed glass beads and 0.2 ml of Tris-EDTA buffer were added to the remaining cell pellets, followed by vortexing for 30 min at room temperature to disrupt the cells completely. Methanol (0.2 ml) and n-pentane (1 ml) were added to the broken cells, followed by vortexing and then collection of the organic layer. The extraction was repeated with 1 ml of pentane, and the organic layer was collected. All of the organic layers extracted from supernatants and broken cells were collected in new test tubes and dried. The squalene, FOH, and GGOH contents were determined as described above.
Jar fermentation.
Yeast precultures were prepared in Erlenmeyer flasks at 30°C with rotary shaking (120 rpm) for 30 h. The main culture medium (initial volume, 3.5 liters) in a 10-liter jar fermentor (model MSJ-U2W; Marubishi Bioengineering Co., Tokyo, Japan) was seeded with a 100-ml aliquot of the preculture. The temperature, agitation speed, flow rate of germ-free air, and pH were controlled at 33°C, 900 rpm, 1 volume of air per volume of culture per minute, and pH 5.5 with 4 N NaOH and 4 N H2SO4. A glucose solution (50%, wt/vol) was fed from 0 to 24 h, and the feed rate was increased with consumption. After 24 h, a mixed solution of glucose and ethanol (25% [wt/vol] and 50% [vol/vol], respectively) was fed at the feed rate of 5.8 g h−1. The density of the feed solution was 0.98 g ml−1. During the fermentation, duplicate aliquots of culture (2 ml each) were collected periodically to determine the amounts of prenyl alcohols without cell disruption, as mentioned above.
RESULTS
Increases in the GGPPase activities on overexpression of the phosphatase genes.
Episomal plasmids pRS436LPP1 and pRS436DPP1 were introduced in YPH499, and the GGPPase activities in the cell extracts of the resulting transformants (LPP1/YPH499 and DPP1/YPH499, respectively) were assayed. The specific GGPPase activity in the host strain (YPH499) was 1.7 U mg−1 of protein (average of two independent experiments). The specific GGPPase activities increased 2.0- and 3.1-fold with the overexpression of the LPP1 and DPP1 genes, respectively (average of two independent clones). These results indicate that the GGPPase activities increased with the overexpression of the LPP1 and DPP1 genes.
Effect of overexpression of the phosphatase genes on GGOH production.
To evaluate the effect of promotion of the GGOH production by phosphatase overexpression, the LPP1 and DPP1 genes were coexpressed with the BTS1 gene in YPH499 with plasmids pRS436LPP1, pRS436DPP1, and pRS435BTS1. The recombinants were incubated in YM broth in test tubes for 4 days, followed by determination of the GGOH content (Fig. 3). Single BTS1 overexpression led to GGOH production (0.11 mg liter−1). Overexpression of additional phosphatase genes had a positive effect, increasing GGOH production. The DPP1 gene gave a better result than the LPP1 gene while single DPP1 gene overexpression did not lead to GGOH production.
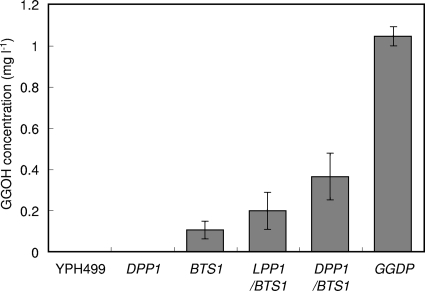
FIG. 3.
GGOH production by recombinant strains overexpressing the DPP1, LPP1, and BTS1 genes and the BTS1-DPP1 (GGDP) fusion with episomal plasmids in test tube cultures. YPH499, host strain; DPP1, pRS436DPP1/YPH499; BTS1, pRS435BTS1/YPH499; LPP1/BTS1, pRS436LPP1/pRS435BTS1/YPH499; DPP1/BTS1, pRS436DPP1/pRS435BTS1/YPH499; GGDP, pRS436GGDP/YPH499. The data represent the averages ± standard deviations for more than three independent clones.
