Abstract
Interspecies hydrogen transfer between organisms producing and consuming hydrogen promotes the decomposition of organic matter in most anoxic environments. Although syntrophic coupling between hydrogen producers and consumers is a major feature of the carbon cycle, mechanisms for energy recovery at the extremely low free energies of reactions typical of these anaerobic communities have not been established. In this study, comparative transcriptional analysis of a model sulfate-reducing microbe, Desulfovibrio vulgaris Hildenborough, suggested the use of alternative electron transfer systems dependent on growth modality. During syntrophic growth on lactate with a hydrogenotrophic methanogen, numerous genes involved in electron transfer and energy generation were upregulated in D. vulgaris compared with their expression in sulfate-limited monocultures. In particular, genes coding for the putative membrane-bound Coo hydrogenase, two periplasmic hydrogenases (Hyd and Hyn), and the well-characterized high-molecular-weight cytochrome (Hmc) were among the most highly expressed and upregulated genes. Additionally, a predicted operon containing genes involved in lactate transport and oxidation exhibited upregulation, further suggesting an alternative pathway for electrons derived from lactate oxidation during syntrophic growth. Mutations in a subset of genes coding for Coo, Hmc, Hyd, and Hyn impaired or severely limited syntrophic growth but had little effect on growth via sulfate respiration. These results demonstrate that syntrophic growth and sulfate respiration use largely independent energy generation pathways and imply that to understand microbial processes that sustain nutrient cycling, lifestyles not captured in pure culture must be considered.
Nutrient cycling on earth is determined primarily by cooperative interactions among microorganisms. The sharing of available energy within communities is particularly important in anaerobic systems, where limited energy is divided among highly specialized and metabolically interdependent populations (36, 37, 39). In the absence of exogenous electron acceptors such as sulfate and nitrate, the mineralization of organic matter in anoxic environments yields primarily carbon dioxide and methane, and this process is controlled by the synergistic activities of multiple anaerobic microbial populations. To better understand the metabolic basis and ecological significance of these syntrophic associations, we constructed an archetypical “community of two” by pairing Desulfovibrio vulgaris Hildenborough with a hydrogenotrophic methanogen, Methanococcus maripaludis strain S2.
D. vulgaris is a representative sulfate-reducing microorganism that couples the oxidation of characteristic substrates, such as H2, lactate, or ethanol, with the reduction of sulfate to sulfide (for a review, see reference 32). In the absence of sulfate, D. vulgaris and sulfate-reducing microorganisms in general ferment organic acids and alcohols, producing hydrogen, acetate, and carbon dioxide, by forming syntrophic associations with hydrogen-consuming populations (3, 23, 40). These alternative lifestyles might be sustained by distinct metabolic systems, possibly reflected in part by the large number of hydrogenases and electron transfer complexes described in previous biochemical studies and more recently revealed in the D. vulgaris Hildenborough genome sequence (14, 32). Thus, even though sulfate respiration and syntrophic growth produce the same oxidized end products (acetate and carbon dioxide), there are probably mechanistic differences in the electron transfer pathways. These differences were evaluated by comparing whole-genome transcriptional profiles of D. vulgaris Hildenborough grown continuously on lactate under two culture conditions: syntrophic cocultures (lacking sulfate) and sulfate-limited monocultures. Complementary mutant studies showed that of the genes highly upregulated during syntrophic growth, at least two genes (coding for the Coo hydrogenase and the high-molecular-weight cytochrome complex) were required for efficient syntrophic growth but not for sulfate respiration.
MATERIALS AND METHODS
Strains.
Transcriptional analyses were performed using D. vulgaris Hildenborough and M. maripaludis S2. Additionally, four mutant strains (three strains described in previous investigations and one strain described here) of D. vulgaris were used during phenotypic growth comparisons. Details for all six strains are shown in Table 1.
TABLE 1.
Strains used in this investigation
| Strain | Mutant | Gene(s) | Description | Source or reference |
|---|---|---|---|---|
| Desulfovibrio vulgaris strains | ||||
| Hildenborough (= ATCC 29579) | NAa | NA | Wild-type strain | ATCC |
| JW3040 | ΔcooL | DVU2288 | Transposon interruption of third gene for CO-induced hydrogenase | This study |
| H801 | Δhmc | DVU0532 to DVU0535 | hmcBCDE deletion mutant | 6 |
| Hyd100 | Δhyd | DVU1769 and DVU1770 | hydAB deletion mutant | 31 |
| NiFe100 | Δhyn | DVU1921 and DVU1922 | hynAB-1 deletion mutant | 11 |
| Methanococcus maripaludis S2 | NA | NA | Wild-type strain | 45a |
NA, not applicable.
Biomass production.
Three biological replicates of cocultures and sulfate-limited D. vulgaris monocultures were grown in a chemostat in coculture medium (CCM) containing 30 mM sodium dl-lactate (cocultures and monocultures) and 10 mM Na2SO4 (monocultures only). CCM also contained a basal salt solution containing (per liter) 2.17 g NaCl, 5.5 g MgCl2·6H2O, 0.14 g CaCl2·2H2O, 0.5 g NH4Cl, and 0.335 g KCl. The medium was buffered using 1.1 mM K2HPO4 and 30 mM NaHCO3 with 1 ml of nonchelated trace elements (46) and 1 ml of a vitamin solution amended with 2.0 g/liter choline chloride (2) added as growth supplements. l-Cysteine·HCl (1 mM) and sulfide (1 mM Na2S·9H2O) were added as reducing agents. Resazurin (1 mg/liter) was added as a redox indicator. Stock solutions of K2HPO4 (1 M), NaHCO3 (6.0 M), l-cysteine·HCl (1 M), Na2S·9H2O (1 M), and the nonchelated trace element and vitamin mixtures were prepared under anoxic conditions. The medium was made by adding salts, sodium dl-lactate, resazurin, and Na2SO4 (if applicable) to water and then autoclaving. After sterilization, the chemostat or medium reservoir bottle was connected to an N2-CO2 (90:10) source and allowed to cool to room temperature before the remaining components were added.
