Abstract
The opportunistic pathogen Pseudomonas aeruginosa utilizes two interconnected acyl-homoserine lactone quorum-sensing (acyl-HSL QS) systems, LasRI and RhlRI, to regulate the expression of hundreds of genes. The QS circuitry itself is integrated into a complex network of regulation by other factors. However, our understanding of this network is still unlikely to be complete, as a comprehensive, saturating approach to identifying regulatory components has never been attempted. Here, we utilized a nonredundant P. aeruginosa PA14 transposon library to identify additional genes that regulate QS at the level of LasRI/RhlRI. We initially screened all 5,459 mutants for loss of function in one QS-controlled trait (skim milk proteolysis) and then rescreened attenuated candidates for defects in other QS phenotypes (LasA protease, rhamnolipid, and pyocyanin production) to exclude mutants defective in functions other than QS. We identified several known and novel genes, but only two novel genes, gidA and pcnB, affected all of the traits assayed. We characterized gidA, which exhibited the most striking QS phenotypes, further. This gene is predicted to encode a conserved flavin adenine dinucleotide-binding protein involved in tRNA modification. Inactivation of the gene primarily affected rhlR-dependent QS phenotypes such as LasA, pyocyanin, and rhamnolipid production. GidA affected RhlR protein but not transcript levels and also had no impact on LasR and acyl-HSL production. Overexpression of rhlR in a gidA mutant partially restored QS-dependent phenotypes. Taken together, these results indicate that GidA selectively controls QS gene expression posttranscriptionally via RhlR-dependent and -independent pathways.
Pseudomonas aeruginosa is a ubiquitous environmental bacterium commonly found in soil and freshwater. It is also an opportunistic human pathogen that infects immunocompromised individuals, including those suffering from cystic fibrosis. It utilizes a quorum-sensing (QS) mechanism to regulate and coordinate virulence gene expression (2, 11, 37, 46). There are two complete acyl-homoserine lactone (acyl-HSL) QS systems in P. aeruginosa, the LasR-LasI (las) system and the RhlR-RhlI (rhl) system. LasI and RhlI produce the signals 3-oxo-dodecanoyl (3OC12)-HSL and butanoyl (C4)-HSL, respectively; these signals bind to and activate their cognate transcriptional regulators, LasR and RhlR. There is a third, orphan regulator, QscR, that also responds to 3OC12-HSL (24). Under standard growth conditions, the las system activates the rhl system (23, 36), and the two systems together control the expression of hundreds of genes (12, 42, 51). A large portion of these QS-controlled genes encode secreted virulence factors such as proteases (LasA protease, LasB elastase, and alkaline protease), biosurfactants (rhamnolipid), and secondary metabolites (hydrogen cyanide and pyocyanin).
P. aeruginosa QS is embedded in a complex network of global regulation (18, 41, 49). Most QS-controlled genes are not induced until the stationary phase of growth even when exogenous acyl-HSL signals are present early in growth (8, 42, 53, 54). Thus, additional factors are required for expression of these QS-controlled genes. Several global regulators and regulatory pathways (e.g., Vfr, GacA/GacS, RsaL, RelA, RpoS, and PqsR [MvfR]/PQS) have already been identified (18, 41). Although much remains to be learned about this complex network of global regulation, it is clear that control of QS is multifaceted and will require an integrative approach to understanding its complexity.
