Abstract
The esat-6 and cfp-10 genes are essential for virulence in Mycobacterium tuberculosis. Among nontuberculous mycobacteria, we found these genes only in M. kansasii, M. szulgai, M. marinum, and M. riyadhense, with unique sequences. This adds a phylogenetic and taxonomical characteristic and may represent a virulence factor for nontuberculous mycobacteria.
The 6-kDa early secretory antigenic target (ESAT-6) and 10-kDa culture filtrate protein (CFP-10) of Mycobacterium tuberculosis are potent T-cell antigens (2, 12). The genes encoding ESAT-6 and CFP-10 are situated within “region of difference 1” (RD1) of Mycobacterium tuberculosis but are also present in some nontuberculous mycobacteria (NTM) (2, 12). Deletion of RD1 in M. tuberculosis significantly decreases its virulence in animal models (8), suggesting that genes residing in RD1 are involved in pathogenesis. In M. tuberculosis, the RD1 proteins effect translocation to the cytosol, a mechanism for survival within macrophages (15). Therefore, RD1 in NTM may also play a crucial role in virulence.
To improve our understanding of the pathogenesis of NTM disease and the possible role of RD1 in virulence, we screened a wide diversity of NTM for the presence of RD1.
From our laboratory database, we retrieved isolates of all five Mycobacterium kansasii subtypes based on 16S-23S internal transcribed spacer sequencing (n = 15), M. szulgai (n = 4), M. marinum (n = 4), M. avium (n = 2), M. conspicuum (n = 4), M. genavense (n = 1), M. bohemicum (n = 2), M. interjectum (n = 2), M. flavescens (n = 5), M. xenopi (n = 2), M. malmoense (n = 2), M. riyadhense (n = 1), and M. tuberculosis H37Rv.
The selection of these strains was based on their phylogenetic relationship with the M. tuberculosis complex in the multigene taxonomical model published by Devulder et al. (3).
To establish the presence of an RD1-like element and sequences of the esat-6 and cfp-10 genes, we used the Esa-12 (CATGACAGAGCAGCAGTG) and Esa-303 (5′-GCCCTATGCGAACATCCC-3′) primers for esat-6 and the opBR78 (5′-GTAGCCCGGGATGGCAGAGATGAAGACCGATGCC-3′) and opBR103 (5′-TCAGAAGCCCATTTGCGAGGACAGC-3′) primers for cfp-10 (1). The M. smegmatis esat-6 and cfp-10 gene sequences were extracted from the whole-genome sequence in the GenBank database (accession number CP000480).
Using these primers, we were able to demonstrate an RD1 for M. tuberculosis H37Rv and for all M. kansasii subtypes, M. szulgai, M. marinum, and M. riyadhense. The PCR was repeatedly negative for isolates of the remaining species, M. avium, M. conspicuum, M. genavense, M. bohemicum, M. interjectum, M. flavescens, M. xenopi, and M. malmoense. For these species, we performed Southern blotting and hybridized DNA membranes using the purified M. tuberculosis H37Rv and M. kansasii type I esat-6 amplicons as probes (1). None of the PCR-negative species hybridized with either probe (data not shown).
The presence of RD1, characterized by an esat-6-like gene and an cfp-10-like gene, is a phylogenetic characteristic among the NTM. It is mainly found only among slowly growing NTM species that are phylogenetically related to the M. tuberculosis complex based on the multigene taxonomical model by Devulder et al. (3) and in the more distantly related rapid grower M. smegmatis. Possibly, the presence of RD1 reflects phylogenetic relationships to the M. tuberculosis complex.
Previous authors have recorded the presence of RD1 in M. flavescens (1). We were unable to demonstrate it in four reference strains (ATCC 23008, ATCC 23033, ATCC 23035, and ATCC 23039) and a clinical isolate. Thus, with its presence of an RD1 region, M. smegmatis still stands out among the rapid growing NTM.
Gey van Pittius et al. have demonstrated the presence of the ESX-5 locus, which they assume is a product of duplication of RD1 and its secretion system (ESX-1), in most slow-growing NTM species (5). Our results seem to suggest that in many slow growers, after this duplication, the original ESX-1 either was lost or has undergone extensive mutation, barring hybridization.