One milliliter of a glycerol stock of a previously grown coculture or monoculture was used to inoculate 100 ml of CCM (amended with sulfate for monocultures) in a 200-ml serum vial. Cultures were incubated in the dark at 37°C with shaking at 250 rpm. When the cultures reached an optical density at 600 nm (OD600) of 0.27 ± 0.01, they were transferred to a 3-liter FairMenTec chemostat (Wald, Switzerland) containing 2 liters of CCM (amended with sulfate for monocultures). Following inoculation, the chemostat was run in batch mode at 37°C with stirring at 250 rpm. The pH was maintained at 7.0 to 7.2 by using bicarbonate buffer and small automated additions of 0.1 M NaOH or HCl as needed. An N2-CO2 (90:10) gas mixture was flushed through a sterile cotton plug before it entered the headspace of the reactor, and the flow rate was maintained at 0.20 ml/min using an Alicat Scientific mass controller (MC-20SCCM-D; Alicat Scientific, Tucson, AZ). The headspace concentrations of CH4, CO2, H2, H2S, O2, and N2 were monitored at 30-min intervals using a Hiden Analytical QIC-20 mass spectrometer (Hiden Analytical, Warrington, United Kingdom). Lactate, acetate, ethanol, glycerol, and formate were measured enzymatically as previously described (40). Continuous-culture operation was initiated after the OD600 reached approximately 0.27. A dilution rate of 0.039 h−1 was maintained, and biomass was harvested when the variance in the OD600 was less than 10% for three retention periods. Samples were taken regularly for direct cell counting and protein measurement. Desulfovibrio/Methanococcus cell ratios were determined using counts of 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells. Total protein levels were determined using the Coomassie Plus assay (Pierce, Rockford, IL). Cells were harvested using an ice-chilled sterile stainless steel tube connected to the chemostat medium exhaust line. Culture fluid was transferred to Falcon tubes (50 ml) that had been stored in an anoxic chamber and prechilled on ice. The tubes were centrifuged for 15 min at 3,220 × g at 4°C. After centrifugation, the supernatant was poured off, and the tubes were immediately frozen at −80°C.
Transcriptional analysis.
Whole-genome microarrays containing 3,482 of the 3,531 protein-encoding sequences of D. vulgaris Hildenborough were synthesized as previously described (4) on UltraGAPS glass slides (Corning Life Sciences, Corning, NY) using a BioRobotics Microgrid II microarrayer (Genomic Solutions, Ann Arbor, MI). Each slide contained duplicate spots for each protein-encoding sequence, and each biological replicate was hybridized to at least three slides. Thus, each log2 expression level described here was obtained using triplicate biological replicates, for each of which there were at least six technical replicates (duplicate on-chip technical replicates and at least three microarray slide replicates).
RNA isolation, quantification, and transcription were performed as previously described (4). Briefly, total cellular RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) and was purified using an RNeasy mini kit (Qiagen, Valencia, CA) with on-column DNase digestion using an RNase-free DNase set (Qiagen, Valencia, CA). The cDNA probes were generated from 10 μg of purified total RNA using reverse transcriptase and then labeled (43). Random hexamers (Invitrogen, Carlsbad, CA) were used for priming, and the fluorophore Cy5-dUT (Amersham Biosciences, Piscataway, NJ) was used for labeling. After labeling, RNA was removed by NaOH treatment, and cDNA was immediately purified using a Qiagen PCR mini kit. Genomic DNA was extracted from cell pellets using a Qbiogene FastDNA spin kit for soil (Mp Biomedicals, Solon, OH). Extracted genomic DNA was labeled with the fluorophore Cy3-dUTP (Amersham Biosciences, Piscataway, NJ). Labeling efficiencies were routinely monitored by measuring the absorbance at 260 nm (for DNA concentrations), 550 nm (for Cy3), or 650 nm (for Cy5).
Cy3-dUTP-labeled genomic DNA of D. vulgaris was aliquoted for triplicate arrays and cohybridized with Cy5-labeled cDNA (41, 47). Cohybridization was performed using dried probes mixed and resuspended in 35 to 40 μl of a hybridization solution containing 50% (vol/vol) formamide, 5× saline sodium citrate (SSC) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0), 0.1% (wt/vol) sodium dodecyl sulfate (SDS), and 0.1 mg/ml herring sperm DNA (Invitrogen, Carlsbad, CA). The hybridization solution was incubated at 95 to 98°C for 5 min, centrifuged briefly, incubated at 50°C, and applied to microarray slides. Hybridization was carried out in hybridization chambers (Corning Life Sciences, Corning, NY) at 45°C overnight (16 to 20 h). At each end of the microarray slide, 10 μl of a 3× SSC solution was added to maintain proper humidity and probe hydration. Slides were washed twice in a solution containing 2× SSC and 0.1% (wt/vol) SDS at 42°C for 5 min, twice in a solution containing 0.1× SSC and 0.1% (wt/vol) SDS at room temperature for 10 min, and twice in 0.1× SSC at room temperature for 1 min. After drying under a stream of N2, the slides were scanned for Cy5 and Cy3 fluorophores using the ScanArray Express microarray analysis system (Perkin Elmner, Waltham, MA). The fluorescence intensities of each spot were determined using 16-bit TIFF scanned images and were quantified with the ImaGene software (v. 6.0; Biodiscovery, Marina Del Rey, CA). Any spot that had fewer than 75% pixels or whose value was more than 3 standard deviations above the local background value in both channels was rejected (10).
For each array, the signal intensity, spot quality, and background intensity of each spot were quantified with the ImaGene software (v. 6.0; Biodiscovery, Marina Del Rey, CA). Computational analyses to determine the expression ratios, log ratios, Z scores, and operon-based estimates of local accuracy were done as previously described (4, 25). Briefly, the log2 expression was normalized globally by calculating the net signal for each spot. This was done by subtracting the background value and adding a pseudosignal of 100, thereby guaranteeing a positive value. If the resulting net signal was less than 50, a value of 550 was used. Following this, the expression levels for each spot were calculated from the ratio of mRNA to genomic DNA (Cy3 channel to Cy5 channel). Expression levels for each replicate were normalized so that the total expression levels for the spots present were identical. Mean expression levels and standard deviations of each spot were estimated, which required an n of >1, where n is the number of scorable replicates. To estimate the difference in gene expression between the control and treatment conditions, normalized log ratios were calculated. Each log ratio was calculated as follows: log2 for coculture − log2 for monoculture. The log ratio was normalized using locally weighted scatter plot smoothing (LOWESS) for the difference versus the sum of the log expression levels (7). Since sector-based artifacts were detected, each log ratio was further normalized by subtracting the median for all spots within each sector. The final normalized log ratio (log2R) was calculated from the average ratio of spots for each gene. The significance of the normalized log ratio was assessed using a Z score calculated as follows (where 0.25 is a pseudovariance term):
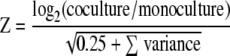 |
Z scores were determined using operon-based estimates of local accuracy as a guide, where each point represented a group of 100 predicted significant changers with similar Z scores. The estimated accuracy of each changer group was derived by inspecting other genes in the same operon as the changers. For random changers, the transcripts for 50% of the genes should have been regulated in the same direction, while for perfect changers 100% of the genes should have been regulated in the same direction. Members of the operons without a consistent signal across replicates (Z < 0.25) were excluded. The operon-based estimates of local accuracy calculated for this experiment suggested that absolute Z scores greater than 1.0 indicated statistically significant up- or downregulation (see Fig. S1 in the supplemental material).
