Abstract
YoeB is a bacterial toxin encoded by the yefM-yoeB toxin-antitoxin system found in various bacterial genomes. Here, we show that Staphylococcus aureus contains two YoeB homologues, both of which function as ribosome-dependent mRNA interferases to inhibit translation initiation in a manner identical to that of YoeB-ec from Escherichia coli.
Escherichia coli contains a number of toxin-antitoxin systems (1, 12). Among these, MazF(19, 20), chpBK (21), and PemK (17) are known to be sequence-specific mRNA interferases (16). Other toxins, such as RelE (4, 9, 14) and YoeB (3, 5, 18), also cleave mRNAs. RelE is a ribosome-associating factor and has no endoribonuclease activity by itself (4, 9, 14). Recently, we have demonstrated that YoeB is a 50S ribosome-associating factor and a potent inhibitor for translation initiation (18). It binds to the A site of the mRNA-ribosome initiation complex to block translation initiation. This results in partial cleavage of the mRNA at 3 bases downstream of the initiation codon regardless of the sequence of the mRNA. YoeB appears to have very weak intrinsic endoribonuclease activity (10), and it has been shown that this activity is not responsible for inhibition of protein synthesis (18). On the basis of these results, YoeB and RelE may be classified as ribosome-dependent mRNA interferases (16). To explore the enzymatic function of YoeB homologues in a clinically significant gram-positive species, we attempted to characterize YoeB homologues from Staphylococcus aureus. Here, we describe that YoeB-sa1 and YoeB-sa2 from S. aureus function as translation initiation inhibitors in an identical manner to that of YoeB-ec from E. coli.
S. aureus is the most significant nosocomial pathogen, and methicillin-resistant S. aureus strains, which caused approximately 19,000 deaths in 2005 in the United States (2, 11), have recently emerged in the community, causing an inordinate number of skin and soft tissue infections in otherwise healthy populations (13, 15).
In contrast to the large array of toxin-antitoxin systems in E. coli and Mycobacterium tuberculosis (1, 12), a BLAST search identified one mazE-mazF-like operon and two yefM-yoeB-like operons in the S. aureus genome (8, 12). A MazF homologue in S. aureus (MazF-sa) has been shown to be an mRNA interferase (7), and recently its pentad recognition cleavage sequence has been determined to be UACAU (22). Its unusual abundance (43 cleavage sites) in the gene sraP, which is one of the crucial adhesive factors involved in adhesion of this pathogen to human platelets, suggests that MazF-sa may be involved in the pathogenicity of this bacterium (22). These yefM-yoeB operons in S. aureus (yefM-yoeB-sa1 and yefM-yoeB-sa2), previously identified as the axe1-txe1 and axe2-txe2 operons, respectively, were shown to be autoregulated and their transcriptions were reported to be stimulated by the addition of antibiotics, such as erythromycin and tetracycline (6).
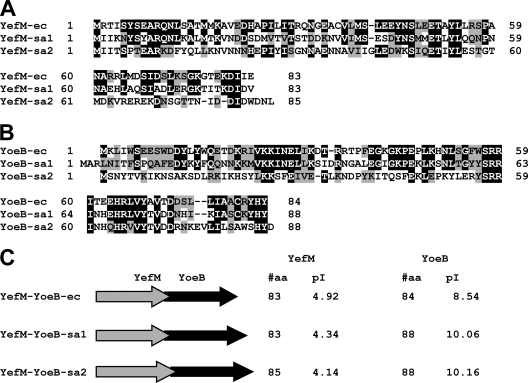
As shown in Fig. 1A, both YefM-sa1 and YefM-sa2 show significant identity (42 and 25%) and homology (57 and 46%) to YefM-ec from E. coli. Similarly, both YoeB-sa1 and YoeB-sa2 also show high identity (52 and 25%) and homology (76 and 44%) to YoeB-ec from E. coli (Fig. 1B). Interestingly, the values for YoeB-sa1 and YoeB-sa2 are higher than those for RelE, a ribosome-dependent transcription initiation inhibitor in E. coli. The identity and homology of RelE to YoeB-ec are 17% and 35%, respectively. Notably, in both operons, the yefM genes and the yoeB genes overlap by 1 base with their respective yoeB genes, yoeB-sa1 and yoeB-sa2 (Fig. 1C). YefM-sa1 consists of 83 amino acid residues, identical to the YefM-ec gene, and YefM-sa2 consists of 85 amino acid residues (Fig. 1C). The pI values of both antitoxins are acidic, like YefM-ec. Similarly, the sizes of YoeB toxins from S. aureus (both 88 residues) are close to that of YoeB-ec (84 residues), and their pI values are basic, as is that of YoeB-ec (Fig. 1C).
