Abstract
In gram-positive bacteria, covalently linked pilus polymers are assembled by a specific transpeptidase enzyme called pilus-specific sortase. This sortase is postulated to cleave the LPXTG motif of a pilin precursor between threonine and glycine and to form an acyl enzyme intermediate with the substrate. Pilus polymerization is believed to occur through the resolution of this intermediate upon specific nucleophilic attack by the conserved lysine located within the pilin motif of another pilin monomer, which joins two pilins with an isopeptide bond formed between threonine and lysine. Here, we present evidence for sortase reaction intermediates in Corynebacterium diphtheriae. We show that truncated SrtA mutants that are loosely bound to the cytoplasmic membrane form high-molecular-weight complexes with SpaA polymers secreted into the extracellular milieu. These complexes are not formed with SpaA pilin mutants that have alanine substitutions in place of threonine in the LPXTG motif or lysine in the pilin motif. The same phenotype is observed with alanine substitutions of either the conserved cysteine or histidine residue of SrtA known to be required for catalysis. Remarkably, the assembly of SpaA pili, or the formation of intermediates, is abolished with a SrtA mutant missing the membrane-anchoring domain. We infer that pilus polymerization involves the formation of covalent pilin-sortase intermediates, which occurs within a molecular platform on the exoplasmic face of the cytoplasmic membrane that brings together both sortase and its cognate substrates in close proximity to each other, likely surrounding a secretion apparatus. We present electron microscopic data in support of this picture.
Adherence to specific host tissue is a key step in bacterial colonization and the establishment of a successful infection by bacterial pathogens. Bacteria express a variety of adhesive cell surface molecules to bind host cells or other substrates in their natural habitat. The proteinaceous filaments known as pili or fimbriae are a clinically important class of these molecules. Both gram-negative and gram-positive bacteria express pili (6, 8). The gram-positive bacterial pili are unique in three respects (12, 25, 31). First, they represent heterodimeric or heterotrimeric protein polymers in which individual pilin subunits are covalently joined to each other (2, 9, 32). Second, the polymer itself is covalently attached to the cell wall (3, 31). Third, unlike pilus assembly in gram-negative bacteria, many of which require chaperones (26), the polymerization of the gram-positive pili and their cell wall attachment require specific transpeptidase enzymes called sortases (31).
Mazmanian and colleagues discovered the sortase SrtA as an enzyme that linked the surface protein A of Staphylococcus aureus to its cell wall (15). Genome sequences revealed that sortases are ubiquitously expressed in gram-positive bacteria, including significant pathogens, such as Actinomyces naeslundii, Bacillus cereus, Corynebacterium diphtheriae, Enterococcus faecalis, Streptococcus agalactiae, and Streptococcus pneumoniae (4, 5, 28). Sortases are classified according to their functions and phylogenic relationships (4, 5). The class that closely matches SrtA of S. aureus in structure and function is now called a housekeeping sortase. Its function is to attach numerous surface proteins to the cell wall (16). Common to each of these cell surface proteins is a cell wall sorting signal with an LPXTG motif that is absolutely necessary for cell wall anchoring (18). Elegant genetic, biochemical, and structural work by the Schneewind laboratory illuminated the universal reaction mechanism of protein sorting in the gram-positive cell wall (14). Cell wall anchoring of surface proteins is catalyzed in two steps. In the first step, SrtA cleaves the TG peptide bond of the LPXTG motif of protein A and forms an acyl enzyme intermediate involving the threonine of protein A and the catalytic cysteine of sortase (22, 27, 29). In the second step, the cleaved protein A is transferred to the cell wall when a nucleophile amine from the lipid II precursor attacks and resolves the acyl enzyme intermediate (20, 21, 30). This seminal work formed the basis of our current model of pilus assembly catalyzed by pilus-specific sortases (12).
We have used C. diphtheriae as a model for studies of the mechanism of pilus biogenesis. The corynebacterial genome encodes six different sortases (32). We now know that while five of these sortases (SrtA to -E) are devoted to pilus assembly, even the housekeeping sortase, SrtF, is required for efficient attachment of pili to the cell wall (23). Corynebacteria produce three distinct types of heterotrimeric pili, which are encoded by three pilus islands, each encoding three pilins (namely, SpaABC, SpaDEF, and SpaGHI) plus one or two cognate sortases essential for the assembly of the respective pilus (7, 24, 32). In each case, the prototype pilus represents a shaft structure made of a specific major pilin (namely, SpaA, SpaD, and SpaH) (12). Each type of pilus also contains a minor pilin at the tip (SpaC, SpaF, and SpaG) and another minor pilin dispersed along the shaft, as well as at the base of the pilus (SpaB, SpaE, and SpaI) (12). How are these polymers assembled, and how are they attached to the cell wall? All pilin proteins are predicted to contain in their amino termini a hydrophobic signal sequence necessary for export to the exoplasm by the Sec machinery. In addition, like the cell wall-anchored protein A of S. aureus, a cell wall sorting signal including the LPXTG motif is also present at the carboxy terminus of each of the Spa proteins of corynebacteria and other pilus proteins found in different gram-positive organisms (17). It is thus logical to imagine that the pilus-specific sortase utilizes the LPXTG motif for pilus polymerization, its cell wall anchoring, or both. Substantial genetic, biochemical, and ultrastructural analyses have proved this prediction. Consequently, Ton-That and Schneewind proposed a model of pilus assembly which posited that the basic mechanism of catalysis is conserved between cell wall sorting of surface proteins and the assembly of the pilus (31).