Mutant construction.
Generation of the hmc, hyd, and hyn deletion mutants has been described previously (6, 11, 31). The cooL transposon mutant was generated by conjugation between D. vulgaris and Escherichia coli BW20767(pRL27) (19). The conjugation protocol used was a modified protocol of Fu and Voordouw (9). Briefly, cultures of D. vulgaris were grown to mid-exponential phase and combined at a 3:1 or 6:1 ratio with the E. coli donor grown to early exponential phase in LC medium (1.0% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl). Mating mixtures were concentrated by centrifugation. The concentrated cells were placed on filter disks (pore diameter, 0.22 μm; GSWP; Millipore, Billerica, MA), and the disks were placed on the surface of solidified LS4 medium (LS4D supplemented with 1% [wt/vol] yeast extract) and incubated for 16 h at 34°C (25). The cells were then washed from the membrane with 2 ml LS4 medium. After 6 h of incubation, antibiotic G418 (400 μg/ml) was added to select for the transposon mutants and nalidixic acid (200 μg/ml) was added to select against the E. coli donor. Cells were then spread onto LS4 agar (100 to 500 μl/plate) containing both antibiotics and incubated in an anaerobic growth chamber at 34°C for at least 4 days for colony growth.
The chromosomal localization of the transposon insertions was determined by sequencing DNA after semirandom PCR amplification using a variation of a previously described protocol (5). One microliter of a 50-μl boiled single-colony suspension in distilled H2O was used as the template DNA in a 20-μl PCR mixture containing primer tpnRL17-1 (5′-AAC AAG CCA GGG ATG TAA CG-3′) and either primer CEKG 2A (5′-GGC CAC GCG TCG ACT AGT AC[N]10 AGA G-3′), primer CEKG 2B (5′-GGC CAC GCG TCG ACT AGT AC[N]10 ACG CC-3′), or primer CEKG 2C (5′-GGC CAC GCG TCG ACT AGT AC[N]10 GAT AT-3′). One microliter of a 1:5 dilution of this reaction mixture was used as the template DNA for a second PCR performed with primers tpnRL17-2 (5′-AGC CCT TAG AGC CTC TCA AAG CAA-3′) and CEKG 4 (5′-GGC CAC GCG TCG ACT AGT AC-3′). The thermocycler conditions used have been described previously (5). Samples that produced distinct PCR products on an agarose gel after the second reaction were cleaned with a PCR purification kit (Qiagen, Valencia, CA) and sequenced by using primer tpnRL17-1. The chromosomal locations of the insertions were identified by BLAST analysis of the sequences adjacent to the transposon.
Phenotypic growth analyses.
All phenotypic growth assays were carried out in 17-ml Hungate tubes equipped with rubber stoppers and screw tops. Cultures were incubated at 37°C in the dark with shaking at 300 rpm. Each tube contained 8 ml of CCM amended with 30 mM of an electron donor (lactate or pyruvate) and 30 mM sulfate (if applicable). The headspace contained N2-CO2 (80:20) at an overpressure of 180 × 102 Pa. Dilutions to 10−8 were prepared using 1-ml glycerol stocks of each Desulfovibrio mutant and Methanococcus strain. Desulfovibrio cultures were grown in CCM amended with 30 mM sulfate. Methanococcus cultures were grown in CCM lacking lactate and amended with 5 mM acetate with a headspace that contained H2-CO2 (80:20) at an overpressure of 250 × 102 Pa. Cocultures were established by combining 1 ml of the highest dilution of an exponentially growing Desulfovibrio culture and 0.5 ml of the highest dilution of an exponentially growing Methanococcus culture. Cocultures were transferred (1%, vol/vol) three times to ensure dilution of any residual sulfate, acetate, or H2 before inoculation of triplicate tubes for growth experiments. The growth in tubes was monitored using OD600; uninoculated medium was used as a blank for reference. Biomass concentrations were estimated using the following previously published conversion values: 1.0 OD600 unit = 0.385 g (dry weight) coculture biomass/liter (40) and 1.0 OD600 unit = 0.309 g (dry weight) monoculture biomass/liter (45). Growth yields were estimated using the maximum OD600, which corresponded to complete consumption of the 30 mM electron donor present. The standard deviations of triplicate samples were also determined. Ratios of the growth yield with lactate to the growth yield with pyruvate, of the growth yield with lactate in a coculture to the growth yield with lactate in a monoculture, and of the growth yield with pyruvate in a coculture to the growth yield with pyruvate in a monoculture were calculated using these estimated yields. The estimated yields and ratios were compared with previously published values for D. vulgaris grown in monoculture and in coculture with Methanosarcina barkeri (44).
Gene expression database accession numbers.
Gene expression data are available at Microbes Online (www.microbesonline.org) and under GEO reference no. GSE17831, GSM444833, and GPL9107.
RESULTS
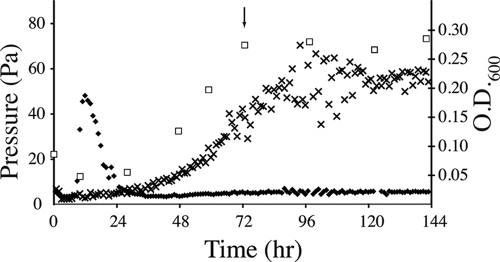
Continuous cultures of D. vulgaris and M. maripaludis syntrophically grown on 30 mM lactate (without sulfate) were maintained using a dilution rate of 0.039 h−1 (Fig. 1). The steady-state concentrations of lactate ranged from 3 to 5 mM during continuous operation, indicating that there was nearly complete utilization of the 30 mM lactate added. Acetate (24 to 27 mM) was the principal by-product produced, although small quantities of ethanol (0.02 to 0.1 mM) were detected throughout batch and continuous culture. Glycerol and formate, which are other potential by-products of lactate fermentation, were not detected (limit of detection, 0.1 mM). Methane production began within 12 h of inoculation and remained relatively stable during steady-state growth, although some fluctuations were routinely observed. These fluctuations occurred immediately after initiation of continuous-culture operation and were likely a result of pressure differentials in the sampling inlet, not biological variation. The gas concentrations shown in Fig. 1 are lower than those for the predicted stoichiometry of lactate oxidation, reflecting loss as dissolved methane in the liquid effluent and in the headspace due to N2-CO2 gas flow during operation of the reactor. The methane values were used primarily to assess steady-state operation. A Desulfovibrio-to-Methanococcus cell ratio of 4:1 was maintained during steady-state growth.
FIG. 1.