In our previous study, the BTS1-ERG20 fusion gene exhibited a good effect, promoting GGOH production. So, in this study we fused the BTS1 gene with the DPP1 gene. Single overexpression of the BTS1-DPP1 fusion gene with plasmid pRS436GGDP further increased GGOH production (Fig. 3, GGDP). GGOH production was 2.9-fold higher with the fusion gene than with coexpression of these genes separately (DPP1/BTS1). Squalene, FOH, and NOH were not detected in any strains used for the experiment shown in Fig. 3. These results suggest that overexpression of the BTS1-DPP1 fusion gene is highly effective for promoting GGOH production.
Construction of a GGOH-producing strain with multicopy integration vectors.
Although episomal plasmids attain high average copy numbers, they suffer from low segregational stability (26). In contrast, vectors integrated into the genome are stable, but since general yeast integration vectors are integrated at only one to two copies per cell, the expression levels of the introduced genes are not so high. To increase the expression levels, multicopy integration vectors targeted to repetitive chromosomal DNA sequences such as the rDNA cluster and long terminal repeats of the Ty element, known as δ sequences, have been developed (18, 19, 30, 31). We constructed multicopy integration vectors targeted to rDNA and Ty δ loci to stably overexpress the genes that can promote GGOH production (Fig. 2B).
First, the HMG1 gene was overexpressed in order to enhance the mevalonate pathway. Multicopy integration vector pRS504HMG1 was introduced into the rDNA loci of YPH499. A significant amount of squalene (191.9 mg liter−1) was produced by the resulting transformant (HMG1/YPH499) (Fig. 4) while the host strain did not produce a detectable amount of squalene (data not shown). GGOH production was only 0.2 mg liter−1 in this strain. The BTS1-DPP1 fusion gene was then introduced into the δ sequences of strain HMG1/YPH499 with multicopy integration vector pDI626GGDP. GGOH production was increased in the resulting transformant (GGDP/HMG1/YPH499) and, in contrast, the squalene production was decreased. Finally, the BTS1-ERG20 fusion gene was introduced into the rDNA loci of strain GGDP/HMG1/YPH499 with multicopy integration vector pRS515GGF. GGOH production by the resulting transformants (GGF/GGDP/HMG1/YPH499) was further increased, and only small amounts of squalene were produced. The average GGOH and squalene production by 43 independent clones was 228.8 mg liter−1 and 6.5 mg liter−1, respectively. FOH and NOH (a positional isomer of FOH formed from FPP under acidic conditions [10, 22]) production was low in all the transformants (less than 0.2 and 0.7 mg liter−1, respectively). In this way, we constructed a haploid strain that overproduces almost only GGOH.
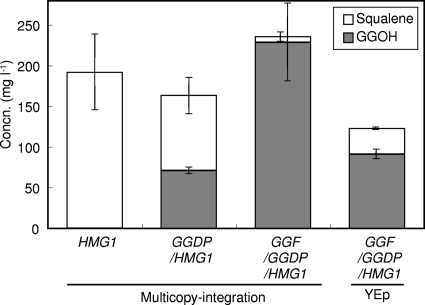
FIG. 4.
Production of squalene and GGOH by recombinant strains overexpressing the HMG1 gene and the BTS1-DPP1(GGDP) and BTS1-ERG20 (GGF) fusion genes in test tube cultures. HMG1, pRS504HMG1/YPH499; GGDP/HMG1, pDI626GGDP/pRS504HMG1/YPH499; GGF/GGDP/HMG1, pRS515GGF/pDI626GGDP/pRS504HMG1/YPH499; GGF/GGDP/HMG1(YEp), pRS434HMG1/pRS435GGF/pRS436GGDP/YPH499. The data represent the averages ± standard deviations for more than three independent clones.
Copy number estimation.
Utilizing quantitative real-time PCR technique, the copy numbers of the HMG1, BTS1-DPP1, and BTS1-ERG20 genes in the genome of strain GGF/GGDP/HMG1/YPH499 were estimated to be 1.5, 4.6, and 26.0 copies, respectively. These results confirm that the multiple vectors were integrated into the genome with the multicopy integration vectors.