According to our current working model (Fig. 1A), the prototype SpaA pilus is assembled as follows. SrtA, which is essential and also specific for SpaA pilus formation, captures and cleaves cognate pilins at the LPXTG motif and forms an acyl enzyme intermediate. To form a dimer of SpaA and SpaC, the proposed tip entity, a conserved lysine in the SpaA pilin motif attacks the Cys-Thr bond of the SpaC-SrtA acyl enzyme intermediate. Shaft formation ensues by the cyclic addition of SpaA to the SpaC-SpaA dimer and the SpaC-SpaAn oligomer formed in the preceding reaction. When a SpaB is attached to the growing pilus terminus by a similar mechanism involving a critical lysine of SpaB, it acts as a switch, terminating pilus polymerization in favor of cell wall anchoring (11). This happens by the classic resolution reaction mentioned above, which involves the lipid II precursor (28), followed by its linkage to the cell wall (11). Alternatively, the SpaB-containing pilus can elongate further by adding a SpaA subunit to SpaB (11). This model explains all the available genetic and biochemical data we have obtained so far in the corynebacterial system, as well as other systems reported by various investigators.
FIG. 1.
(A) Working model of pilus assembly in C. diphtheriae. Spa pilins are synthesized in the cytoplasm and transported across the cytoplasmic membrane by the Sec machinery. The membrane-bound pilus-specific sortase SrtA cleaves the Spa pilins at the LPXTG motif and forms an acyl enzyme intermediate with the substrates. Pilus polymerization occurs when this intermediate is resolved by a nucleophilic attack by the lysine residue within the pilin motif of an adjacent intermediate. Cell wall anchoring terminates pilus polymerization when SpaB is incorporated into the pilus base by the housekeeping sortase, SrtF (see the text for details). (B) Membrane localization of the pilus-specific sortase SrtA. Corynebacteria grown to mid-log phase were separated from the culture medium (M) by centrifugation. The cell wall (W) was removed from its protoplast by muramidase treatment of the cells. The protoplasts were lysed, and membrane (P*) and cytoplasmic (C) compartments were obtained by ultracentrifugation. Protein samples were separated on 4 to 12% Tris-glycine gradient gels and detected by immunoblotting them with the specific antisera α-SrtA, α-SecA, and α-SpaA. The positions of molecular mass markers (kDa) are indicated. WT, wild type.
Significantly, there has been no report demonstrating the proposed intermediates of pilus assembly, to our knowledge. The present study was initiated to explore this key element of our model of pilus assembly, as well as the localization of the sortase in the membrane and its organization in the exoplasmic membrane in relation to the cognate pilins and the general secretion machinery.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The C. diphtheriae strains (Table 1) used in this study were grown on heart infusion agar or in heart infusion broth (HIB). Escherichia coli strains were grown on Luria broth or agar. Where needed, kanamycin and ampicillin were added at concentrations of 50 μg/ml and 100 μg/ml, respectively. All reagents were obtained from Sigma unless stated otherwise. The plasmids used in the study are listed in Table 1.
TABLE 1.
Strains of C. diphtheriae and plasmids used in this study
| Strain or plasmid | Genotype and description | Reference |
|---|---|---|
| Strains | ||
| NCTC13129 | Wild type | 32 |
| HT2 | ΔsrtA | 32 |
| HT11/pSpaA | ΔspaA; expressing wild-type SpaA from C. diphtheriae | 32 |
| HT52 | ΔspaA/ΔsrtA | This study |
| Plasmids | ||
| pSrtA | pCGL0243 containing wild-type SrtA from C. diphtheriae | 32 |
| pSrtAΔC10 | Plasmid expressing corynebacterial SrtA lacking the C-terminal 10 residues | This study |
| pSrtAΔC13 | Plasmid expressing corynebacterial SrtA lacking the C-terminal 13 residues | This study |
| pSrtAΔC19 | Plasmid expressing corynebacterial SrtA lacking the C-terminal 19 residues | This study |
| pSrtAΔC29 | Plasmid expressing corynebacterial SrtA lacking the C-terminal 29 residues | This study |
| pSpaA-SrtAΔC13 | Plasmid expressing a full length SpaA and SrtA lacking the C-terminal 13 residues | This study |
| pSpaA-K190A-SrtAΔC13 | pSpaA-SrtAΔC13 containing SpaA with a mutation in lysine 190 | This study |
| pSpaA-T494S-SrtAΔC13 | pSpaA-SrtAΔC13 containing SpaA with a mutation in threonine 494 | This study |
| pSpaA-SrtAΔC13-C222A | pSpaA-SrtAΔC13 containing SrtA with a mutation in cysteine 222 | This study |
| pSpaA-SrtAΔC13-H160A | pSpaA-SrtAΔC13 containing SrtA with a mutation in histidine 160 | This study |
Generation of recombinant plasmids.
(i) SrtA truncations Primers SpaA-promoter-5 and SrtAΔC-3 (Table 2) were used to PCR amplify the spaA promoter region and truncated open reading frames of srtA with template DNA of pSrtA (32). The generated fragments were digested with BglII and ligated into the Bglll-cut E. coli/Corynebacterium shuttle vector pCGL0243. The recombinant plasmid was then electroporated into C. diphtheriae by a standard protocol (32).
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| ΔspaA-A | CGCAAGCTTACTAACGTGAAAACAAAA |
| ΔspaA-B | CCCATCCACTAAACTTAAACAACACCCCCGTGCAGTCAT |
| ΔsrtA-C | TGTTTAAGTTTAGTGGATGGGCTTCGCAGGAAGAGGACA |
| ΔsrtA-D | CGCAGATCTTTGTATCCGTCATCATTA |
| SpaA-promoter-5 | AAAAGATCTTGAGTTCGATTGGCTTTTTTTC |
| SrtAΔC10-3 | CGCAGATCTTTAAAGCACGAGGGTGATGATGA |
| SrtAΔC13-3 | CGCAGATCTTTAGGTGATGATGATGGCGAT |
| SrtAΔC19-3 | CGCAGATCTTTAGGCGGTGAGTGCTAGGG |
| SrtAΔC29-3 | CGCAGATCTTTACATCCAGGGTTGCCAGATTT |
| SrtA-C222A-5 | CAAATAACACTCATCACCGCCACCCCCTACGCCGTCAAC |
| SrtA-C222A-3 | GTTGACGGCGTAGGGGGTGGCGGTGATGAGTGTTATTTG |
| SrtA-H160A-5 | CACCCCGTGATCACCGGAGCCAGCGGCCTTGCCAACGCC |
| SrtA-H160A-3 | GGCGTTGGCAAGGCCGCTGGCTCCGGTGATCACGGGGTG |
| SpaA-K190A-5 | GACGTGCACGTGTATCCCGCGCACCAGGCTTTGTCTGAG |
| SpaA-K190A-3 | CTCAGACAAAGCCTGGTGCGCGGGATACACGTGCACGTC |
| SpaA-T494S-5 | GGATTTGAACTGCCACTCTCGGGTGGTTCGGGGCGCATC |
| SpaA-T494S-3 | GATGCGCCCCGAACCACCCGAGAGTGGCAGTTCAAATCC |
The restriction sites in the primers are underlined.