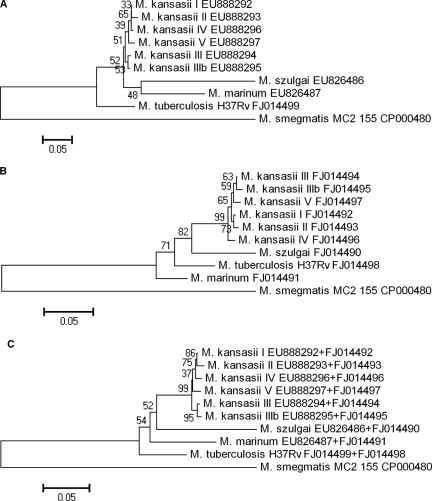
All five subtypes of M. kansasii had distinct esat-6 and cfp-10 sequences; type three was subdivided into two separate lineages based on both esat-6 and cfp-10 sequences. Among M. marinum and M. szulgai strains, no difference was noted, and the sequences obtained for M. riyadhense were unique. We aligned the esat-6 and cfp-10 gene sequences separately and concatenated, using the Clustal X software program (14). The resulting topology and tree, inferred by neighbor joining and visualized using the MEGA 4.0 software package (13), were evaluated by bootstrap analyses based on 1,000 resamplings. The resulting trees are shown in Fig. 1A (esat-6), B (cfp-10), and C (esat-6 and cfp-10 concatenated). From these trees, it is obvious that the slow-growing RD1-positive NTM have esat-6 and cfp-10 sequences much more closely related to those of M. tuberculosis than to those of M. smegmatis (Fig. 1A to C).
FIG. 1.
Phylogenetic trees based on multiple sequence alignment of esat-6 (A), cfp-10 (B), and concatenated esat-6 and cfp-10 (C) sequences. The neighbor-joining tree was created and bootstrapped 1,000× with CLUSTALX and visualized with MEGA 4.0 (13, 14). Bootstrap values are indicated at the nodes.
Thus, the presence of RD1 in these slow growers marks a genetically closely related Mycobacterium grouping, and sequencing of RD1 is a tool for (sub)species identification. Therefore, we propose that future introduction of new species phylogenetically related to the RD1-positive grouping should include an investigation of RD1 presence and gene sequences.
Deletion of RD1 lowers the virulence of M. tuberculosis complex bacteria (8, 9, 11). Although the presence of RD1 may thus be important for virulence, we were not able to detect this genomic region in well-known causative agents of disease in humans, including M. avium and M. malmoense (7, 12). In M. smegmatis, ESAT-6 and CFP-10 secretion has a role in conjugation, rather than translocation to the cytosol, by which M. tuberculosis survives within macrophages (4, 15). The remaining NTM species that harbor RD1 are phylogenetically more closely related to M. tuberculosis than to M. smegmatis (3), which is also expressed in their esat-6 and cfp-10 sequences (Fig. 1). Moreover, M. kansasii, M. szulgai, and M. marinum are considered the most pathogenic among the NTM (1, 6, 16). Therefore, RD1 may play a role in virulence of these NTM.
The RD1 sequences differed between M. kansasii type 1, an important causative agent of NTM disease, and the other types, which are less involved or not involved in human disease (1). Clinically relevant and nonrelevant M. szulgai isolates, determined using the American Thoracic Society diagnostic criteria (6, 16), shared identical sequences. The RD1 presence and gene sequences and thus the protein structure do not provide a complete explanation of the virulences of the different NTM. Presumably, host factors and pathogen factors other than RD1 presence are also important. In vitro infection experiments are necessary to clarify the role of RD1-containing slow-growing NTM.
Demonstration and characterization of RD1 in NTM have gained significance with the advent of the gamma interferon release assays (IGRAs) for the diagnosis of (latent) tuberculosis. These assays measure gamma interferon production and release by patients' T lymphocytes after incubation with the ESAT-6 and CFP-10 antigens of M. tuberculosis (10). The presence of similar antigens in NTM and thus recognition of these antigens by patients infected by these NTM theoretically lowers the specificity of the IGRAs in diagnosing latent tuberculosis.
In conclusion, an RD1 element, similar to that of M. tuberculosis, is present in M. kansasii, M. szulgai, M. marinum, and M. riyadhense. The presence of RD1 in general is a phylogenetic and taxonomical characteristic of NTM, which hints at a phylogenetic relationship with the M. tuberculosis complex. The RD1 sequence analysis enables distinction to the species or subspecies level. Future studies describing related new species should investigate RD1 presence and gene sequences. The role of RD1 as a virulence factor and the impact of RD1-containing NTM on functioning of the IGRAs should be the subjects of further studies.