In this study, we employed a saturation mutagenesis approach, making use of a recently constructed nonredundant transposon mutant library of P. aeruginosa strain PA14 (25), to identify additional regulators of QS gene expression. We characterized one such novel regulator, GidA, which shares homology with conserved flavin adenine dinucleotide-binding proteins involved in tRNA modification (27, 55). Initially isolated as a factor involved in a glucose-inhibited cell division phenotype in Escherichia coli (50), more recent studies have established that GidA, together with MnmE, is responsible for the addition of a carboxymethylaminomethyl group to uridine 34 of a subset of tRNAs (55). This process is important for the appropriate decoding of two-family box triplet codons. Despite the potential to indiscriminately affect cellular gene expression, GidA homologs rather specifically regulate virulence gene expression in Aeromonas hydrophila (45), Pseudomonas syringae (21), Shigella flexneri (9), and Streptococcus pyogenes (5), with little to no impact on growth. In Myxococcus xanthus, GidA is involved in fruiting-body development (52). Our data show that GidA specifically controls rhl-dependent QS gene expression in P. aeruginosa by modulating RhlR expression posttranscriptionally.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains used in this study are listed in Table 1. The MAR2 × T7 mutants used in the study were taken from a subset of the parental PA14 mariner transposon insertion library (25). The mutants were handled and stored as described in the library's user manual (25). Strains were initially plated from frozen stocks onto Luria-Bertani (LB) agar plates and grown for 24 h at 37°C. Unless noted otherwise, P. aeruginosa liquid cultures were grown at 37°C with agitation (250 rpm) in Lennox LB broth buffered with 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.0. Escherichia coli DH5α (Invitrogen, CA) was cultured in LB at 37°C. Where appropriate, the antibiotics gentamicin and tetracycline (10 μg/ml for E. coli and 100 μg/ml for P. aeruginosa) were added to the growth media.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PA14 | Wild type | 25 |
| PA14 gidA7 | MAR2 × T7 gidA mutant derived from PA14 (mutant 44643) | 25 |
| PA14 lasR | TnphoA lasR mutant derived from PA14 | 47 |
| PA14 rhlR | MAR2 × T7 rhlR mutant derived from PA14 (mutant 37943) | 25 |
| PA14_73400 | MAR2 × T7 mnmE mutant derived from PA14 (mutant 38726) | 25 |
| Additional MAR2 × T7 mutants | See Table S1 in the supplemental material and http://ausubellab.mgh.harvard.edu/cgi-bin/pa14/home.cgi | 25 |
| PA14 ΔgidA | Markerless in-frame deletion mutant derived from PA14 | This study |
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| Plasmids | ||
| pCF430 | Broad-host-range vector containing araC-PBAD promoter (tetracycline resistant) | 28 |
| pCF430G | gidA open reading frame and ribosome-binding site of PA14 in HindIII/XbaI of pCF430 | This study |
| pCF430R | NheI/SacI fragment containing rhlR open reading frame and ribosome-binding site of PAO1 subcloned from pJN105.rhlR | This study |
| pEX19Tc | Gene replacement vector with sacB counterselectable marker (tetracycline resistant) | 15 |
| pEX19Tc.ΔgidA | pEX19Tc containing gidA with a markerless in-frame deletion | This study |
For high-throughput assays, colonies grown on rectangular LB plates in a 12-by-8 format were replicate plated using a 96-pin replicator. Subsequent assays were either plate based or liquid based (see below). For the latter, 600 μl of LB-MOPS in 96-deep-well titer blocks were inoculated from plates and grown for 18 h. For liquid culture assays performed with few individual strains, including assays for LasA protease activity, pyocyanin production, Western blotting, 3OC12-HSL and C4-HSL quantitation, and real-time PCR, experimental P. aeruginosa cultures were started by inoculating 25 ml LB-MOPS in 250-ml Erlenmeyer flasks with mid-logarithmic-phase bacteria to an optical density at 600 nm (OD600) of 0.02. Culture aliquots were harvested after 7 to 8 h of growth (stationary phase). For real-time PCR analysis, culture aliquots were also harvested after 4 to 5 h of growth (transition from logarithmic to stationary phase). The culture supernatants were filter sterilized (pore size, 0.2 μm) for individual phenotypic assays.
For complementation analysis, LB-MOPS medium containing l-arabinose (50 mM final concentration) and tetracycline was inoculated with a colony from a freshly streaked culture. The cultures were grown for 24 h (stationary phase) prior to analysis.
DNA manipulations.
General molecular cloning techniques were based on standard protocols (38). Chromosomal DNA was isolated using the Puregene Core Kit A (Gentra Systems). Plasmids used for complementation were constructed as follows. A 1,893-bp DNA fragment containing the gidA coding sequence was amplified from PA14 chromosomal DNA using primers 5′-N6AAGCTTCGCTAAATCCTTATACTGTCCG-3′ and 5′-N6TCTAGAGGTGTTGGGTTACCGCAGAC-3′. The fragment was then digested with HindIII and XbaI (the respective restriction enzyme recognition sites are underlined) and ligated to HindIII/XbaI-digested pCF430 to generate pCF430G (Table 1). Proper construction was verified by DNA sequencing. Plasmid pCF430R (Table 1) was generated by subcloning a 700-bp NheI/SacI fragment from pJN105.rhlR (40).