Construction of a GGOH-producing strain with episomal plasmids.
In comparison, we also attempted to coexpress these three genes (HMG1, BTS1-ERG20, and BTS1-DPP1 genes) in YPH499 with three episomal plasmids, pRS434HMG1, pRS435GGF, and pRS436GGDP, respectively. After successive introduction of these three plasmids, the resulting transformants [GGF/GGDP/HMG1/YPH499(YEp)] produced 91.0 mg liter−1 of GGOH, along with 31.5 mg liter−1 of squalene (Fig. 4). The GGOH production with the episomal plasmids was less than half that with the multicopy integration vectors.
Stability of the vectors.
The mitotic stability of the multicopy integration vectors was confirmed by GGOH productivity with strain GGDP/HMG1/YPH499 in test tube cultures. After four sequential batch cultivations (with at least seven generations per culture) of strain GGDP/HMG1/YPH499 in nonselective complex medium (YPD), GGOH productivity was measured in test tube cultures. GGOH production was reduced by only 17% compared to before the serial cultivations. On the other hand, with strain GGF/GGDP/HMG1/YPH499(YEp) that harbors three episomal plasmids, GGOH productivity was almost completely lost (less than 0.8%) after the same serial cultivations. These results indicated that the multicopy integration vectors were stably retained and that, in contrast, the episomal plasmids were not stable in nonselective cultures.
Construction of a diploid prototrophic strain.
Diploids and prototrophs are industrially attractive because of the advantage of good growth in a low-cost medium. In order to complement auxotrophic markers, DNA fragments of the HIS3, LYS2, and ADE2 coding regions were prepared by PCR from S288C genomic DNA. The primer pairs of HIS3-F and HIS3-R, LYS2-F and LYS2-R, and ADE2-F and ADE2-R listed in Table 2 were used for the PCR, respectively, of HIS3, LYS2, and ADE2. The HIS3 fragment was introduced into GGF/GGDP/HMG1/YPH499, and a histidine prototroph was selected on an SD-His plate. The resulting transformant was named GGF/GGDP/HMG1/YPH499-H.
YPH500 was sequentially transformed with multicopy integration vectors pRS524HMG1, pDI626GGDP, and pRS515GGF to produce a transformant, GGF/GGDP/HMG1/YPH500. This strain produced 152.1 mg liter−1 and 2.5 mg liter−1 of GGOH and squalene, respectively, in test tube cultures. The LYS2 and ADE2 fragments were introduced into this strain, followed by selection of a lysine and adenine prototroph on an SD-Lys-Ade plate. The resulting transformant was named GGF/GGDP/HMG1/YPH500-AK. Fresh cultures of GGF/GGDP/HMG1/YPH499-H and GGF/GGDP/HMG1/YPH500-AK were mixed and mated in YPD medium overnight, followed by selection on an SD plate without adenine and complete supplement mixture. The resulting diploid prototrophic strain was named strain 5X10. Diploid construction was confirmed by microscopic analysis of the spore formation by strain 5X10 on a sporulation plate.
GGOH production with the diploid prototrophic strain.
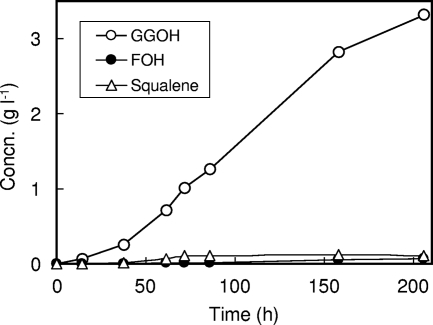
The growth rate was improved for diploid prototrophic strain 5X10 compared with haploid auxotrophic strains (data not shown). Strain 5X10 produced a larger amount of GGOH (283.1 mg liter−1) than the haploid strains and only slight amounts of FOH, NOH, and squalene on test tube cultivation. This strain was then cultivated in 10-liter jar fermentors under aerobic conditions as described in Materials and Methods. A mixed solution of glucose and ethanol was fed, and this increased GGOH production to 3.31 g liter−1 after a 206-h incubation (Fig. 5). Squalene, FOH, and NOH production amounted to 0.11, 0.07, and 0.01 g liter−1, respectively. In this way, we constructed a diploid prototrophic strain that overproduces almost only GGOH. GGOH and squalene production by the recombinant yeasts examined in this study is summarized in Table 3.