FIG. 1.
Sequence alignments of YoeB and YefM homologues from S. aureus with YoeB and YefM from E. coli. (A) Alignment of YefM-sa1 and YefM-sa2 with E. coli YefM-ec. Identical and homologous residues are shown in black and shaded backgrounds, respectively. Identities and homologies of these proteins are as follows: between YefM-ec and YefM-sa1, identity of 42% (35/83) and homology of 57% (47/83); between YefM-ec and YefM-sa2, identity of 25% (21/83) and homologyof 46% (38/83); between YefM-sa1 and YefM-sa2, identity of 25% (21/83) and homology of 47% (39/83). (B) Alignment of YoeB-sa1 and YoeB-sa2 with YoeB-ec from E. coli. Identity and homology of these proteins were as follows: between YoeB-ec and YoeB-sa1, identity of 52% (44/84) and homology of 76% (64/84); between YoeB-ec and YoeB-sa2, identity of 25% (21/84) and homology of 44% (37/84); between YoeB-sa1 and YoeB-sa2, identity of 30% (26/88) and homology of 45% (40/88). (C) All three yefM genes overlap by 1 base with the downstream yoeB genes. Total residue numbers and pI values are shown on the right.
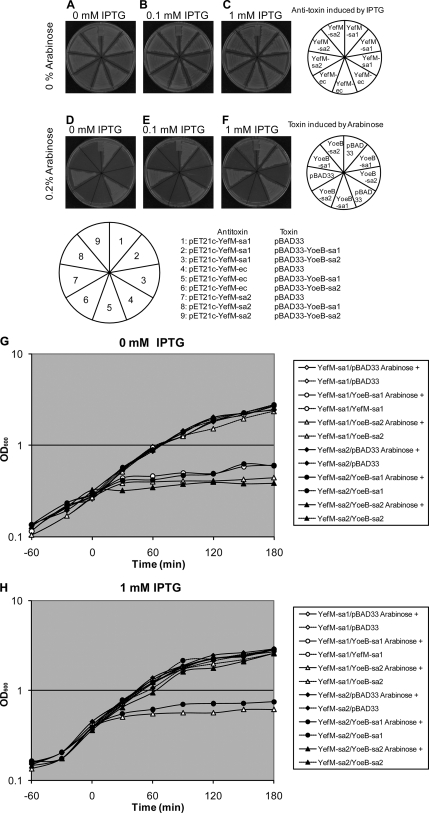
yefM-sa1 and yefM-sa2 from S. aureus ATCC 25923 were cloned in the pET21c plasmid (Novagen, Inc., WI), and yoeB-sa1 and yoeB-sa2 were cloned in pBAD33. These plasmids were designated as pET21c-YefM-sa1, pPET21c-YefM-sa2, pBAD33-YoeB-sa1, and pBAD33-YoeB-sa2, respectively. The DNA sequences of these genes were confirmed to be identical to those of SA2196-SA2195 and SA2246-SA2245 of S. aureus strain N315. The yefM and yoeB genes from E. coli were also cloned into pET21c or pBAD33 as previously reported (18) and designated as pET21c-YefM-ec and pBAD33-YoeB-ec. E. coli C43(DE3) cells were cotransformed with different inducible plasmids for the toxin and antitoxin and streaked on M9 agar plates supplemented with 0.2% Casamino Acids and 0.2% glycerol, with or without 0.2% arabinose and/or 0, 0.1, or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated at 37°C for 14 h (Fig. 2A to F). Cotransformants of pET21c-YefM-sa and pBAD33-YoeB-sa could not form colonies in the presence of 0.2% arabinose without IPTG (Fig. 2D, sections 2 and 9). When YefM-sa1 was induced by 0.1 mM IPTG, colony formation was recovered only for YoeB-sa1 (Fig. 2E, section 2). When the IPTG concentration