The location of the transposon (MAR2 × T7) insertion in the PA14 gidA mutant was confirmed by PCR and subsequent agarose gel electrophoresis. Primers were designed based on the presumed location of the transposon 1,716 bp downstream of the gidA translational start (25). Primers 5′-CATTACAGTTTACGAACCGAACAG-3′ (forward) and 5′-GCAAGGCAAGGACAGCTGGTG-3′ (reverse) amplified a fragment of the appropriate size (661 bp in length; 179 bp of MAR2 × T7 and 482 bp of gidA).
Construction of a gidA deletion mutant.
The gene deletion strategy of Hoang et al. was used to construct a markerless gidA deletion mutant of P. aeruginosa strain PA14 (15, 44). Splicing by overlap extension-PCR was used to construct a 981-bp in-frame deletion in gidA. The resulting protein contains the first 14 amino acids fused in frame with the last 289 amino acids. The following primers were used (sequences in the 5′ to 3′ direction): primer 1, N6TCTAGACATGGGATGGATATCCACCTGG; primer 2, ACCGCCGATCACGATTACGTC; primer 3, GACGTAATCGTGATCGGCGGTGCGATCGAGTACGACTTCTTC; and primer 4, N6AAGCTTCCATTTGATCAGCAGGGCCAAG. Primers 1 and 4 are flanking primers containing engineered XbaI and HindIII restriction sites, respectively (underlined), and primers 2 and 3 are partially overlapping, internal primers to create the in-frame deletion. The fusion product was digested with XbaI and HindIII and ligated to the equally digested allelic exchange plasmid pEX19Tc (15). The resulting construct, pEX19Tc.ΔgidA, was sequenced prior to transformation into E. coli SM10. Mating with P. aeruginosa PA14 and appropriate selection steps yielded a gidA deletion mutant. Proper construction was confirmed by PCR amplification of chromosomal DNA.
Growth curves.
Growth analysis of each strain listed in Table S1 in the supplemental material was conducted simultaneously in a 96-well format using a microplate reader (Infinite M200; Tecan). Strains were grown at 37°C with shaking in 200 μl of LB-MOPS medium and sealed with 50 μl of mineral oil. The OD600 was determined every 15 min for 8 h, and the doubling time in exponential phase for each strain was calculated with the respective data. The presence of mineral oil did not restrict growth during logarithmic phase (data not shown). Growth of PA14 gidA::MAR2 × T7 and the PA14 wild-type parent was also quantified in standard culture flasks. Twenty-five milliliters of LB-MOPS medium (in 250-ml flasks) was inoculated with mid-logarithmic-phase cells to an OD600 of 0.02 and incubated at 37°C with shaking (250 rpm). The OD600 was measured every hour for 8 h.
Skim milk proteolysis, rhamnolipid production, and adenosine utilization.
Skim milk proteolysis (39), rhamnolipid production (22) and adenosine utilization (13, 39) were determined through the use of agar plate assays. Isolated colonies, freshly grown on LB plates, were patched individually with a toothpick or replica plated onto the respective agar test plates. Rhamnolipid plates were incubated at 37°C for 24 h, followed by 24 h at 4°C or until a blue halo appeared. Relative rhamnolipid production was determined by the size of the blue halo around each colony. Skim milk plates contained not only 4% (wt/vol) skim milk but also one-quarter-strength LB. The addition of LB allowed QS-deficient mutants to grow as well as the wild-type parent.
Pyocyanin production and LasA protease activity.
Pyocyanin production was measured in two different ways. For high-throughput analysis, 200 μl of culture supernatant was transferred to a 96-well microtiter plate and the OD was determined at 310 nm with a microplate reader (31). For accurate quantitation of QS-dependent products from individual cultures grown in flasks, pyocyanin was extracted from liquid culture supernatant and absorbance was read at 520 nm as described previously (10). LasA protease activity was determined by measuring the rate of Staphylococcus aureus cell lysis by P. aeruginosa culture supernatant (decrease in OD600 per minute) (8, 20). Twenty microliters of culture supernatant was transferred to microtiter plates containing 180 μl of boiled S. aureus suspension (OD600 of 0.8) buffered with 10 mM K2HPO4 (pH 7.5). OD600 readings were taken in a microplate reader every 4 min for 1 h.