FIG. 5.
Fermentation profile of diploid prototrophic strain 5X10 in a 10-liter jar fermentor. After the glucose feeding was stopped, as described in Materials and Methods, a mixed solution of glucose and ethanol (25% [wt/vol] and 50% [vol/vol], respectively) was fed after 24 h at the feed rate of 5.8 g h−1. The data represent the averages for duplicate samples.
TABLE 3.
Summary of GGOH and squalene production by the recombinant yeasts in test tube cultures and jar fermentor
| Culture method and yeast strain | GGOH (mg liter−1 [SD])a | Squalene (mg liter−1 [SD])a |
|---|---|---|
| Test tube cultures | ||
| YPH499 | ND | ND |
| HMG1/YPH499 | 0.2 (0.2) | 191.9 (0.2) |
| GGDP/HMG1/YPH499 | 71.1 (4.1) | 91.8 (22.2) |
| GGF/GGDP/HMG1/YPH499 | 228.8 (47.6) | 6.5 (5.4) |
| GGF/GGDP/HMG1/YPH499(YEp) | 91.0 (5.6) | 31.5 (1.5) |
| GGF/GGDP/HMG1/YPH500 | 152.1 (19.4) | 2.5 (1.4) |
| 5X10 | 283.1 (21.1) | 0.08 (0.03)b |
| Jar fermentor culture | ||
| 5X10 | 3,310.0 | 106.9b |
The data are averages (standard deviations) for more than three independent clones in test tube cultures except for the 5X10 culture from the jar fermentor, for which the averages are for two samples in a 10-liter jar fermentor. ND, not detected.
Squalene concentrations might have been underestimated because the cells were extracted without cell disruption.
DISCUSSION
Isoprenoids comprise one of the most structurally diverse groups of natural products, and they have a number of different and essential functions in living cells (3, 16). They are of particular interest for microbial production because they exist at low concentrations in their host organisms and are structurally complex, with multiple chiral centers, which makes them difficult subjects for both biological extraction and chemical synthesis (6). In this study, we demonstrated the overproduction of GGOH, a dephosphorylated derivative of an isoprenoid, on metabolic engineering of the yeast S. cerevisiae. Enhancement of the mevalonate pathway and redirection of the carbon flux to GGOH through the overexpression of several key enzymes made it possible to construct a high-level GGOH-producing yeast. A phosphatase that can effectively dephosphorylate GGPP to yield GGOH proved to be one of the key enzymes. We demonstrated that the overexpression of the diacylglycerol diphosphate phosphatase (DPP1) gene enhanced GGOH production (Fig. 3). The higher GGOH production with the DPP1 gene than with the LPP1 gene may be partly accounted for by the higher GGPPase activity with the DPP1 gene. Dpp1p is predicted to be an integral membrane protein with six transmembrane-spanning domains (37). It is likely that Dpp1p can gain access to substrates that exhibit high affinity for membrane lipids such as GGPP. Indeed, it has been reported that Dpp1p preferred GGPP as a substrate to its precursors such as IPP (9). Such a substrate preference is suitable for GGOH production because the precursors of GGOH tend not to be dephosphorylated by overproduced Dpp1p before their use for GGOH synthesis.
We also found that overexpression of the artificial fusion of the BTS1 and DPP1 genes was more efficient for producing GGOH than coexpression of these genes separately (Fig. 3). It is possible that the accessibility of Bts1p to a lipid-soluble substrate (i.e., FPP) is enhanced by colocalization with Dpp1p in the membrane. We also think that the two sequential enzymatic reactions can be catalyzed more efficiently by one molecule of the bifunctional fusion protein than by the two enzymes separately. This is the same as for the BTS1-ERG20 fusion gene. Such a proximity effect and substrate channeling for fusion enzymes have been reported to increase the overall catalytic activity of the reaction in vitro (11, 15, 17, 33). We demonstrated that the use of bifunctional fusion genes was also highly effective for metabolic engineering.