(ii) pSpaA-SrtAΔC13 Primers SpaA-promoter-5 and SrtAΔC13-3 (Table 2) were used to PCR amplify the promoter region and coding sequence of spaA and the truncated open reading frame of srtA with chromosomal template DNA of C. diphtheriae NCTC13129. The amplified fragment was digested with BglII and ligated into the Bglll-cut vector pCGL0243. The recombinant plasmid was then electroporated into a C. diphtheriae strain lacking spaA and srtA (Table 1), which was generated as previously described (32).
(iii) Site-directed mutagenesis of recombinant plasmids PCR-based site-directed mutagenesis of double-stranded DNA was employed in this study (28). Plasmid DNA of pSpaA-SrtAΔC13 was used as a template for PCR amplification with Pfu DNA polymerase using primer sets (5′ and 3′) flanking six codons on both sides of the desired site of mutation (Table 2). Mutant plasmids were verified by sequencing them and then transformed into C. diphtheriae by electroporation.
Cell fractionation and Western blotting.
Corynebacteria grown overnight were used to inoculate a fresh culture (1:50 dilution) grown to mid-log phase at 37°C in HIB. All aliquots with the same number of cells were fractionated into medium and cell pellets by centrifugation. The washed cell pellets were treated with muramidase in sucrose buffer SMM (0.5 M sucrose, 10 mM MgCl2, and 10 mM malate, pH 6.8) for 1 h (for cell fractionation) or 6 h or overnight at 37°C. After the muramidase treatment, the soluble cell wall fraction was separated from the protoplasts by centrifugation. The protoplasts (pellets) were suspended in 50 μl of SMM and lysed by the addition of 200 μl of ice-cold water and three cycles of freeze-thawing in a dry ice-ethanol bath. The membranes were separated from the cytoplasm by ultracentrifugation for 30 min at 100,000 rpm and 4°C in an ultracentrifuge. The culture medium; cell wall, membrane, and cytoplasmic fractions; and protoplasts were subjected to trichloroacetic acid precipitation and an acetone wash. Samples were then boiled in sodium dodecyl sulfate (SDS)-containing sample buffer, separated by 4 to 12% Tris-glycine gradient gels (Invitrogen), and subjected to immunoblotting with rabbit antisera as previously described (23). A histidine antibody was used to detect a histidine-containing protein that was secreted and cell associated as a loading control (reference 11 and data not shown).
Thin sections and electron microscopy.
Corynebacteria grown until log phase in HIB were pelleted and washed twice with phosphate-buffered saline (PBS). The cell pellets were resuspended in a fixative solution containing 3% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. After 2 h of fixation, followed by centrifugation, the bacterial pellets were incubated with 1% paraformaldehyde in 0.1 M cacodylate buffer, and the samples were stored overnight at 4°C. The pellets were rinsed with 0.1 M cacodylate buffer, dehydrated with graded concentrations of cold methanol, and infiltrated with LR Gold resin (Polysciences, Warrington, PA)-methanol mixtures at −20°C. The pellets were embedded in LR Gold resin with benzoin methyl ether catalyst and polymerized with UV light (365 nm) at −20°C. Thin sections (∼70 nm) were cut with a diamond knife using a Reichert Ultracut E ultramicrotome and collected on Formvar-carbon-coated 200-mesh nickel grids.
Immunolabeling experiments were carried out as previously described (17). Briefly, thin sections on nickel grids were washed three times with PBS containing 2% bovine serum albumin (BSA) and blocked for 1 h in PBS with 0.1% gelatin. The samples were stained with primary antibody diluted 1:100 in PBS with 2% BSA for 1 h, followed by washing and blocking. Pili were stained with 12-nm gold-goat anti-rabbit immunoglobulin G (IgG) (Jackson ImmunoResearch) diluted 1:20 in PBS with 2% BSA for 1 h, followed by washing in PBS with 2% BSA. For double-labeling experiments, the same procedure was applied using another primary antibody and goat anti-rabbit IgG conjugated with gold particles of different sizes. The grids were washed five times with water before being stained with 1% uranyl acetate. Samples were viewed in a Jeol 100CX electron microscope.
RESULTS
The pilus-specific sortase SrtA is a stable component of the corynebacterial membrane.
A basic feature of the current working model of gram-positive pilus assembly is that the covalent polymerization of pilin precursors in the bacterial exoplasm is catalyzed by a pair of pilus-specific sortase enzymes that are anchored to the membrane in sufficiently close proximity to each other to allow the stepwise transpeptidation reactions to occur rapidly and efficiently (Fig. 1A). Is sortase membrane associated? Alignment of the primary structures of all known pilus-specific sortases revealed that they harbor not only the amino-terminal signal sequence required for the translocation of the protein from the cytoplasm to the exoplasm by the general secretion machinery (the SecA/SecYEG translocon), but also a well-defined carboxy-terminal hydrophobic region that is immediately followed by a basic patch (5). This small domain made of about 25 amino acids (Fig. 2A), which can potentially form a transmembrane helix, thus provides a conserved mechanism for the pilus-specific sortase proteins to form the proposed pilus assembly center anchored to the membrane. To obtain evidence for this postulate, we first performed simple cell fractionation experiments and tested the membrane association of the single pilus-specific sortase SrtA of corynebacteria that is known to be essential for the assembly of the heterotrimeric SpaA-type pilus. In these experiments (Fig. 1B), we used quantitative cell fractionation procedures to isolate from fresh corynebacterial cultures three different subcellular fractions: the cell wall upon solubilization by enzymatic hydrolysis, the membrane pellet, and the soluble cytoplasmic fractions (see Materials and Methods). Next, by SDS-polyacrylamide gel electrophoresis resolution of proteins in these fractions and quantitative Western blot analysis with specific antibodies (see Materials and Methods), we monitored the presence of SrtA in these fractions, as well as in the culture medium, to give us a complete picture (Fig. 1B).