Representative biomass (□), hydrogen (⧫), and methane (×) profiles for a single biological coculture replicate. The continuous culture was started at 72 h (arrow).
Whole-genome transcriptional analysis revealed highly divergent profiles for Desulfovibrio growing in a coculture and Desulfovibrio growing in a sulfate-limited monoculture (see Tables S1 and S2 in the supplemental material). Syntrophically grown D. vulgaris had 169 upregulated open reading frames (ORFs) and 254 downregulated ORFs compared with sulfate-limited monocultures grown with the same generation time. ORFs were considered to be statistically significantly up- or downregulated if the absolute value of the Z score was greater than 1.0, as determined using operon-based estimates of local accuracy (see Fig. S1 in the supplemental material). Clustering into orthologous groups showed that the ORFs associated with energy production and conservation were the most highly upregulated, both in terms of proportion and in terms of number (20% and 42, respectively) (see Fig. S2 in the supplemental material). The largest numbers of downregulated genes were in the signal transduction group (46 of 273 genes) and inorganic ion transport and metabolism group (23 of 105 genes). Every cluster of orthologous groups contained at least one statistically significant changer, based on an absolute Z score of >1.
Among the largest increases in expression were those in operons coding for three multisubunit transmembrane proteins associated with electron transfer reactions: the high-molecular-weight cytochrome (Hmc; DVU0531 to DVU0536; log2R = 1.5 to 2.2), a cytoplasmic hydrogenase (Coo; DVU2286 to DVU2293; log2R = 1.1 to 1.7), and a putative heterodisulfide reductase (Hdr; DVU2399 to DVU2405; log2R = 1.0 to 1.6). Significant increases in expression were also observed for genes coding for a transmembrane three-subunit molybdopterin oxidoreductase (DVU0692 to DVU0694; log2R = 1.0), two periplasmic hydrogenases (hydAB [DVU1769 and DVU1770] and hynAB-1 [DVU1921 and DVU1922]; log2R = 1.0 to 2.2), an alcohol dehydrogenase (Adh; DVU2405; log2R = 3.0), and an aldehyde-ferredoxin oxidoreductase (Aor; DVU1179; log2R = 1.9). Only two genes known to be associated with electron transfer reactions were significantly downregulated, a flavodoxin gene (DVU2680; log2R = −5.2) and an adjacent hypothetical gene (DVU2681).
Syntrophy was also associated with changes in the transcription of genes in a predicted operon (DVU3024 to DVU3033) coding for lactate uptake and oxidation. The enzymes in this pathway are predicted to produce acetate, CO2, ATP, and reduced electron carriers and include lactate permease (DVU3026), a putative lactate dehydrogenase related to the membrane-bound glycolate oxidase of E. coli (DVU3027 and DVU3028), a monomeric pyruvate:ferredoxin oxidoreductase (DVU3025), phosphate acetyltransferase (DVU3029), and acetate kinase (DVU3030). The lactate permease and pyruvate oxidase genes in this operon-like arrangement were clearly upregulated (log2R = 1.2 and log2R = 2.0, respectively) during syntrophic growth, as was a second lactate permease gene (DVU2285; log2R = 1.4) located upstream and directly adjacent to the gene for the Coo hydrogenase. The genes coding for the lactate permease (DVU3026) and a contiguous lactate dehydrogenase (DVU3027 and DVU 3028) are conserved in another lactate-oxidizing syntroph (Pelotomaculum thermopropionicum) in an operon-like arrangement with the same gene order as that in D. vulgaris Hildenborough (18).
Despite trace concentrations of sulfate in the growth medium, key genes for sulfate respiration (genes encoding ATP sulfurylase, adenylyl-sulfate reductase, dissimilatory sulfite reductase, pyrophosphatase, and thiosulfate reductase) were also upregulated during syntrophic growth, consistent with previous observations of constitutive expression (13, 49). However, none of the sulfate permease genes were upregulated, and one (DVU0053) was significantly downregulated.
Although most differentially expressed genes (ca. 400 genes) have no assigned function (lipoproteins, hypothetical proteins, and conserved hypothetical proteins), some of the more highly upregulated genes have homologs (or possible orthologs) in the genomes of characterized bacterial syntrophs. For example, comparative analysis of one upregulated hypothetical gene cluster (DVU2648 to DVU2655) found no informative BLAST matches except for DVU2655 encoding a putative d-alanyl-d-alanine carboxypeptidase (dacA). D. vulgaris DacA shares the highest levels of similarity with proteins in Syntrophobacter fumaroxidans and Syntrophus aciditrophicus (1). Members of another cluster of upregulated and highly expressed ORFs with unknown functions (DVU0144 to DVU0150) share high levels of similarity with genes found in S. fumaroxidans (SFUM0625 to SFUM0629).
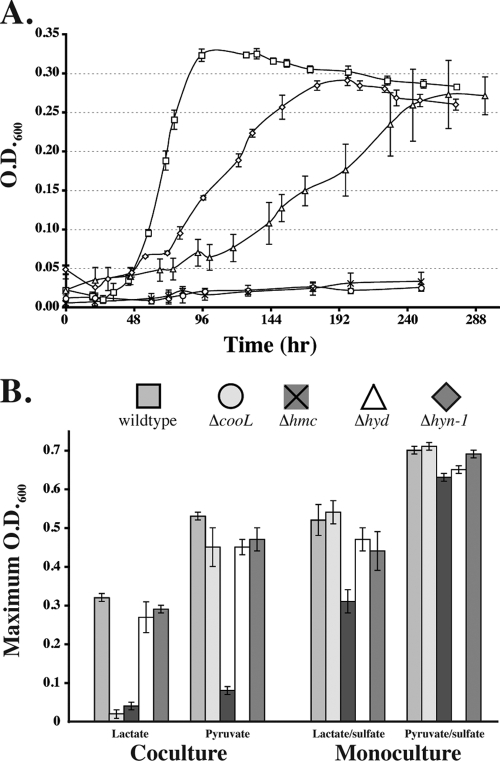
Complementary analyses of D. vulgaris mutants with deletions or disruptions in the coo, hmc, hyd, and hyn operons (all upregulated during growth in coculture) provided direct evidence for functions in syntrophy. The mutations in these mutants affected almost exclusively the capacity for syntrophic growth, either greatly inhibiting (ΔcooL and Δhmc) or slowing and slightly inhibiting (Δhyd and Δhyn-1) growth in coculture (Fig. 2 and Table 2). Notably, among these mutants, only the Δhmc strain showed slightly impaired respiratory growth (approximately 60% of the maximum cell density compared with the wild type). The ΔcooL and Δhmc mutants were capable of only sparse growth in cocultures on lactate, and the maximum cell density was approximately 10% that of the wild type. However, when the mutants were grown in cocultures on pyruvate, only the Δhmc mutant was impaired (the cell density was <15% of the maximum cell density of the wild type). Cocultures established with the Δhyd and Δhyn-1 mutants achieved cell densities comparable to those of the wild type on both lactate and pyruvate, but the growth rates were reduced. Recovery of Methanococcus from all cocultures by addition of H2 and acetate demonstrated that there was an active hydrogenotrophic population, confirming the attribution of the observed growth defects to mutations in Desulfovibrio.