Finally, we constructed a high-level GGOH-producing yeast that is available for industrial processes involving multicopy integration vectors. We confirmed that multicopy integration vectors were more stable than episomal plasmids in a nonselective complex medium after more than 25 generations. We demonstrated that coexpression of the two fusion genes (BTS1-DPP1 and BTS1-ERG20) along with the HMG1 gene resulted in an almost complete shift of the metabolic flux from squalene to GGOH production (Fig. 4 and Table 3). Then, we constructed a diploid prototrophic strain, 5X10. The GGOH productivity and growth rate of strain 5X10 were higher than values for the haploid strains. Using this diploid prototrophic strain, we have established a means of GGOH overproduction in laboratory-scale jar fermentors, as shown in Fig. 5. Gradual feeding of a mixed solution of glucose and ethanol further increased GGOH production up to 3.31 g liter−1. This corresponds to the carbon conversion efficiency of 1.65% (GGOH produced per carbon sources consumed) and a GGOH content of 70.9 mg g−1 of dry cell weight. These values were very high compared with the production of carotenoids (7) and isoprenoids (16) by recombinant yeasts, such as lycopene (7.8 mg g−1) (34), β-carotene (5.9 mg g−1) (38), amorphadiene (149 mg liter−1) (29), and artemisinic acid (1.06 g liter−1) (28). This study clearly demonstrated high-level GGOH overproduction by a metabolically engineered S. cerevisiae strain that may be available for industrial processes.
Acknowledgments
We thank Kumi Terada, Chiharu Mori, and Keiko Uemura for their technical assistance.
Footnotes
Published ahead of print on 10 July 2009.
REFERENCES
- 1.Arnezeder, C., and W. A. Hampel. 1990. Influence of growth rate on the accumulation of ergosterol in yeast cells. Biotechnol. Lett. 12:277-282. [Google Scholar]
- 2.Bansal, V. S., and S. Vaidya. 1994. Characterization of two distinct allyl pyrophosphatase activities from rat liver microsomes. Arch. Biochem. Biophys. 315:393-399. [DOI] [PubMed] [Google Scholar]
- 3.Barkovich, R., and J. C. Liao. 2001. Metabolic engineering of isoprenoids. Metab. Eng. 3:27-39. [DOI] [PubMed] [Google Scholar]
- 4.Benford, H. L., J. C. Frith, S. Auriola, J. Monkkonen, and M. J. Rogers. 1999. Farnesol and geranylgeraniol prevent activation of caspases by aminobisphosphonates: biochemical evidence for two distinct pharmacological classes of bisphosphonate drugs. Mol. Pharmacol. 56:131-140. [DOI] [PubMed] [Google Scholar]
- 5.Chambon, C., V. Ladeveze, A. Oulmouden, M. Servouse, and F. Karst. 1990. Isolation and properties of yeast mutants affected in farnesyl diphosphate synthetase. Curr. Genet. 18:41-46. [DOI] [PubMed] [Google Scholar]
- 6.Chang, M. C., and J. D. Keasling. 2006. Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2:674-681. [DOI] [PubMed] [Google Scholar]
- 7.Das, A., S. H. Yoon, S. H. Lee, J. Y. Kim, D. K. Oh, and S. W. Kim. 2007. An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl. Microbiol. Biotechnol. 77:505-512. [DOI] [PubMed] [Google Scholar]
- 8.Donald, K. A., R. Y. Hampton, and I. B. Fritz. 1997. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 63:3341-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faulkner, A., X. Chen, J. Rush, B. Horazdovsky, C. J. Waechter, G. M. Carman, and P. C. Sternweis. 1999. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphate phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 274:14831-14837. [DOI] [PubMed] [Google Scholar]