FIG. 2.
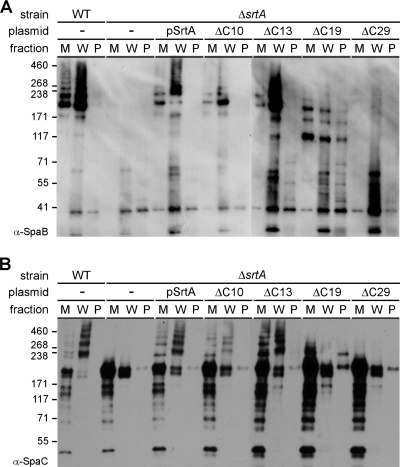
The TM domain of SrtA is required for the assembly of SpaA pili and the formation of high-molecular-weight pilin-sortase complexes. (A) Presentation of wild-type SrtA predicted to have the N-terminal signal peptide sequence (hatched) and the C-terminal TM domain (black), followed by a positively charged tail. A putative catalytic pocket is composed of His160 and Cys222. The positions where truncations were made are underlined. (B and C) Cells of the wild-type (WT) strain and its isogenic derivative carrying deletions of srtA or the derivative expressing SrtA mutants grown in HIB were fractionated into culture medium (M), cell wall (W), and protoplast (P) fractions. Protein samples were separated on 4 to 12% Tris-glycine gradient gels and detected by immunoblotting them with the specific antisera α-SpaA (B) and α-SrtA (C). The positions of molecular mass markers (kDa), high-molecular-weight polymers (HMW), and sortase monomers (P) are indicated.
When wild-type corynebacterial cells were analyzed by subcellular fractionation, nearly all of the ∼30-kDa SrtA protein was recovered in the pelleted membrane fraction, with little or none detectable in the solubilized cell wall, the soluble cytoplasmic fraction, or the culture medium (Fig. 1B, top, left lanes). By comparison, while the SecA component of the secretion machinery used here as a control showed an identical membrane localization pattern (Fig. 1B, α-SecA), the SpaA pili were fractionated mostly in the cell wall fraction, as expected, and very few or no SpaA polymers were detected in the membrane fraction (Fig. 1B, α-SpaA). In parallel experiments, fractions made from the ΔsrtA mutant revealed no bands when probed with the antibody against SrtA (α-SrtA). Importantly, the ΔsrtA mutant also showed no pilus polymers, as expected, but revealed the presence of SpaA monomers that were detected not only in the cytoplasmic fraction, but also in the culture medium. The significant amount of secreted SpaA observed in the absence of SrtA suggests that SpaA's association with the membrane may not be as stable as that of the sortase SrtA. Lastly, we tested how plasmid-expressed SrtA associated with the membrane and found that it behaved the same way as that expressed from the chromosome (Fig. 1B, right). Taken together, these data authenticated the important conclusion that SrtA is membrane localized. This then enabled us to perform the genetic experiments presented below, which were designed to decipher the structural determinants that anchor SrtA to the membrane.
The predicted transmembrane (TM) domain of SrtA is required for efficient pilus assembly.
The stable association of SrtA with the membrane could be mediated by the insertion of the predicted TM domain in the lipid bilayer. If so, the deletion of this domain should affect membrane anchoring and, in turn, might diminish pilus polymerization. However, the available structural data showed that sortases adopt a stable catalytically active structure in the absence of the predicted TM domain in the carboxy terminus (13, 19). To determine whether the TM domain and the charged tail are required for SpaA polymerization in vivo, we generated a series of truncated SrtA mutants in the expression plasmid (Fig. 2A). The plasmids expressing SrtA mutants were transformed into a corynebacterial strain that lacks srtA. We then monitored pilus polymerization in the various strains grown in liquid culture to mid-exponential phase by Western blotting, using equivalent samples representing the bacterium-free culture medium, the solubilized bacterial cell wall, and the protoplast pellet (see Materials and Methods). Note that the cell wall fraction was generated by muramidase treatment of cells and spinning out the protoplasts. The culture medium, cell wall, and protoplast fractions were all precipitated with trichloroacetic acid and dissolved in SDS-sample buffer for gel electrophoresis (Fig. 2B and C).
When the subcellular fractions of wild-type bacteria were probed with α-SpaA antibody (Fig. 2B), we observed most of the SpaA polymers (designated SpaAHMW) in the cell wall fraction and none in the protoplast-associated fraction. A very small amount of pilus polymers was also detected in the culture medium of the wild-type strain. In contrast, no pilus polymers were found in any of the fractions made from the mutant corynebacterial strain that lacks srtA but expresses each of the other five sortases, thereby reproducing the known essential role of SrtA in pilus assembly (32). This defect was complemented by the plasmid that expresses a full-length SrtA protein. However, the complemented strain secreted more SpaA polymers into the culture medium (Fig. 2B), which could result from an imbalance in the amount of SrtA relative to SrtF, the housekeeping sortase that mediates the efficient attachment of pilus polymers to the cell wall (23).
Interestingly, deletion of the positively charged tail of SrtA or three additional residues encroaching on the predicted TM domain in the plasmid-borne srtA gene did not abrogate SpaA pilus polymerization (Fig. 2B, ΔC10 and ΔC13 mutants). Nonetheless, SpaA polymerization was completely abolished when both the charged tail and the TM domain were deleted (ΔC29 mutant). Moreover, shortening of the TM domain also greatly diminished pilus polymerization, as the ΔC19 mutant produced a very small amount of low-molecular-mass (∼250 kDa) SpaA polymers. Therefore, the TM domain somehow plays a vital role in SpaA polymerization in vivo.