FIG. 2.
(A) Growth curves for wild-type and mutant D. vulgaris cocultures on lactate. The error bars indicate standard deviations of triplicate cultures. (B) Maximum cell densities (as determined by OD600) of wild-type and mutant D. vulgaris cocultures and monocultures. The maximum cell densities are indicated by the highest OD600 values observed after exponential growth. Symbols indicate values on growth curves in panel A; shading indicates cell densities in panel B.
TABLE 2.
Estimated growth yields for wild-type cocultures and monocultures grown on lactate or pyruvate (30 mM)
| Strain(s) | Coculture
|
Monoculture
|
Coculture yield/monoculture yield ratio for:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Estimated growth yield (g [dry wt] cells/mol substrate) on lactate | Estimated growth yield (g [dry wt] cells/mol substrate) on pyruvate | Lactate yield/pyruvate yield ratio | Estimated growth yield (g [dry wt] cells/mol substrate) on lactate | Estimated growth yield (g [dry wt] cells/mol substrate) on pyruvate | Lactate yield/pyruvate yield ratio | Lactate | Pyruvate | |
| Wild type | 4.2 ± 0.1 | 6.8 ± 0.3 | 0.61 ± 0.05 | 5.0 ± 0.4 | 7.2 ± 0.1 | 0.69 ± 0.08 | 0.83 ± 0.08 | 0.94 ± 0.05 |
| Coculture from Traore et al. | 5.3 ± 0.9 | 6.3 ± 0.9 | 0.84 ± 0.22 | 6.7 ± 1.2 | 10.1 ± 1.7 | 0.66 ± 0.25 | 0.79 ± 0.25 | 0.62 ± 0.22 |
a Monocultures were grown with 30 mM sulfate. The coculture obtained from Traore et al. (44) contained D. vulgaris and M. barkeri. All lactate yield/pyruvate yield, lactate coculture yield/lactate monoculture yield, and pyruvate coculture yield/pyruvate monoculture yield ratios were calculated using the growth yields indicated. The values are means ± standard deviations of triplicate samples.
DISCUSSION
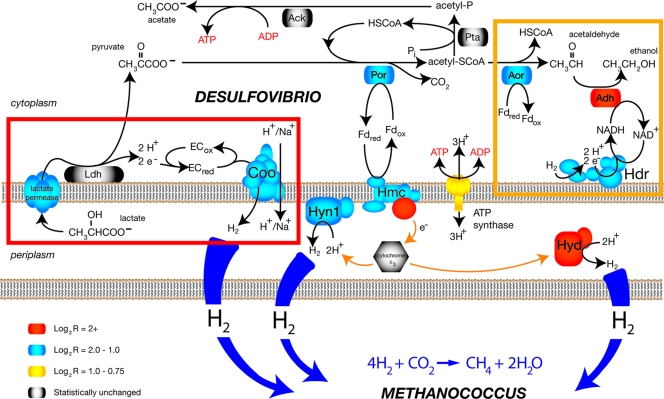
A conceptual model for electron transfer during syntrophic growth that captures the transcription and mutant data is shown in Fig. 3, and this model provides a framework for the following discussion of the electron transfer reactions and energetics of syntrophic growth. Lactate is transported from the periplasm via a dedicated lactate permease (DVU3026) and oxidized to pyruvate by a putative lactate dehydrogenase (DVU3027 and DVU3028), which likely functions primarily during syntrophic growth. The extracted electrons reduce an unknown electron carrier and are shuttled to the Coo hydrogenase, which subsequently forms H2 while concomitantly translocating protons (or sodium) across the cytoplasmic membrane. Pyruvate is oxidized by the pyruvate oxidoreductase, generating reduced ferredoxin. The membrane-associated Hmc complex then couples the oxidation of reduced ferredoxin to the reduction of a periplasmic cytochrome and/or hydrogenases (Hyn-1 and Hyd), yielding hydrogen as a final product.
FIG. 3.
Proposed metabolic model for syntrophic growth of D. vulgaris Hildenborough. Colors indicate transcriptional changes in individual genes during coculture growth compared with a sulfate-limited monoculture. The lactate permease is encoded by DVU3026. Abbreviations: Ldh, lactate dehydrogenase (likely DVU3027); ECred and ECox, reduced and oxidized unknown electron carrier interacting with Ldh, respectively; Por, pyruvate:ferredoxin oxidoreductase (DVU3025); Pta, phosphate acetyltransferase (DVU3029); Ack, acetate kinase (DVU3030); Aor, aldehyde:ferredoxin oxidoreductase (DVU1179); Adh, alcohol dehydrogenase (DVU2405); Hdr, putative heterodisulfide reductase (DVU2399 to DVU2404); Fdred and Fdox, reduced and oxidized ferredoxin, respectively; Coo, cytoplasmic hydrogenase (DVU2286 to DVU2293); Hmc, high-molecular-weight cytochrome complex (DVU0531 to DVU0536); Hyn1, [Ni-Fe] hydrogenase isozyme 1 (DVU1921 and DVU1922); Hyd, [Fe] hydrogenase (DVU1769 and DVU1770); CoA, coenzyme A. The red box indicates unique lactate oxidation enzymes that function during syntrophic growth. The orange box indicates the proposed hypothetical pathway for ethanol production (via hydrogen consumption).
The standard free energy yields for syntrophic growth on either lactate or pyruvate are well above the generally accepted minimum energy needed to support the two populations (36), but these yields are lower for growth on lactate than for growth on pyruvate (equations 4 and 5).
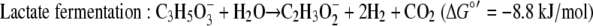 |
(1) |
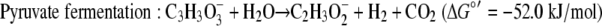 |
(2) |
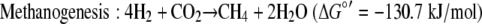 |
(3) |
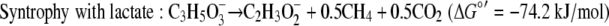 |
(4) |
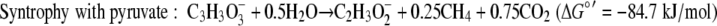 |
(5) |
Using the concentrations observed during steady state (4 mM lactate, 26 mM acetate, 2.5 × 10−5 atm H2, 0.05 atm CO2, 0.0006 atm CH4; temperature, 310 K), the free energy yields for lactate fermentation (−67.3 kJ/mol) (equation 1) and syntrophic growth on lactate (−82.8 kJ/mol) (equation 4) are more favorable. The lower free energy available for growth on lactate (equation 4) is determined primarily by the energy cost of the two-electron oxidation of lactate to pyruvate and hydrogen (equation 8).