Assays for 3OC12-HSL and C4-HSL.
Two milliliters of ethyl acetate-extracted P. aeruginosa culture supernatant was used to measure 3OC12-HSL and C4-HSL levels with E. coli bioassays as previously described (32-34). Synthetic 3OC12-HSL and C4-HSL were used to generate standard curves.
Western blot analysis.
Western blotting was performed as described previously (40). Culture aliquots were harvested by centrifugation. Cell pellets were resuspended in LasR protein buffer and sonicated. The resulting lysates were centrifuged to remove insoluble material. Protein concentrations in the soluble fraction were determined by Bradford assay. Equal amounts of protein were separated by 12.5% polyacrylamide gel electrophoresis. The separated proteins were blotted onto a nylon membrane and probed with polyclonal anti-LasR and anti-RhlR rabbit antibodies as described previously (40).
Complementation and suppression analysis.
Complementation and suppression of the PA14 gidA transposon mutant was carried out with the low-copy-number plasmids pCF430G and pCF430R, which express gidA and rhlR from the arabinose-inducible araBAD promoter. l-Arabinose was added to the cultures at a concentration of 50 mM. The rhlR allele was from P. aeruginosa PAO1 rather than PA14. Both alleles differ by only a single silent mutation at position 717; however, the affected codon is not in a mixed-codon family box and is therefore not recognized by a GidA-modified tRNA.
Real-time PCR.
Total RNA isolation and cDNA synthesis with semirandom primers were performed as described previously (42). RNA quality was determined using an Agilent Bioanalyzer 2100. Real-time PCR was performed as described previously (40) with a 7300 real-time PCR system (Applied Biosystems) using specific primers for rhlA and rhlR, 1 ng of purified cDNA as measured by Nanodrop (Thermo Scientific), and Power SYBR green PCR master mix reagents (Applied Biosystems). The primers were designed using Primer Express software (Applied Biosystems). Serially diluted genomic DNA was used as the template to obtain a relative standard curve. Instead of normalizing transcript levels to those of a calibrator gene, as is commonly done in one-step real-time reverse transcription-PCRs, we chose a two-step reaction and calibrated the amount of input cDNA prior to PCR (39). Thus, data are normalized to the total amount of cDNA.
RESULTS
High-throughput screening for QS regulators identifies gidA.
We used a P. aeruginosa PA14 nonredundant transposon insertion library (25) to screen for mutants deficient in QS-dependent phenotypes. The library is a collection of 5,459 mutant strains where virtually every strain has a unique gene disabled by transposon insertion. The library allowed us to perform a comprehensive genome-wide search for genes that regulate QS in P. aeruginosa. To identify global regulators that affect multiple QS-regulated traits, we employed a two-step screening procedure. Screen 1 involved screening of all mutants for defects in skim milk proteolysis. Screen 2 involved screening the proteolysis-deficient mutants identified in screen 1 for additional QS-controlled phenotypes, namely, staphylolytic (LasA) activity, rhamnolipid production, and pyocyanin production. This second, more focused screen allowed us to eliminate mutants from screen 1 that are defective in processes other than QS, such as protease production or protease secretion. The different QS phenotypes assayed are dependent on the LasRI and/or RhlRI systems to various degrees (see below).