To further characterize the pilus assembly defect observed with the TM-less SrtA mutants, we next probed the same samples described above with antibodies against the minor subunit SpaB (α-SpaB) and SpaC (α-SpaC) (Fig. 3). Similar to the results obtained for SpaA, no SpaB and SpaC polymers were observed in the SrtA mutant devoid of the charged tail and the TM domain (Fig. 3, ΔC29). In agreement with our previous results showing that SrtF catalyzes the cell wall anchoring of minor pilins (23), monomeric SpaC (predicted mass, 200 kDa) was clearly present in the cell wall fraction of the ΔsrtA mutant. Interestingly, large amounts of monomeric SpaC and cleavage products were also found in the culture medium of each strain tested; this extracellular SpaC antigen was especially prominent with the strains harboring each of the truncated SrtA mutants (Fig. 3B, ΔC29). It is also interesting that there were several distinctive SpaB high-molecular-weight species in the ΔC19 mutant, whereas the predicted mass of a SpaB monomer is 21 kDa. We do not know their exact molecular nature, though a possible scenario is that these bands represent some intermediate forms of SpaB assembly (see below).
FIG. 3.
Requirement for the SrtA TM domain for the assembly of minor pilins SpaB and SpaC. The samples prepared for Fig. 2 were immunoblotted with the specific antisera α-SpaB (A) and α-SpaC (B). The positions of molecular mass markers (kDa) are shown on the left. WT, wild type.
Mutant SrtA enzymes form stable covalent complexes with pilin polymers.
The pilus polymerization defect observed with our TM-truncated SrtA mutants could be due to several reasons. Foremost of these is the fact that the polymerization defect results from the instability of the truncated proteins or from defective retention of these proteins within the exoplasm. To resolve these issues, we performed quantitative Western blotting to monitor the mutant sortases with α-SrtA antibody probing the same subcellular fractions described above, which revealed an intriguing picture of sortase localization (Fig. 2C). We showed above that the wild-type SrtA protein is associated with the isolated membrane fraction (Fig. 1B). Consistent with that result, the wild-type SrtA protein with a predicted mass of 32 kDa was recovered almost exclusively in the bacterial protoplasts (Fig. 2C, left lanes). In sharp contrast, while some amount of each of the ΔC10, ΔC13, ΔC19, and ΔC29 mutant sortase proteins was recovered in the protoplast fraction, a significant fraction of the mutant proteins was also detected in novel high-molecular-weight forms (SrtAHMW) that were secreted in the culture medium. This extracellular secretion of the mutant sortase proteins suggests defective membrane localization resulting from truncation of the basic patch and the TM domains. Strikingly, the ΔC13 mutant that is devoid of the basic patch and contains a short TM motif produces a ladder of high-molecular-mass SrtA species reminiscent of the ladder typically seen for pilus polymers (>200 to 500 kDa). Evidently, the SrtA ΔC13 mutant is quite proficient in catalyzing the polymerization of the SpaA pili. However, because the mutant sortase is only loosely bound to the cytoplasmic membrane, virtually all of the polymers are ultimately secreted in the culture medium in the form of covalent intermediates containing the SrtA enzyme.
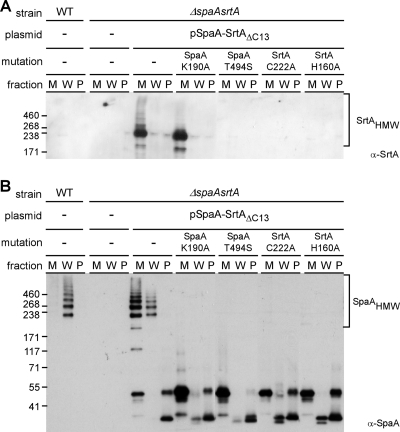
If our hypothesis is correct, the formation of these intermediates should be abolished by mutations that make sortase catalytically inactive. Recall that according to our working model, SrtA is postulated to cleave the SpaA LPXTG motif between threonine and glycine and to form an acyl enzyme intermediate via its catalytic cysteine residue. Pilus polymerization occurs when this intermediate is resolved by another adjacent intermediate through a nucleophilic attack involving the conserved lysine residue of the pilin motif (Fig. 1A). Also, it was shown previously in staphylococci that the activity of the sortase enzyme is dependent on a histidine residue, which forms the catalytic pocket, together with the cysteine residue (29). We therefore investigated whether these important catalytic elements are essential for the formation of the substrate enzyme intermediates. To do so, we constructed a plasmid that expresses both the full-length SpaA and the SrtA ΔC13 mutant (pSpaA-SrtAΔC13). Site-directed mutagenesis was then performed on this plasmid to generate an alanine substitution for the lysine in the SpaA pilin motif, serine substitution in place of threonine in the SpaA LPXTG motif, and alanine substitutions in place of cysteine and histidine in SrtA. The resulting plasmids were then introduced into a corynebacterial strain that lacks both spaA and srtA genes. In preliminary experiments of immunoblotting with antibodies against SpaA or SrtA, we established that the expression of various spaA and srtA mutants was comparable to that of the parental plasmid (data not shown). We next examined pilus polymerization and SrtA localization, as shown in Fig. 2.
The results obtained with the parental plasmid expressing wild-type SpaA and the ΔC13 SrtA mutant were similar to those reported above and reproduced our observation that the truncated SrtA mutant forms SpaA polymer intermediates that are largely secreted into the culture medium (Fig. 4, compare the lanes showing Western blots with α-SrtA [A] and α-SpaA [B]). As predicted, no SpaA or SrtA polymers were observed with plasmids expressing SpaA-T494S, SrtA-C222A, or SrtA-H160A (Fig. 4A). Also, no SpaA polymer was observed with the plasmid expressing SpaA-K190A, as expected. Remarkably, however, this plasmid led to the production and secretion of a major high-molecular-mass component (with an apparent mass of 240 kDa) that contained SrtA and SpaC, but no SpaA, thus representing the SpaC-SrtA covalent intermediate (Fig. 4A and data not shown). Together, these results document the existence of sortase enzyme intermediates and establish the vital role of proper membrane localization in the biogenesis of pilus polymers and subsequent cell wall anchoring.