Nucleotide sequence accession numbers.
All obtained sequences have been deposited in the GenBank database; accession numbers are detailed in Table 1.
TABLE 1.
GenBank accession numbers of isolates sequenced in this study
| Species | Sequence accession no.
|
|
|---|---|---|
| esat-6 | cfp-10 | |
| M. szulgai | EU826486 | FJ014490 |
| M. marinum | EU826487 | FJ014491 |
| M. riyadhense | EU552926 | EU552927 |
| M. kansasii I | EU888292 | FJ014492 |
| M. kansasii II | EU888293 | FJ014493 |
| M. kansasii III | EU888294 | FJ014494 |
| M. kansasii IIIb | EU888295 | FJ014495 |
| M. kansasii IV | EU888296 | FJ014496 |
| M. kansasii V | EU888297 | FJ014497 |
| M. tuberculosis H37Rv | FJ014499 | FJ014498 |
Footnotes
Published ahead of print on 17 July 2009.
REFERENCES
- 1.Arend, S. M., P. E. W. de Haas, E. Leyten, I. Rosenkrands, L. Rigouts, P Andersen, W. Mijs, J. T. van Dissel, and D. van Soolingen. 2005. ESAT-6 and CFP-10 in clinical versus environmental isolates of Mycobacterium kansasii. J. Infect. Dis. 1911301-1310. [DOI] [PubMed] [Google Scholar]
- 2.Berthet, F. X., P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low molecular-mass culture filtrate protein (CPF-10). Microbiology 1443195-3203. [DOI] [PubMed] [Google Scholar]
- 3.Devulder, G., M. Perouse de Montclos, and J. P. Flandrois. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55293-302. [DOI] [PubMed] [Google Scholar]
- 4.Flint, J. L., J. C. Kowalski, P. K. Karnati, and K. M. Derbyshire. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 10112598-12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gey van Pittius, N. C., S. L. Sampson, H. Lee, Y. Kim, P. D. van Helden, and R. M. Warren. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffith, D. E., T. Aksamit, B. A. Brown-Elliot, A. Catanzaro, C. Daley, F. Gordin, S. M. Holland, R. Horsburgh, G. Huitt, M. F. Iademarco, M. Iseman, K. Olivier, S. Ruoss, C. Fordham von Reyn, R. J. Wallace, Jr., and K. Winthrop. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175367-416. [DOI] [PubMed] [Google Scholar]
- 7.Hoefsloot, W., M. J. Boeree, J. van Ingen, W. C. M. de Lange, P. N. R. Dekhuijzen, and D. van Soolingen. 2008. The rising incidence and clinical relevance of Mycobacterium malmoense; a review of the literature. Int. J. Tuberc. Lung Dis. 12987-993. [PubMed] [Google Scholar]
- 8.Lewis, K. N., R. Liao, K. M. Guinn, M. J. Hickey, S. Smith, M. A. Behr, and D. R. Sherman. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis. 187117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mostowy, S., D. Cousins, and M. A. Behr. 2004. Genomic interrogation of the dassie bacillus reveals it as a unique RD1 mutant within the Mycobacterium tuberculosis complex. J. Bacteriol. 186104-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai, M., L. W. Riley, and J. M. Colford, Jr. 2004. Interferon-γ assays in the immunodiagnosis of tuberculosis: a systematic review. Lancet Infect. Dis. 4761-776. [DOI] [PubMed] [Google Scholar]
- 11.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46709-717. [DOI] [PubMed] [Google Scholar]
- 12.Sorensen, A. L., S. Nagai, G. Houen, P. Andersen, and A. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 631710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 14.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Wel, N., D. Hava, D. Houben, D. Fluitsma, M. van Zon, J. Pierson, M. Brenner, and P. J. Peters. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 1291287-1298. [DOI] [PubMed] [Google Scholar]
- 16.van Ingen, J., M. J. Boeree, W. C. M. de Lange, P. E. W. de Haas, P. N. R. Dekhuijzen, and D. van Soolingen. 2008. Clinical relevance of Mycobacterium szulgai in the Netherlands. Clin. Infect. Dis. 461200-1205. [DOI] [PubMed] [Google Scholar]