Screen 1 identified 59 mutants with reduced skim milk proteolysis (see Table S1 in the supplemental material). This number excludes mutants with severely reduced growth on skim milk plates and mutants whose doubling time in liquid LB culture, assayed in microtiter plate format, was >30% of that of the PA14 wild type. We rediscovered several genes known to control QS in P. aeruginosa PAO1. These were vfr, gacA, rpoN, dksA, in addition to the QS core genes lasRI and rhlRI. Vfr is a catabolite repressor homolog that directly induces lasR transcription (1). GacA is part of a regulatory pathway that controls QS posttranscriptionally (14, 19). RpoN is an alternative sigma factor that regulates rhlI expression (48), and DksA also controls expression of several QS-dependent phenotypes posttranscriptionally (17). Several other known QS regulatory genes were not rediscovered (see Table S1 in the supplemental material) but were nevertheless included in the second screen for comparison. Screen 2 revealed that many of the 59 mutants were also deficient in additional QS-controlled phenotypes (see Table S1 in the supplemental material). However, only three mutants were at least partially deficient in all of the QS phenotypes tested (several others were defective in skim milk proteolysis, LasA activity, and pyocyanin production but did not grow on rhamnolipid minimal medium, suggesting a QS-independent pleiotrophic defect such as amino acid auxotrophy). The three mutants harbor transposon insertions in vfr and in two genes not previously known to be involved in QS regulation, pcnB and gidA. pcnB encodes poly(A) polymerase, which is involved in destabilization of mRNA in bacteria, a property that in some cases can directly regulate gene expression (16). In this study, we chose the gidA mutant for further characterization as it exhibited the most compelling QS phenotypes, being completely deficient in three of the four traits assayed. We confirmed the location of the transposon insertion in the gidA mutant by PCR as described in Materials and Methods.
Previous work indicated a role for gidA in virulence gene regulation in several bacterial pathogens (5, 9, 21, 45). A recent mechanistic study demonstrated its involvement in tRNA modification in association with mnmE in E. coli (4). To investigate the contribution of mnmE to the observed QS phenotypes in P. aeruginosa, we attempted to characterize a PA14 mutant with an insertion in a gene that is 67% identical to E. coli mnmE. However, this mutant exhibited severe growth defects in all of the standard media employed, thus not allowing us to investigate QS-specific deficiencies.
A gidA mutant is defective primarily in rhl-dependent QS.
As mentioned above, high-throughput screening revealed that gidA causes defects in several QS-dependent phenotypes. To confirm and extend the results from high-throughput screening, we repeated assays for the gidA mutant and compared the results to those for the wild type, a lasR mutant, and a rhlR mutant. We also included a new assay, growth on adenosine, to better determine the relative contributions of las and rhl QS systems in gidA-mediated regulation. Utilization of adenosine as the sole carbon source requires expression of nucleoside hydrolase (Nuh), which is dependent solely on the las system (13, 39). We found no adenosine utilization defect in the gidA mutant (Fig. 1A), indicating that las QS is not affected by gidA. Consistent with the high-throughput data, the gidA mutant also had only a modest defect in skim milk proteolysis (Fig. 1C). Skim milk proteolysis is primarily las dependent (involving las-specific alkaline protease and las- and rhl-dependent LasB elastase), because a rhlR mutant exhibited only a slight proteolytic deficiency. However, the gidA mutant was more strongly affected in two different rhl-specific QS phenotypes, rhamnolipid and pyocyanin production (Fig. 1B and D). LasA protease expression, which depends on both las and rhl systems for full activity, was also severely affected in the gidA mutant (Fig. 1E). We define a phenotype or gene as las specific if it is affected in a lasR (or lasI) mutant but not in a rhlR (or rhlI) mutant. On the other hand, we define a phenotype or gene as rhl specific if it is affected in a rhlR (or rhlI) mutant and it is affected to the same degree in a lasR (or lasI) mutant. Under most growth conditions, a rhl-specific trait is equally affected in a lasR (or lasI) mutant because the las system controls expression of the rhl system. However, under some conditions, for example, those employed for the rhamnolipid assay, the rhl system is independent of the las system, and a lasR (or lasI) mutant displays a wild-type phenotype.
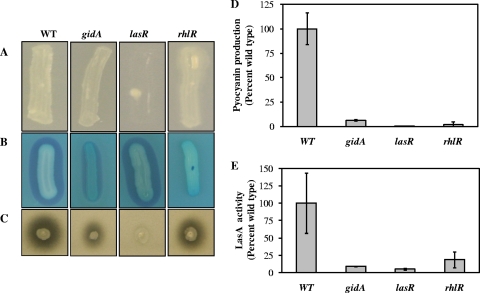
FIG. 1.