- 10.George-Nascimento, C., R. Pont-Lezica, and O. Cori. 1971. Non enzymic formation of nerolidol from farnesyl pyrophosphate in the presence of bivalent cations. Biochem. Biophys. Res. Commun. 45:119-124. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert, M., R. Bayer, A. M. Cunningham, S. DeFrees, Y. Gao, D. C. Watson, N. M. Young, and W. W. Wakarchuk. 1998. The synthesis of sialylated oligosaccharides using a CMP-Neu5Ac synthetase/sialyltransferase fusion. Nat. Biotechnol. 16:769-772. [DOI] [PubMed] [Google Scholar]
- 12.Hampton, R., D. Dimster-Denk, and J. Rine. 1996. The biology of HMG-CoA reductase: the pros of contra-regulation. Trends Biochem. Sci. 21:140-145. [PubMed] [Google Scholar]
- 13.Hyatt, J. A., G. S. Kottas, and J. Effler. 2002. Development of synthetic routes to d,l-α-tocopherol (vitamin E) from biologically produced geranylgeraniol. Org. Process Res. Dev. 6:782-787. [Google Scholar]
- 14.Jiang, Y., P. Proteau, D. Poulter, and S. Ferro-Novick. 1995. BTS1 encodes a geranylgeranyl diphosphate synthase in Saccharomyces cerevisiae. J. Biol. Chem. 270:21793-21799. [DOI] [PubMed] [Google Scholar]
- 15.Kim, G. J., D. E. Lee, and H. S. Kim. 2000. Construction and evaluation of a novel bifunctional N-carbamylase-d-hydantoinase fusion enzyme. Appl. Environ. Microbiol. 66:2133-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirby, J., and J. D. Keasling. 2008. Metabolic engineering of microorganisms for isoprenoid production. Nat. Prod. Rep. 25:656-661. [DOI] [PubMed] [Google Scholar]
- 17.Liang, P. H., and K. S. Anderson. 1998. Substrate channeling and domain-domain interactions in bifunctional thymidylate synthase-dihydrofolate reductase. Biochemistry (Mosc.) 37:12195-12205. [DOI] [PubMed] [Google Scholar]
- 18.Lopes, T. S., G. J. Hakkaart, B. L. Koerts, H. A. Raue, and R. J. Planta. 1991. Mechanism of high-copy-number integration of pMIRY-type vectors into the ribosomal DNA of Saccharomyces cerevisiae. Gene 105:83-90. [DOI] [PubMed] [Google Scholar]
- 19.Lopes, T. S., J. Klootwijk, A. E. Veenstra, P. C. van der Aar, H. van Heerikhuizen, H. A. Raue, and R. J. Planta. 1989. High-copy-number integration into the ribosomal DNA of Saccharomyces cerevisiae: a new vector for high-level expression. Gene 79:199-206. [DOI] [PubMed] [Google Scholar]
- 20.Muramatsu, M., C. Ohto, S. Obata, E. Sakuradani, and S. Shimizu. 2008. Accumulation of prenyl alcohols by terpenoid biosynthesis inhibitors in various microorganisms. Appl. Microbiol. Biotechnol. 80:589-595. [DOI] [PubMed] [Google Scholar]
- 21.Nikawa, J., and M. Kawabata. 1998. PCR- and ligation-mediated synthesis of marker cassettes with long flanking homology regions for gene disruption in Saccharomyces cerevisiae. Nucleic Acids Res. 26:860-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishino, T., N. Suzuki, and H. Katsuki. 1982. Enzymatic formation of nerolidol in cell-free extract of Rhodotorula glutinis. J. Biochem. 92:1731-1740. [DOI] [PubMed] [Google Scholar]
- 23.Ogura, K., and T. Koyama. 1998. Enzymatic aspects of isoprenoid chain elongation. Chem. Rev. 98:1263-1276. [DOI] [PubMed] [Google Scholar]
- 24.Ohizumi, H., Y. Masuda, S. Nakajo, I. Sakai, S. Ohsawa, and K. Nakaya. 1995. Geranylgeraniol is a potent inducer of apoptosis in tumor cells. J. Biochem. (Tokyo) 117:11-13. [DOI] [PubMed] [Google Scholar]