was increased to 1 mM, both YoeB-sa1 and YoeB-sa2 toxicities were attenuated (Fig. 2F, sections 2 and 9). Even if there was substantial homology in both pairs of these toxins and antitoxins (Fig. 1A and B), no neutralization was observed with heterogeneous antitoxins from S. aureus (Fig. 2D, E, and F, sections 3 and 8) and those from E. coli (Fig. 2D, E, and F, sections 5 and 6 for YoeB-sa1 and YoeB-sa2, respectively). The growth of cells with pBAD33-YoeB-sa2 was less robust compared with other transformants when the cells did not carry pET21c-YefM-sa2 (Fig. 2A, B, and C, sections 3, 6, and 9). Restoration of cell growth seemed to be incomplete even with 1 mM IPTG in the plates (Fig. 2F, section 9). The cotransformants with pPET21c-YefM-sa and pBAD33-YoeB-sa were also incubated in LB medium with (Fig. 2H) or without (Fig. 2G) 1 mM IPTG from time −30 min and with or without 0.2% arabinose from time zero. When only YoeB-sa was induced with arabinose, clear growth inhibition was observed in cells carrying either pBAD33-YoeB-sa1 or pBAD33-YoeB-sa2 (Fig. 2G). In the case of YefM-sa induction with IPTG, cell growth reached a level equivalent to that for cells without pBAD33-YoeB-sa (Fig. 2H). No neutralization was observed with heterogeneous antitoxins from S. aureus (Fig. 2H,YefM-sa1/YoeB-sa2 and YefM-sa2/YoeB-sa1), as seen in the complementation assay on M9 plates.
FIG. 2.
Induction of YoeB-sa and YefM-sa in E. coli. (A to F) The columns of the plates show different concentrations of IPTG (0 [A and D], 0.1 [B and E], and 1 mM [C and F]), which induces the expression of antitoxins YefM-sa1 and YefM-sa2 from S. aureus and YefM-ec from E. coli; the rows of the plates show different concentrations of arabinose (0 [A, B, and C] and 0.2% [D, E, and F]), which induces the expression of the toxins YoeB-sa1 and YoeB-sa2 from S. aureus. (G and H) Growth curves of E. coli C43(DE3) cotransformed with pPET21c-YefM-sa1 or pBAD33-YoeB-sa1 (○), pPET21c-YefM-sa1 or pBAD33-YoeB-sa2 (▵), pPET21c-YefM-sa1 or pBAD33 (⋄), pPET21c-YefM-sa2 or pBAD33-YoeB-sa1(•), pPET21c-YefM-sa2 or pBAD33-YoeB-sa2 (▴), and pPET21c-YefM-sa2 or pBAD33 (⧫). Cells were incubated with (H) or without (G) 1 mM IPTG from −30 min and with (solid lines) or without (broken lines) 0.2% arabinose from time zero.