FIG. 4.
Essential residues required for the formation of pilin-sortase intermediates. Cells of the wild-type (WT) strain and its isogenic derivative carrying deletions of spaA and srtA or the derivative expressing various SrtA mutants grown in HIB were fractionated into culture medium (M), cell wall (W), and protoplast fractions (P) as described in the legend to Fig. 2. Proteins were separated on 4 to 12% Tris-glycine gradient gels and detected by immunoblotting them with the specific antisera α-SrtA (A) and α-SpaA (B). The positions of molecular mass markers (kDa) and high-molecular-weight polymers (HMW) are indicated.
Sortase and cognate pilins are colocalized together with the SecA translocase on the membrane.
Like the pilus-specific sortases, the pilins also contain a presumptive TM domain that permits efficient pilus assembly (reference 11 and data not shown). To probe whether a sortase is colocalized with the cognate pilins on the membrane, essentially forming a pilus assembly center, we performed thin-section immunoelectron microscopy. Thin sections of corynebacteria were first probed with α-SrtA antibody, followed by staining with 18-nm gold particles. After extensive washes, the thin sections were next labeled with a specific antibody against individual SpaA-type pilins, followed by staining with 12-nm gold particles. With the ΔspaA/pSpaA strain, which expresses long SpaA pili, we observed SpaA-labeled gold particles in close proximity to those of the membrane-bound SrtA (Fig. 5A). This was not observed in the strain that lacks SrtA (Fig. 5B). In similar experiments, we labeled thin sections with α-SpaA/12-nm gold particles and α-SpaB/18-nm gold particles (Fig. 5C) or α-SpaC/18-nm gold particles (Fig. 5D) and observed the colocalization of SpaA with SpaB and SpaC (Fig. 5C and D). The remarkably close clustering of gold particles labeling the SrtA enzyme and the cognate pilins on the membrane strongly suggests the existence of organized pilus assembly centers.
FIG. 5.
The major pilin SpaA is colocalized with its cognate sortase SrtA and minor pilins SpaB and SpaC. Corynebacterial thin sections on nickel grids were stained with specific antiserum against SpaA (α-SpaA) and goat anti-rabbit IgG conjugated to 12-nm gold particles (filled arrows). Extensively washed samples were then reacted with α-SrtA (A and B), α-SpaB (C), or α-SpaC (D), followed by IgG-conjugated 18-nm gold particles (open arrows). Samples were viewed by transmission electron microscopy after being stained with 1% uranyl acetate. Scale bars, 0.5 μm.
To determine how the clusters of sortase and pilins are spatially organized with the secretion machinery, we next probed thin sections with α-SecA antibody. Representative images presented in Fig. 6 show that SrtA clusters (12 nm) are present in the vicinity of SecA (18 nm) (Fig. 6B). Clusters of SpaA (12 nm) are also present in close proximity to SecA (Fig. 6D). The fact that such close appositions of SecA and SpaA are also visible in the ΔsrtA mutant hints that the localization of SpaA in the pilus assembly center does not rely upon specific capture by the cognate sortase (Fig. 6E and F). We conclude that the membrane-embedded pilin subunits form a pilus assembly center in conjunction with cognate sortase, as well as the general secretion machinery.
FIG. 6.
Sortase and its cognate substrates colocalize with the Sec translocase. Except for the one shown in panel A, which was labeled with α-SecA and 12-nm gold particles, corynebacterial thin sections on nickel grids were stained as described in the legend to Fig. 5 with α-SecA and 18-nm gold particles (B, D, and E) or 12-nm gold particles (C), followed by α-SrtA (B and C) or α-SpaA (D and E) and 12-nm gold particles (B, D, and E) or 18-nm gold particles (C). An enlarged area of panel E (boxed) is shown in panel F. The samples were viewed by transmission electron microscopy. Scale bars, 0.2 μm.
DISCUSSION
The covalent attachment of extracellular proteins to the cell wall peptidoglycan is a fundamental feature of the envelope biogenesis of gram-positive bacteria. Central to this process are transpeptidation reactions that are catalyzed by sortases ubiquitously expressed in gram-positive bacteria. Sortases catalyze two different types of transpeptidation reactions described to date (12). One class of sortases, referred to as the housekeeping sortases, which include the founding member, SrtA, from S. aureus and SrtF of corynebacteria, incorporate numerous protein substrates into the cell wall via their linkages to the lipid II precursor. In contrast, a distinct group of sortase enzymes that are dedicated to the assembly of gram-positive pili perform the covalent cross-linking of pilin proteins that are specific for an individual sortase. The attachment of the resulting pilus polymer to the cell wall is then catalyzed by the housekeeping sortase or the pilus-specific sortase itself. In this paper, we have described a series of genetic, biochemical, and immunocytochemical studies of the pilus-specific sortase SrtA of corynebacteria as a model illuminating two basic aspects of this process: (i) the spatial organization of the sortase, the cognate pilins, and the secretion machinery on the bacterial cell membrane and (ii) the sortase enzyme intermediates that are generated during the catalysis of pilus polymerization.
The prototype SpaA pilus assembled by corynebacterial SrtA represents a heterogeneous set of polymers in which the major pilin SpaA makes up the pilus shaft of various lengths with the minor pilin SpaC at the pilus tip and SpaB at the pilus terminus and also interspersed along long pilus fibers that are formed when SpaA is overproduced in the cell (11). Our current model of pilus assembly envisions that the transpeptidation reaction that joins two pilin monomers (and a pilin monomer to the growing oligomer) is brought about by the formation of specific acyl enzyme intermediates involving the conserved catalytic cysteine residue of the sortase and the conserved threonine residue of the pilin protein's sorting signal (LPXTG). The model further postulates that the covalent linkage takes place when the lysine residue in the conserved pilin motif of one SpaA-SrtA intermediate attacks the Thr-Cys isopeptide bond of another SrtA acyl enzyme intermediate molecule, thereby ligating the cleaved pilin protein to another intermediate and regenerating the active catalytic center of the latter. In support of this scheme, we have presented here for the first time physical evidence for the existence of SrtA acyl enzyme intermediates and have also shown that the formation of these intermediates requires the catalytic cysteine of SrtA and the pilin motif lysine of SpaA. Importantly, we demonstrated that SrtA is not only membrane associated, but also colocalized with the cognate pilins, and that the proper localization of SrtA on the membrane is vital for pilus polymerization. Together, these results support a unified picture of pilus biogenesis in gram-positive bacteria within a highly organized assembly center, a “pilusosome,” coupled to a protein secretion apparatus located nearby.