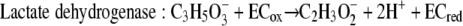 |
(6) |
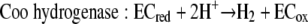 |
(7) |
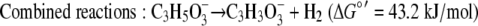 |
(8) |
where ECox is the oxidized electron carrier and ECred is the reduced electron carrier. Reverse electron flow is thought to be necessary to sustain lactate oxidation to pyruvate during respiratory growth (42). This cost is also reflected by the lower biomass yield on lactate than on pyruvate for either growth modality (Table 2), as previously observed for D. vulgaris Hildenborough paired with a different methanogen (44). Since syntrophic growth on lactate provides considerably less energy than the energy that is available through respiration, we anticipated mechanistic differences in electron transfer reactions governing the initial two-electron oxidation of lactate.
These mechanistic differences were further suggested by the global upregulation of genes associated with energy conservation and electron transfer during syntrophic growth. Upregulation of genes in a predicted operon coding for lactate uptake and oxidation suggested that the immediate fates of electrons derived from lactate oxidation differ for syntrophic growth and respiratory growth. Notably, the lactate dehydrogenase is homologous to a membrane-bound glycolate oxidase in E. coli that is directly coupled to the electron transport chain (20, 26, 34). Additionally, the coo genes are upregulated and encode a protein homologous to those found in other Bacteria and Archaea that function as proton (or sodium)-translocating hydrogenases (8, 16, 22, 35), strongly suggesting that they have a similar electrogenic role in Desulfovibrio. Another highly upregulated transmembrane protein (Hmc) likely shuttles electrons from the cytoplasm to and from soluble periplasmic carriers, such as cytochrome c3 (28, 33), thus providing a possible link between cytoplasmic oxidation and periplasmic hydrogenases. There is no evidence that Hmc has a function in direct proton translocation.
Strains with mutations in a subset of the upregulated genes were used to confirm that there was direct involvement in syntrophy. The growth phenotype of the ΔcooL mutant is of particular significance for the proposed mechanism of syntrophic growth. This mutant affected only the lactate-grown coculture, and it had no effect on respiratory growth with either lactate or pyruvate and no significant effect on the pyruvate-grown coculture. As shown by the model in Fig. 3 and equations 6 and 7, electrons derived from the oxidation of lactate may be shuttled via an undefined electron carrier (likely the quinone pool) to the Coo hydrogenase. The combined reaction (equation 8) is favorable only at very low concentrations of H2 and pyruvate. Continuous consumption of these compounds either internally (pyruvate) or externally via the methanogen (hydrogen) contributes to more energetically favorable conditions for continued lactate oxidation. Since the internal concentrations of lactate and pyruvate likely remain relatively stable, fluctuations in the hydrogen concentration primarily govern the thermodynamic feasibility of lactate oxidation. Increases in the hydrogen concentration inhibit lactate oxidation by preventing the Coo hydrogenase from reoxidizing the electron carrier. For example, at the steady-state hydrogen concentrations observed (5 to 6 Pa) and assuming that there was no contribution by reverse electron flow, lactate oxidation ceases at an intracellular lactate/pyruvate ratio of approximately 1,000. However, without measurement of intracellular metabolite concentrations it is not possible to constrain the requirement for reverse electron flow in the initial oxidation of lactate. If reverse electron flow is required for lactate oxidation, the Coo hydrogenase might provide a mechanism, if this hydrogenase uses the proton motive force to produce hydrogen (rather than functioning to export protons as shown in Fig. 3).
Since a mutation in the Coo hydrogenase impairs only growth on lactate in coculture, this hydrogenase appears to be part of a dedicated system for syntrophic growth on lactate. A similar function is indicated for Hmc, which is required for the reoxidation of reduced ferredoxin generated by the activity of a pyruvate-ferredoxin oxidoreductase (Por; DVU3025) (30). This activity accounts for the defective growth of the mutant in coculture on both lactate and pyruvate, along with the reduced impact on respiratory growth previously attributed to compensation by alternative transmembrane electron carriers (e.g., the Tmc and Rnf complexes) apparently specific to sulfate reduction (6, 29, 45).
Both the Δhyd and Δhyn-1 mutants exhibited small differences in the maximum cell density during syntrophic growth on both lactate and pyruvate, possibly because alternative periplasmic hydrogenases (Hyn-2 and Hys) masked the lesions. Their impact on the overall coculture growth rate, but not on the monoculture growth rate, suggests that there was incomplete compensation by the two alternative hydrogenases under these growth conditions. The different coculture growth rates observed likely resulted from the various affinities and activities of the Hyd and Hyn-1 hydrogenases that affected only the hydrogen production rate (27).
Some evidence suggests that Hdr has a role in ethanol production and consumption (13). In association with Adh and Aor, Hdr may reduce acetyl coenzyme A and produce the small quantities of ethanol that are produced during syntrophic growth (Fig. 3). As observed in other sulfate-reducing microorganisms, this pathway could transiently serve as an alternative electron transport mechanism to maintain the redox balance during periods when the hydrogen concentration is elevated (21). However, both the accumulation of toxic by-products, such as ethanol or acetaldehyde, and the lack of energy recovered from acetyl coenzyme A reduction to ethanol make this alternative pathway unfavorable for continued growth.
Although many genes encoding electron transfer functions are upregulated, the single mostly highly downregulated gene in cocultures codes for a flavodoxin (log2R = −5.2). The downregulation of this gene appears to be related to a general downregulation of genes involved in iron and metal uptake when D. vulgaris is grown in coculture. This is almost certainly an indirect consequence of the absence of appreciable sulfide production by syntrophically grown Desulfovibrio. The formation of metal sulfides during growth by sulfate respiration greatly reduces metal availability, as reflected by the higher expression levels in monocultures of genes for iron transport (feoA, feoB, tonB), molybdenum uptake (modB), and zinc uptake (znuAB) (see Table S2 in the supplemental material). A related flavodoxin in the cyanobacterium Anabaena replaces ferredoxin as an electron carrier from photosystem I to ferredoxin-NADP+ reductase under iron-deficient conditions (12). Thus, although the Desulfovibrio flavodoxin has been implicated as a constitutive component of the sulfate reduction pathway, its highly repressed expression in coculture suggests that it may function primarily under iron limitation conditions (17). Genes encoding chemotaxis-related functions also comprise a general category of genes that tend to be downregulated in coculture (e.g., cheA, cheY, and multiple genes coding for methyl-accepting chemotaxis proteins [MCPs] [see Table S2 in the supplemental material]), although one MCP gene (DVU0344) was upregulated about fivefold. Similar downregulation of genes for MCPs was observed for Rhizobium leguminosarum following its differentiation into a plant-associated symbiotic bacteroid, suggesting that the transition from monoculture to coculture by Desulfovibrio is also associated with a comparable physiological change (48).