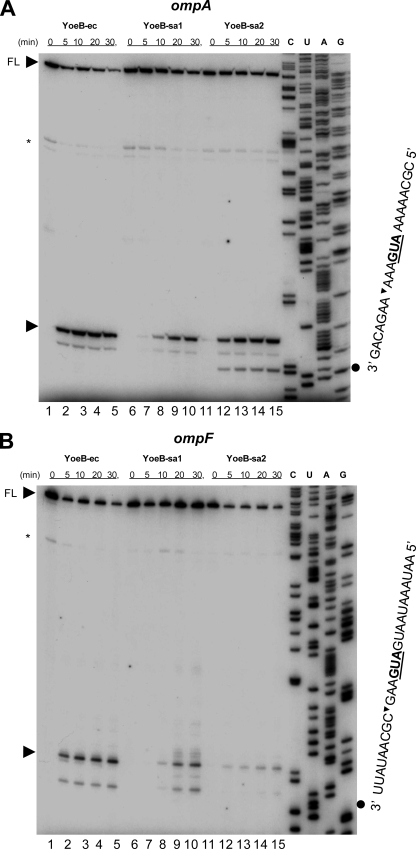
To elucidate the mechanism of action of YoeB-sa1 and YoeB-sa2, we carried out the in vivo primer extension experiments to identify the cleavage sites of the chromosomally encoded ompA and ompF mRNA in the same manner as for YoeB-ec (18). For the primer extension in vivo, pBAD33-YoeB-sa1, pBAD33-YoeB-sa2, and pBAD33-YoeB-ec were transformed into E. coli BW25113 cells. yoeB was induced by addition of arabinose (final concentration, 0.2%), and then total RNA was isolated by the hot phenol method at the time intervals indicated in Fig. 3 (18, 24). Primer extension was carried out at 47°C for 1 h in 20 μl of the reaction mixture containing 15 μg of the total RNA, 10 mM deoxynucleoside triphosphates, 10 U avian myeloblastosis virus reverse transcriptase (Roche Diagnostic GmbH, Mannheim, Germany) using the supplemented reaction buffer containing 0.8 U of RNase inhibitor (Roche) as described previously (22, 23). The reactions were stopped by adding 6 μl of sequence loading buffer (95% deionized formamide, 10 mM EDTA, 10 mM NaOH, 0.05% bromophenol blue, and 0.05% xylene cyanol). The samples were incubated at 90°C for 2 min prior to electrophoresis on a 6% polyacrylamide gel containing 8 M urea. The following primers were 5′-labeled with [γ-32P]ATP using T4 polynucleotide kinase: for ompA, 5′-TGT ACC AGG TGT TAT CTT TC-3′, and for ompF, 5′-TTG CCA TCT TTG TTA TAG AT-3′. The sequence ladders shown in Fig. 3 were obtained using the same primers as for the primer extension, with their corresponding genes cloned in pCR2.1-TOPO as the templates (Invitrogen, Carlsbad, CA) as described previously (18). In the reaction mixtures containing YoeB-sa1 (Fig. 3, lanes 6 to 10) or YoeB-sa2 (lanes 11 to 15), a major cleavage band appeared 3 bases downstream of the initiation codon AUG, as also seen in the reaction mixture containing YoeB-ec (lanes 1 to 5). The intensity of the major bands for both ompA (Fig. 3A) and ompF (Fig. 3B) increased from 5 to 30 min after the induction of both YoeB-sa1 and YoeB-sa2. The intensity at 5 min after induction of YoeB-sa2 was higher than that of YoeB-sa1 in both ompA and ompF mRNA (Fig. 3A and B, lane 12); however, waiting 20 to 30 min following induction produced the opposite finding, as the higher-intensity bands of major products were observed in YoeB-sa1 in ompF mRNA (Fig. 3B, lanes 9, 10, 14 and 15). In the case of YoeB-ec, the major band intensities were saturated at 10 to 20 min after its induction as previously reported (18). Minor bands appearing in the 5′-untranslated region under the full-length mRNAs are due to the secondary structures in the mRNAs, as the same minor bands were observed even before induction of YoeB-ec, YoeB-sa1, and YoeB-sa2 (Fig. 3A and B, lanes 1, 6, and 11). All minor bands downstream of the major band with YoeB-sa1 and YoeB-sa2 were also observed with YoeB-ec, except for the band with YoeB-sa2 and the ompA mRNA at 9 bases downstream of the initiation codon (Fig. 3A, lanes 12 to 15). It is not clear at present why this band was formed only with YoeB-sa2. It should be noted that with both YoeB-sa1 and YoeB-sa2, the intensities of the bands representing the full-length ompA (Fig. 3A) and ompF (Fig. 3B) mRNAs were constant throughout the reaction. In contrast, with YoeB-ec, the intensity of the full-length bands for both ompA (Fig. 3A) and ompF (Fig. 3B) decreased while the intensity of the band downstream of the initiation codon increased. These results indicate that YoeB- ec appears to have strong ribosome-dependent mRNA interferase activity in comparison with YoeB toxins from S. aureus.