QS-dependent phenotypes of a P. aeruginosa PA14 gidA mutant. Assays were performed with the wild type (WT) and the gidA, lasR, and rhlR mutants. (A) Growth on adenosine. (B) Rhamnolipid production. The presence of a blue halo surrounding the bacterial streak indicates rhamnolipid production. (C) Skim milk proteolysis. Zones of clearance surrounding the bacterial streak indicate proteolytic activity. (D) Pyocyanin production, expressed as percentages of the wild-type level. (E) LasA activity, expressed as percentages of the wild-type level. Each experiment was performed at least three times. Plate images depict representative experiments. Values shown in graphs are means from three independent biological experiments, normalized to culture density (OD600). Error bars indicate ±1 standard deviation.
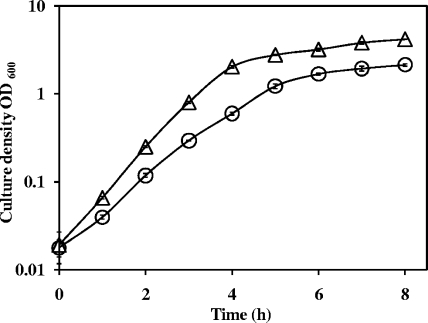
Individual growth curves showed that the gidA mutant was attenuated in logarithmic growth compared with the wild type (doubling times of 45 and 36 min, respectively) and that it entered stationary phase at a slightly lower density (Fig. 2). While the gidA mutant grew more slowly, this relatively small difference alone is not sufficient to explain the dramatic QS phenotypes observed. In addition, these growth differences were taken into account for all assays performed in liquid culture. Taken together, our phenotypic data indicate that the gidA mutant is deficient primarily in rhl-dependent QS phenotypes.
FIG. 2.
Growth of the P. aeruginosa PA14 gidA mutant (circles) and the wild-type parent (triangles). Values are means from three biologically independent experiments. Error bars indicate ± 1standard deviation.
Complementation fully restores the wild-type phenotype in a gidA mutant.
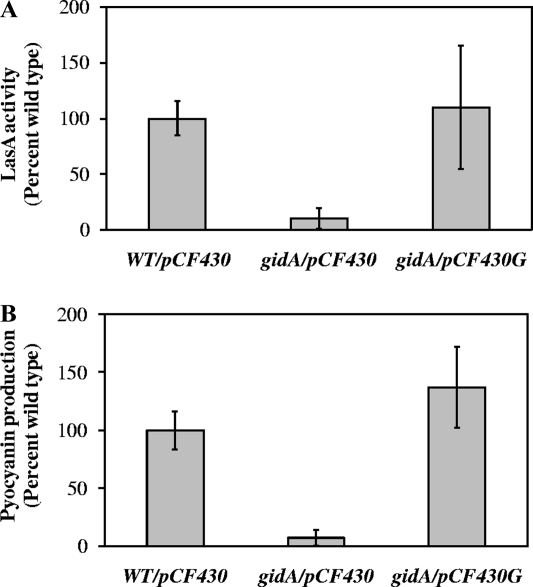
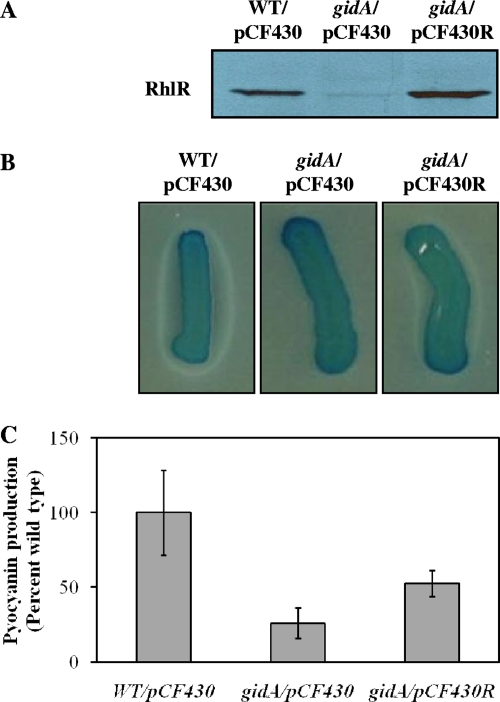
We performed a complementation analysis to exclude the possibility of polar effects of the transposon insertion. This was important because gidA is the first gene in a four-gene operon and because two genes in the operon, soj and spo0J, are involved in chromosome partitioning and cell division. Introduction of an intact copy of the gidA gene in the mutant restored QS-dependent phenotypes to wild-type levels (Fig. 3). This indicates that QS-deficient phenotypes conferred by the transposon insertion in the gidA gene are solely due to loss of function of gidA itself.