Until now, the proposed involvement of an acyl enzyme intermediate in sortase-catalyzed transpeptidation reactions and its resolution by a nucleophilic amine moiety was mainly based on classic work on S. aureus SrtA (1, 27, 30). In that case, the existence of the intermediate was inferred from the susceptibility of the complex to hydroxylamine (27, 30), which is known to cleave the isopeptide bond forming the intermediate. As for transpeptidation catalyzed by a pilus-specific sortase, the essential role of a conserved lysine has been well documented (32). However, to our knowledge, the existence of the intermediate has not been documented in any pilus system until now. In fact, for unknown reasons, our own limited efforts to find intermediates of the SpaA pilus assembly using the traditional methods have not been successful so far. Nevertheless, in our attempts to characterize the structural and topological requirements of the cognate sortase, SrtA, we serendipitously uncovered the intermediates in a corynebacterial mutant strain, which expresses a truncated form of sortase that is catalytically active but somehow defective in membrane localization.
Sequence alignment of known pilus-specific sortase proteins showed the conservation of a small hydrophobic domain followed by a basic patch in the carboxy terminus (5). We reasoned that this hydrophobic region might serve as a membrane-anchoring domain by forming a transmembrane helix, while the adjacent basic patch might help to secure the helix within the membrane bilayer. Consistent with this view, we showed that the SrtA protein is tightly associated with the bacterial membrane (Fig. 1). If the membrane association of SrtA is important for its function, and the carboxy terminus of SrtA promotes its membrane association, then truncation of the putative membrane-anchoring domain should affect pilus polymerization. Our results obtained with a series of truncated SrtA mutants are entirely consistent with this prediction (Fig. 2 and 3). We showed that while a short truncation mutation (ΔC10) that removed the adjacent basic patch did not affect pilus polymerization, a larger truncation mutation (ΔC29) removing both the basic patch and the hydrophobic region virtually abolished SpaA polymerization (Fig. 2). Importantly, the ΔC29 SrtA mutant was stably expressed and exported to the exoplasm to a level comparable to that of the wild-type SrtA (Fig. 2C); however, the polymerization ability of the protein was greatly compromised (Fig. 2B). The presence of a fraction of this mutant SrtA in the culture medium points to a membrane localization defect that likely causes the defect in SpaA polymerization. Interestingly, a similar membrane retention defect is also evident with the basic-patch-truncated SrtA mutants ΔC10 and ΔC13, which indicates the functional importance of the basic patch and offers a basis for the conservation of this region among the pilus-specific sortases, as demonstrated recently (10). The fact that pilus polymerization is hardly affected with these mutants (Fig. 2B), however, indicates that upon export to the exoplasm, the mutant proteins remain membrane bound for a sufficiently long time to allow several cycles of catalysis in forming polymers that are comparable in length to that produced by the wild-type SrtA.
Remarkably, our analysis of the culture medium originating from the cultures of the mutant strains unveiled the elusive acyl enzyme intermediates that we have been seeking. Although the substrate enzyme intermediates are secreted by each of the truncated SrtA mutants we constructed and analyzed, which lack either the basic patch or both the basic patch and the hydrophobic domain, it was only the basic-patch-lacking mutants that displayed enzyme complexes containing pilus polymers of heterogeneous sizes (see the ladders in lanes M for ΔC13 in Fig. 2B and C). That these secreted complexes are bona fide intermediates of the pilus polymerization reaction was established by the fact that their formation was entirely dependent on not only the catalytic cysteine residue of SrtA, but both the threonine residue of the LPXTG sorting signal and the lysine residue of the pilin motif of SpaA (Fig. 4). Moreover, these complexes contain SpaC and SpaB (Fig. 3). The fact that these intermediates are released into the extracellular medium demonstrates the critical function of proper membrane topology of SrtA in the cell wall-anchoring step, which is believed to involve a nucleophilic attack by the lipid II precursor (20, 21). We imagine that the basic patch might facilitate the proper apposition of the membrane-bound sortase with the membrane-bound lipid II precursor for an efficient transfer of the pilus from the sortase to the lipid II precursor. Also, because the housekeeping sortase, SrtF, catalyzes the cell wall attachment of a majority of the pilus polymers (23), the optimal transfer of the pilus polymer from SrtA to SrtF might also depend on the proper membrane colocalization of the two sortases facilitated by the basic patch. It is important to investigate these issues further in future work.
The fact that an acyl enzyme intermediate was virtually undetectable in the wild-type corynebacterial cells in a rapidly growing culture (Fig. 2C) demonstrates that the entire process of pilus polymerization and cell wall attachment is extremely rapid. To be so, it makes sense that the sortase and the pilin precursor subunits are present in close proximity to each other, forming an assembly line in the exoplasm that is also in the vicinity of the membrane export channel that translocates these proteins from the cytoplasm to the exoplasm. The thin-section immunoelectron microscopic studies we presented here strongly support the existence of such an assembly center. First, we detected SpaA, SpaB, and SpaC in close clusters distributed near the membrane (Fig. 5). Second, clusters of SrtA and clusters of SpaA pilins are colocalized with the SecA component of the secretion machinery believed to export these proteins (Fig. 6). Importantly, the SpaA clusters localize with SecA even when SrtA is absent (Fig. 6). This indicates that both SrtA and pilin precursor substrates are efficiently embedded in the membrane in the vicinity of the export apparatus. Pilus polymerization starts with the rapid formation of sortase enzyme intermediates as soon as the pilin precursor emerges from the secretion apparatus, and the polymerization continues as oligomers are built in the form of acyl enzyme intermediates; new molecules of pilins are continuously captured by a sortase whose catalytic center was regenerated in the process of joining one pilin to another. We suspect that SrtF is also a part of the assembly center; however, a test of this will require a high-quality antibody that is presently being generated. Additional work is currently under way in our laboratory to verify the proposed linkages and to determine whether SrtA forms oligomers, whether it interacts with SrtF, and how the specificity of pilus polymerization is achieved.