In contrast to the downregulation of many genes for chemotaxis, several genes coding for parts the flagellar system basal body and filament (flgC, flgB, flgL) (see Table S2 in the supplemental material) are significantly upregulated in coculture. Although we have no immediate explanation for these expression trends, recent studies of a similar syntrophic couple between a bacterium (P. thermopropionicum) and an archaeon (Methanothermobacter thermautotrophicus) have shown that the bacterial flagellum mediates physical association between the two species (38). In addition to promoting a close physical association thought to enhance syntrophy, the presence of the filament cap protein (FliD) alone induced expression of methanogen genes required for syntrophic growth. Thus, the available data suggest that there are specific metabolic and protein-mediated systems for communication between evolutionarily unrelated species of microorganisms.
The existence of independent electron transfer systems for syntrophic growth and respiratory growth in Desulfovibrio also has relevance for the evolution of microorganisms that function primarily as syntrophs. The evolutionary history of “obligate” syntrophs is closely intertwined with that of sulfate reducers, and syntrophs appear to have diverged on more than one occasion from sulfate-reducing ancestors. Independent energy conservation pathways functioning during sulfate respiration and syntrophy may have permitted this evolution. The genomes of S. aciditrophicus (a gram-negative deltaproteobacterium) and P. thermopropionicum (related to gram-positive Desulfotomaculum species) tentatively support this hypothesis as both of them contain homologues of genes encoding enzymes that function in the syntrophic growth of Desulfovibrio (including Coo and Hmc, electron transfer, and ferredoxin recycling) (18, 24). More importantly, P. thermopropionicum and several related species contain vestiges of an ancestral sulfate-reducing pathway, suggesting that there was relatively recent adaptation to low-sulfate environments (15). While environmental fluctuations of sulfate likely contributed to evolution of independent “syntrophic” metabolism, stable anaerobic environments lacking terminal electron acceptors may promote loss of functional abilities in the genome, such as those observed in the facultative syntrophs P. thermopropionicum and Pelotomaculum schinkii. The stable environments may assist in the development of more specialized ecological niches, increasingly segregating independent energy generation pathways in divergent microbial species.
Supplementary Material
Acknowledgments
This work was part of work by the Virtual Institute for Microbial Stress and Survival (http://VIMSS/lbl.gov) supported by the U.S. Department of Energy Office of Science Office of Biological and Environmental Research Genomics:GTL program through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
M. maripaludis strain S2 and culturing assistance were kindly provided by John Leigh (University of Washington). We thank Bernard Schink (University of Constance) and Everett Shock (Arizona State University) for thoughtful editorial assistance.
Footnotes
Published ahead of print on 6 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandis, A., and R. K. Thauer. 1981. Growth of Desulfovibrio species on hydrogen and sulfate as sole energy-source. J. Gen. Microbiol. 126249-252. [Google Scholar]
- 3.Bryant, M. P., L. L. Campbell, C. A. Reddy, and M. R. Crabill. 1977. Growth of Desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl. Environ. Microbiol. 331162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chhabra, S. R., Q. He, K. H. Huang, S. P. Gaucher, E. J. Alm, Z. He, M. Z. Hadi, T. C. Hazen, J. D. Wall, J. Zhou, A. P. Arkin, and A. K. Singh. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 1881817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13233-240. [DOI] [PubMed] [Google Scholar]
- 6.Dolla, A., B. K. J. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174143-151. [DOI] [PubMed] [Google Scholar]
- 7.Dudoit, S., and J. Fridlyand. 2002. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol. 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, J. D., R. L. Kerby, G. P. Roberts, and P. W. Ludden. 1996. Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J. Bacteriol. 1781515-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu, R. D., and G. Voordouw. 1997. Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Arch. Microbiol. 1431815-1826. [DOI] [PubMed] [Google Scholar]
- 10.Gao, H. C., Y. Wang, X. D. Liu, T. F. Yan, L. Y. Wu, E. Alm, A. Arkin, D. K. Thompson, and J. Z. Zhou. 2004. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 1867796-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goenka, A., J. K. Voordouw, W. Lubitz, W. Gartner, and G. Voordouw. 2005. Construction of a NiFe-hydrogenase deletion mutant of Desulfovibrio vulgaris Hildenborough. Biochem. Soc. Trans. 3359-60. [DOI] [PubMed] [Google Scholar]
- 12.Goni, G., B. Herguedas, M. Hervas, J. R. Peregrina, M. A. De la Rosa, C. Gomez-Moreno, J. A. Navarro, J. A. Hermoso, M. Martinez-Julvez, and M. Medina. 2009. Flavodoxin: a compromise between efficiency and versatility in the electron transfer from photosystem I to ferredoxin-NADP+ reductase. Biochim. Biophys. Acta 1787144-154. [DOI] [PubMed] [Google Scholar]
- 13.Haveman, S. A., V. Brunelle, J. K. Voordouw, G. Voordouw, J. F. Heidelberg, and R. Rabus. 2003. Gene expression analysis of energy metabolism mutants of Desulfovibrio vulgaris Hildenborough indicates an important role for alcohol dehydrogenase. J. Bacteriol. 1854345-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., R. Seshadri, S. A. Haveman, C. L. Hemme, I. T. Paulsen, J. F. Kolonay, J. A. Eisen, N. Ward, B. Methe, L. M. Brinkac, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, D. Fouts, D. H. Haft, J. Selengut, J. D. Peterson, T. M. Davidsen, N. Zafar, L. Zhou, D. Radune, G. Dimitrov, M. Hance, K. Tran, H. Khouri, J. Gill, T. R. Utterback, T. V. Feldblyum, J. D. Wall, G. Voordouw, and C. M. Fraser. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22554-559. [DOI] [PubMed] [Google Scholar]
- 15.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Loy, Y. L. Qiu, P. Hugenholtz, N. Kimura, M. Wagner, A. Ohashi, and H. Harada. 2006. Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl. Environ. Microbiol. 722080-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerby, R. L., S. S. Hong, S. A. Ensign, L. J. Coppoc, P. W. Ludden, and G. P. Roberts. 1992. Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J. Bacteriol. 1745284-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knauf, M. A., F. Lohr, M. Blumel, S. G. Mayhew, and H. Ruterjans. 1996. NMR investigation of the solution conformation of oxidized flavodoxin from Desulfovibrio vulgaris—determination of the tertiary structure and detection of protein-bound water molecules. Eur. J. Biochem. 238423-434. [DOI] [PubMed] [Google Scholar]