- 25.Ohto, C., M. Muramatsu, S. Obata, E. Sakuradani, and S. Shimizu. 2008. Overexpression of the gene encoding HMG-CoA reductase in Saccharomyces cerevisiae for production of prenyl alcohols. Appl. Microbiol. Biotechnol. 82:837-845. [DOI] [PubMed] [Google Scholar]
- 26.Parekh, R. N., M. R. Shaw, and K. D. Wittrup. 1996. An integrating vector for tunable, high copy, stable integration into the dispersed Ty δ sites of Saccharomyces cerevisiae. Biotechnol. Prog. 12:16-21. [DOI] [PubMed] [Google Scholar]
- 27.Polakowski, T., U. Stahl, and C. Lang. 1998. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl. Microbiol. Biotechnol. 49:66-71. [DOI] [PubMed] [Google Scholar]
- 28.Ro, D. K., M. Ouellet, E. M. Paradise, H. Burd, D. Eng, C. J. Paddon, J. D. Newman, and J. D. Keasling. 2008. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 8:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ro, D. K., E. M. Paradise, M. Ouellet, K. J. Fisher, K. L. Newman, J. M. Ndungu, K. A. Ho, R. A. Eachus, T. S. Ham, J. Kirby, M. C. Chang, S. T. Withers, Y. Shiba, R. Sarpong, and J. D. Keasling. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940-943. [DOI] [PubMed] [Google Scholar]
- 30.Sakai, A., F. Ozawa, T. Higashizaki, Y. Shimizu, and F. Hishinuma. 1991. Enhanced secretion of human nerve growth factor from Saccharomyces cerevisiae using an advanced δ-integration system. Biotechnology 9:1382-1385. [DOI] [PubMed] [Google Scholar]
- 31.Sakai, A., Y. Shimizu, and F. Hishinuma. 1990. Integration of heterologous genes into the chromosome of Saccharomyces cerevisiae using a delta sequence of yeast retrotransposon Ty. Appl. Microbiol. Biotechnol. 33:302-306. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Seo, H. S., Y. J. Koo, J. Y. Lim, J. T. Song, C. H. Kim, J. K. Kim, J. S. Lee, and Y. D. Choi. 2000. Characterization of a bifunctional enzyme fusion of trehalose-6-phosphate synthetase and trehalose-6-phosphate phosphatase of Escherichia coli. Appl. Environ. Microbiol. 66:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimada, H., K. Kondo, P. D. Fraser, Y. Miura, T. Saito, and N. Misawa. 1998. Increased carotenoid production by the food yeast Candida utilis through metabolic engineering of the isoprenoid pathway. Appl. Environ. Microbiol. 64:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, L. 2003. Detection of farnesyl diphosphate accumulation in yeast ERG9 mutants. Anal. Biochem. 317:180-185. [DOI] [PubMed] [Google Scholar]
- 36.Takeda, Y., K. Nakao, K. Nakata, A. Kawakami, H. Ida, T. Ichikawa, M. Shigeno, Y. Kajiya, K. Hamasaki, Y. Kato, and K. Eguchi. 2001. Geranylgeraniol, an intermediate product in mevalonate pathway, induces apoptotic cell death in human hepatoma cells: death receptor-independent activation of caspase-8 with down-regulation of Bcl-xL expression. Jpn. J. Cancer Res. 92:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toke, D. A., W. L. Bennett, D. A. Dillon, W. I. Wu, X. Chen, D. B. Ostrander, J. Oshiro, A. Cremesti, D. R. Voelker, A. S. Fischl, and G. M. Carman. 1998. Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding diacylglycerol pyrophosphate phosphatase. J. Biol. Chem. 273:3278-3284. [DOI] [PubMed] [Google Scholar]
- 38.Verwaal, R., J. Wang, J. P. Meijnen, H. Visser, G. Sandmann, J. A. van den Berg, and A. J. van Ooyen. 2007. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 73:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]