FIG. 3.
In vivo primer extension. (A) Primer extension analysis of the ompA mRNA. (B) Primer extension analysis of the ompF mRNA. Total RNAs were extracted at different time points after arabinose induction as shown. The major cleavage site is indicated by an arrowhead on the left side of the respective gel. The sequences of major cleavage sites in ompA (A) and ompF (B) are shown on the right side of the respective gel. On the top of the gels, the full-length RNA bands are indicated with arrowheads and the FL notation.
In summary, both YoeB toxins from S. aureus, YoeB-sa1 and YoeB-sa2, may primarily inhibit translation initiation, as shown with E. coli YoeB-ec (18). The actual roles and functions of these toxins in S. aureus remain to be elucidated, and additional work is needed in the strains of this pathogen in which the toxin-antitoxin system has been deleted.
Acknowledgments
We thank Sangita Phadtare for critical reading of the manuscript.
This work was partially supported by NIH grant 1RO1GM081567-01A2 and a research fund from Takara-Bio, Inc., Japan.
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Brown, J. M., and K. J. Shaw. 2003. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 1856600-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camargo, I. L., and M. S. Gilmore. 2008. Staphylococcus aureus—probing for host weakness? J. Bacteriol. 1902253-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen, S. K., G. Maenhaut-Michel, N. Mine, S. Gottesman, K. Gerdes, and L. Van Melderen. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 511705-1717. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, S. K., and K. Gerdes. 2003. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 481389-1400. [DOI] [PubMed] [Google Scholar]
- 5.Christensen-Dalsgaard, M., and K. Gerdes. 2008. Translation affects YoeB and MazF messenger RNA interferase activities by different mechanisms. Nucleic Acids Res. 366472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donegan, N. P., and A. L. Cheung. 2009. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 1912795-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu, Z., N. P. Donegan, G. Memmi, and A. L. Cheung. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 1898871-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 471419-1432. [DOI] [PubMed] [Google Scholar]
- 9.Hayes, C. S., and R. T. Sauer. 2003. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell 12903-911. [DOI] [PubMed] [Google Scholar]
- 10.Kamada, K., and F. Hanaoka. 2005. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19497-509. [DOI] [PubMed] [Google Scholar]
- 11.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 12.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel, M. 2009. Community-associated methicillin-resistant Staphylococcus aureus infections: epidemiology, recognition and management. Drugs 69693-716. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112131-140. [DOI] [PubMed] [Google Scholar]
- 15.Wiersma, P., M. T. D'Angelo, W. R. Daley, J. Tuttle, K. E. Arnold, S. M. Ray, J. L. Ladson, S. N. Bulens, and C. L. Drenzek. 15 April 2009, posting date. Surveillance for severe community-associated methicillin-resistant Staphylococcus aureus infection. Epidemiol. Infect. [Epub ahead of print.] doi: 10.1017/S0950268809002490. [DOI] [PubMed]
- 16.Yamaguchi, Y., and M. Inouye. 2009. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 85467-500. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, J., Y. Zhang, L. Zhu, M. Suzuki, and M. Inouye. 2004. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 27920678-20684. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, Y., and M. Inouye. 2009. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 2846627-6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, Y., J. Zhang, H. Hara, I. Kato, and M. Inouye. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2803143-3150. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12913-923. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, Y., L. Zhu, J. Zhang, and M. Inouye. 2005. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 28026080-26088. [DOI] [PubMed] [Google Scholar]
- 22.Zhu, L., K. Inoue, S. Yoshizumi, H. Kobayashi, Y. Zhang, M. Ouyang, F. Kato, M. Sugai, and M. Inouye. 2009. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 1913248-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu, L., S. Phadtare, H. Nariya, M. Ouyang, R. N. Husson, and M. Inouye. 2008. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu, L., Y. Zhang, J. S. Teh, J. Zhang, N. Connell, H. Rubin, and M. Inouye. 2006. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 28118638-18643. [DOI] [PubMed] [Google Scholar]