- 18.Kosaka, T., S. Kato, T. Shimoyama, S. Ishii, T. Abe, and K. Watanabe. 2008. The genome of Pelotomaculum thermopropionicum reveals niche-associated evolution in anaerobic microbiota. Genome Res. 18442-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178193-201. [DOI] [PubMed] [Google Scholar]
- 20.Lord, J. M. 1972. Glycolate oxidoreductase in Escherichia coli. Biochim. Biophys. Acta 267227-237. [DOI] [PubMed] [Google Scholar]
- 21.Ma, K., and M. W. Adams. 1999. An unusual oxygen-sensitive, iron- and zinc-containing alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1811163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maness, P. C., J. Huang, S. Smolinski, V. Tek, and G. Vanzin. 2005. Energy generation from the CO oxidation-hydrogen production pathway in Rubrivivax gelatinosus. Appl. Environ. Microbiol. 712870-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McInerney, M. J., and M. P. Bryant. 1981. Anaerobic degradation of lactate by syntrophic associations of Methanosarcina barkeri and Desulfovibrio species and effect of H2 on acetate degradation. Appl. Environ. Microbiol. 41346-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McInerney, M. J., L. Rohlin, H. Mouttaki, U. Kim, R. S. Krupp, L. Rios-Hernandez, J. Sieber, C. G. Struchtemeyer, A. Bhattacharyya, J. W. Campbell, and R. P. Gunsalus. 2007. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc. Natl. Acad. Sci. USA 1047600-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay, A., Z. L. He, E. J. Alm, A. P. Arkin, E. E. Baidoo, S. C. Borglin, W. Q. Chen, T. C. Hazen, Q. He, H. Y. Holman, K. Huang, R. Huang, D. C. Joyner, N. Katz, M. Keller, P. Oeller, A. Redding, J. Sun, J. Wall, J. Wei, Z. M. Yang, H. C. Yen, J. Z. Zhou, and J. D. Keasling. 2006. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 1884068-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ornston, L. N., and M. K. Ornston. 1969. Regulation of glyoxylate metabolism in Escherichia coli K-12. J. Bacteriol. 981098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peck, H. D., Jr. 1994. Molecular biology of the sulfate-reducing bacteria, p. 41-75. In J. M. Odom and J. R. Singleton (ed.), The sulfate-reducing bacteria: contemporary perspectives. Springer Verlag, New York, NY.
- 28.Pereira, I. A. C., C. V. Romao, A. V. Xavier, J. LeGall, and M. Teixeira. 1998. Electron transfer between hydrogenases and mono- and multiheme cytochromes in Desulfovibrio ssp. J. Biol. Inorg. Chem. 3494-498. [Google Scholar]
- 29.Pereira, P. M., M. Teixeira, A. V. Xavier, R. O. Louro, and I. A. C. Pereira. 2006. The Tmc complex from Desulfovibrio vulgaris Hildenborough is involved in transmembrane electron transfer from periplasmic hydrogen oxidation. Biochemistry 4510359-10367. [DOI] [PubMed] [Google Scholar]
- 30.Pieulle, L., B. Guigliarelli, M. Asso, F. Dole, A. Bernadac, and E. C. Hatchikian. 1995. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim. Biophys. Acta 125049-59. [DOI] [PubMed] [Google Scholar]
- 31.Pohorelic, B. K. J., J. K. Voordouw, E. Lojou, A. Dolla, J. Harder, and G. Voordouw. 2002. Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabus, R., T. Hansen, and F. Widdle. 3 October 2006, posting date. Dissimilatory sulfate- and sulfur-reducing prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.3 ed. Springer-Verlag, New York, NY.
- 33.Rossi, M., W. B. R. Pollock, M. W. Reij, R. G. Keon, R. D. Fu, and G. Voordouw. 1993. The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J. Bacteriol. 1754699-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sallal, A. K., and N. A. Nimer. 1989. The intracellular localization of glycolate oxidoreductase in Escherichia coli. FEBS Lett. 258277-280. [DOI] [PubMed] [Google Scholar]
- 35.Sapra, R., M. Verhagen, and M. W. W. Adams. 2000. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1823423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schink, B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schink, B. 2002. Synergistic interactions in the microbial world. Antonie van Leeuwenhoek 81257-261. [DOI] [PubMed] [Google Scholar]
- 38.Shimoyama, T., S. Kato, S. Ishii, and K. Watanabe. 2009. Flagellum mediates symbiosis. Science 3231574. [DOI] [PubMed] [Google Scholar]
- 39.Stams, A. J. M., C. M. Plugge, F. A. M. de Bok, B. van Houten, P. Lens, H. Dijkman, and J. Weijma. 2005. Metabolic interactions in methanogenic and sulfate-reducing bioreactors. Water Sci. Technol. 5213-20. [PubMed] [Google Scholar]
- 40.Stolyar, S., S. Van Dien, K. L. Hillesland, N. Pinel, T. J. Lie, J. A. Leigh, and D. A. Stahl. 2007. Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol. 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talaat, A. M., S. T. Howard, W. Hale, I. V., R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, K. H. Nealson, J. Lim, J. Yates, C. C. Brandt, J. M. Tiedje, and J. Z. Zhou. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: possible involvement of Fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traore, A. S., M. L. Fardeau, C. E. Hatchikian, J. Legall, and J. P. Belaich. 1983. Energetics of growth of a defined mixed culture of Desulfovibrio vulgaris and Methanosarcina barkeri: interspecies hydrogen transfer in batch and continuous cultures. Appl. Environ. Microbiol. 461152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voordouw, G. 2002. Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 1845903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Whitman, W. B., J. Shieh, S. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic Methanococci. Syst. Appl. Microbiol. 7235-240. [Google Scholar]
- 46.Widdel, F., and F. Bak. 1992. Gram-negative sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes: a handbook on the biology of bacteria. Ecophysiology, isolation, identification, applications, 2nd ed. Springer- Verlag, New York, NY.
- 47.Williams, B. A., R. M. Gwirtz, and B. J. Wold. 2004. Genomic DNA as a cohybridization standard for mammalian microarray measurements. Nucleic Acids Res. 32e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yost, C. K., K. L. Del Bel, J. Quandt, and M. F. Hynes. 2004. Rhizobium leguminosarum methyl-accepting chemotaxis protein genes are down- regulated in the pea nodule. Arch. Microbiol. 182505-513. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, W. W., D. E. Culley, J. C. M. Scholten, M. Hogan, L. Vitiritti, and F. J. Brockman. 2006. Global transcriptomic analysis of Desulfovibrio vulgaris on different electron donors. Antonie van Leeuwenhoek 89221-237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.