Acknowledgments
We thank Donald Oliver (Wesleyan University) for Bacillus SecA antibodies, Arthur R. Hand (University of Connecticut Health Center) for help with electron microscopy, and Anjali Mandlik (Harvard Medical School) and members of our laboratory for critical review of the manuscript and discussion.
This work was supported by U.S. Public Health Service grant AI061381 from the National Institute of Allergy and Infectious Diseases to H.T.-T.
Footnotes
Published ahead of print on 10 July 2009.
REFERENCES
- 1.Aulabaugh, A., W. Ding, B. Kapoor, K. Tabei, L. Alksne, R. Dushin, T. Zatz, G. Ellestad, and X. Huang. 2007. Development of an HPLC assay for Staphylococcus aureus sortase: evidence for the formation of the kinetically competent acyl enzyme intermediate. Anal. Biochem. 36014-22. [DOI] [PubMed] [Google Scholar]
- 2.Budzik, J. M., L. A. Marraffini, P. Souda, J. P. Whitelegge, K. F. Faull, and O. Schneewind. 2008. Amide bonds assemble pili on the surface of bacilli. Proc. Natl. Acad. Sci. USA 10510215-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budzik, J. M., S. Y. Oh, and O. Schneewind. 2008. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J. Biol. Chem. 28336676-36686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comfort, D., and R. T. Clubb. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 722710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156289-297. [DOI] [PubMed] [Google Scholar]
- 6.Duguid, J. P., I. W. Smith, G. Dempster, and P. N. Edmunds. 1955. Non-flagellar filamentous appendages (fimbriae) and haemagglutinating activity in Bacterium coli. J. Pathol. Bacteriol. 70335-348. [DOI] [PubMed] [Google Scholar]
- 7.Gaspar, A. H., and H. Ton-That. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 1881526-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girard, A. E., and B. H. Jacius. 1974. Ultrastructure of Actinomyces viscosus and Actinomyces naeslundii. Arch. Oral Biol. 1971-79. [DOI] [PubMed] [Google Scholar]
- 9.Kang, H. J., F. Coulibaly, F. Clow, T. Proft, and E. N. Baker. 2007. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 3181625-1628. [DOI] [PubMed] [Google Scholar]
- 10.Kline, K. A., A. L. Kau, S. L. Chen, A. Lim, J. S. Pinkner, J. Rosch, S. R. Nallapareddy, B. E. Murray, B. Henriques-Normark, W. Beatty, M. G. Caparon, and S. J. Hultgren. 2009. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J. Bacteriol. 1913237-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandlik, A., A. Das, and H. Ton-That. 2008. The molecular switch that activates the cell wall anchoring step of pilus assembly in Gram-positive bacteria. Proc. Natl. Acad. Sci. USA 10514147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 1633-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manzano, C., C. Contreras-Martel, L. El Mortaji, T. Izore, D. Fenel, T. Vernet, G. Schoehn, A. M. Di Guilmi, and A. Dessen. 2008. Sortase-mediated pilus fiber biogenesis in Streptococcus pneumoniae. Structure 161838-1848. [DOI] [PubMed] [Google Scholar]
- 14.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285760-763. [DOI] [PubMed] [Google Scholar]
- 16.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 401049-1057. [DOI] [PubMed] [Google Scholar]
- 17.Mishra, A., A. Das, J. O. Cisar, and H. Ton-That. 2007. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J. Bacteriol. 1893156-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neiers, F., C. Madhurantakam, S. Falker, S. Normark, B. Henriques-Normark, and A. Achour. 2009. Cloning, expression, purification, crystallization and preliminary X-ray analysis of the pilus-associated sortase C from Streptococcus pneumoniae. Acta Crystallogr. F 6555-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry, A. M., H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 27716241-16248. [DOI] [PubMed] [Google Scholar]
- 21.Ruzin, A., A. Severin, F. Ritacco, K. Tabei, G. Singh, P. A. Bradford, M. M. Siegel, S. J. Projan, and D. M. Shlaes. 2002. Further evidence that a cell wall precursor [C(55)-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J. Bacteriol. 1842141-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268103-106. [DOI] [PubMed] [Google Scholar]
- 23.Swaminathan, A., A. Mandlik, A. Swierczynski, A. Gaspar, A. Das, and H. Ton-That. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 66961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swierczynski, A., and H. Ton-That. 2006. Type III pilus of corynebacteria: pilus length is determined by the level of its major pilin subunit. J. Bacteriol. 1886318-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telford, J. L., M. A. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in Gram-positive pathogens. Nat. Rev. Microbiol. 4509-519. [DOI] [PubMed] [Google Scholar]
- 26.Thanassi, D. G., and S. J. Hultgren. 2000. Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system. Methods 20111-126. [DOI] [PubMed] [Google Scholar]
- 27.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 9612424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 1694269-278. [DOI] [PubMed] [Google Scholar]
- 29.Ton-That, H., S. K. Mazmanian, L. Alksne, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J. Biol. Chem. 2777447-7452. [DOI] [PubMed] [Google Scholar]
- 30.Ton-That, H., S. K. Mazmanian, K. F. Faull, and O. Schneewind. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J. Biol. Chem. 2759876-9881. [DOI] [PubMed] [Google Scholar]
- 31.Ton-That, H., and O. Schneewind. 2004. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 12228-234. [DOI] [PubMed] [Google Scholar]
- 32.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 501429-1438. [DOI] [PubMed] [Google Scholar]