FIG. 3.
Restoration of the wild-type phenotype in a P. aeruginosa PA14 gidA mutant. Wild-type (WT) and gidA mutant (gidA) strains were supplied with a plasmid expressing gidA from PBAD (pCF430G) or with empty vector (pCF430). Cultures were grown in the presence of 50 mM arabinose to induce gene expression. (A) LasA activity. (B) Pyocyanin production. Values are means from three independent biological experiments, normalized to culture density (OD600) and expressed as percentages of the wild-type value. Error bars indicate ±1 standard deviation.
Because the transposon insertion was near the 3′ end of the gidA gene, the resulting protein could have retained residual activity. To test whether this was in fact the case, we constructed a markerless in-frame gidA deletion mutant (see Materials and Methods). Its QS phenotypes and growth characteristics were indistinguishable from those of the transposon mutant (data not shown). This suggests that the transposon insertion completely inactivated GidA and further confirms that the insertion does not exert any polar effects on downstream genes. As the properties of the deletion and the transposon insertion mutants were identical, we performed further experiments with the transposon insertion mutant.
GidA controls RhlR expression at the posttranscriptional level.
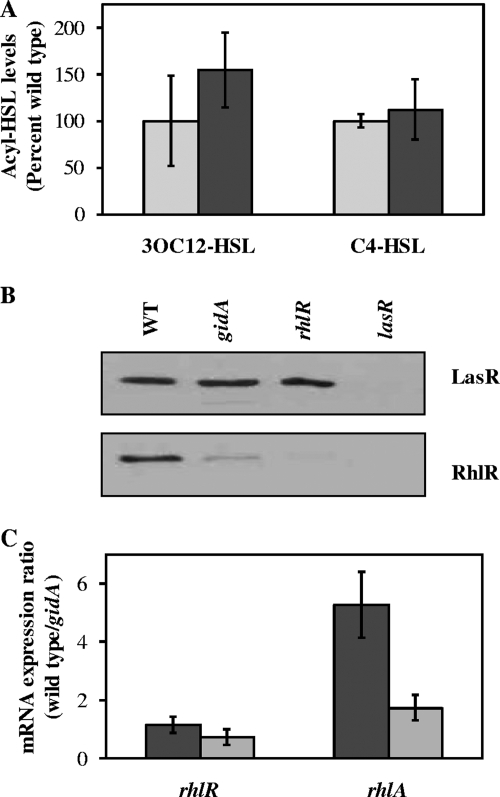
Next, we sought to obtain further insights into how GidA controls QS. Based on the presumed function of GidA, we predicted that it might affect the expression of central QS regulatory proteins posttranscriptionally, in particular, RhlI or RhlR. To investigate whether GidA affects acyl-HSL synthase levels, we measured 3OC12-HSL and C4-HSL levels in the wild type and the gidA mutant. We found no differences in the levels of either acyl-HSL (Fig. 4A). Next, we evaluated LasR and RhlR protein levels by Western blotting. Protein levels of RhlR were greatly reduced in a gidA mutant, while LasR levels were comparable to those in the wild type (Fig. 4B). RhlR protein levels were restored to wild-type levels in the gidA mutant when complemented with gidA in trans (data not shown). These results indicate that GidA influences the expression of RhlR, which is consistent with our phenotypic data.
FIG. 4.
Acyl-HSL levels and QS gene expression in the P. aeruginosa PA14 gidA mutant and the wild-type parent. (A) 3OC12-HSL and C4 HSL levels, expressed as percentages of the wild-type level. Dark gray bars represent the gidA mutant, and light gray bars represent the wild-type (WT) parent. (B) Western blot analysis of LasR (top panel) and RhlR (bottom panel) proteins. For comparison, protein levels in the lasR and rhlR mutants are shown. Equal amounts of clarified lysates were loaded for each strain. Smaller bands, indicative of protein degradation products, were not detected. (C) Real-time PCR. mRNA levels of the rhlA and rhlR genes were measured in the gidA mutant and the wild-type parent at the transition from logarithmic to stationary phase (4 to 5 h, dark gray) and in stationary phase (7 to 8 h, light gray). Values represent ratios of the transcript levels for the wild type versus the gidA mutant. Equal amounts of cDNA were used for each amplification reaction. Values are means from three independent biological experiments. Error bars indicate ±1 standard deviation.
To distinguish between transcriptional and posttranscriptional control, we measured transcript levels of rhlR by real-time PCR (Fig. 4C). We also measured transcript levels of the RhlR-dependent gene rhlA, which is involved in rhamnolipid biosynthesis (29, 30). We expected downregulation of the rhlA gene in a gidA mutant, as it has low RhlR protein levels. In the gidA mutant, rhlR mRNA levels were near wild-type levels, whereas rhlA mRNA levels, as expected, were significantly decreased, resulting in an elevated ratio of wild-type to gidA mutant transcripts (Fig. 4C). Taken together, these results indicate that gidA modulates expression of RhlR posttranscriptionally and that this in turn affects transcription of RhlR-dependent genes.
Induction of rhlR expression partially restores the wild-type phenotype.
Because GidA affected RhlR protein levels, we asked whether RhlR-dependent phenotypes could be rescued by increased expression of RhlR in a gidA mutant. We introduced a plasmid carrying rhlR under the control of an arabinose-inducible promoter into a gidA mutant. When induced fully, this strain expressed RhlR protein at levels slightly higher than those of the wild type and showed partial restoration of RhlR-dependent phenotypes (Fig. 5). This partial suppression suggests that gidA, in addition to affecting RhlR expression, also regulates these phenotypes in an RhlR-independent fashion.
FIG. 5.
Suppression of gidA mutant phenotypes by overexpression of rhlR. P. aeruginosa PA14 wild-type (WT) and gidA mutant strains were supplied with a plasmid expressing rhlR from PBAD (pCF430R) or with empty vector (pCF430). Cultures were grown in the presence of 50 mM arabinose to induce gene expression. (A) Western blot analysis of RhlR protein. Equal amounts of clarified lysates were loaded for each strain. Smaller bands, indicative of RhlR degradation products, were not detected. (B) Rhamnolipid production. (C) Pyocyanin production, expressed as percentages of the wild-type value. Values are means from three independent biological experiments, normalized to culture density (OD600). Error bars indicate ±1 standard deviation.
DISCUSSION
P. aeruginosa QS signaling is highly complex and involves a network of interconnected pathways (18, 41, 49). Several factors that regulate QS have been identified, but a comprehensive screen, such as a saturation mutagenesis, has not been attempted. In this study, we utilized a nonredundant P. aeruginosa transposon library to identify global regulators of QS gene expression. We rediscovered several known regulators but missed others (see Table S1 in the supplemental material). The fact that rsaL was missed is not surprising as it is a repressor, not an activator, of QS (6). However, it is not clear why other regulators that function as activators were not identified. One reason for this could be strain-specific regulatory differences. We used P. aeruginosa PA14 in our study, while most of the QS regulators have been identified and characterized in strain PAO1. Overall, the fact that not many new regulatory genes conferring multiple QS deficiencies were discovered suggests that the capacity for global regulation of QS, at the level of LasRI and/or RhlRI, is limited.
One such gene, whose inactivation conferred substantial deficiencies in several QS phenotypes, was gidA. A P. aeruginosa PA14 gidA mutant was primarily deficient in rhl-dependent phenotypes (Fig. 1). GidA in P. aeruginosa is 630 amino acids in length and is 70% identical to the E. coli protein. Two forms of the protein exist, a long form and a short form. Both forms contain an N-terminal flavin adenine dinucleotide-binding domain, but only the long form contains a C-terminal tRNA-binding domain required for tRNA modification (27). P. aeruginosa gidA is the long form. GidA requires another protein, MnmE, for tRNA modification (4). Unfortunately, we were unable to investigate the effect of an mnmE mutation on QS gene expression, thus precluding further evidence for a synergistic role of the two proteins in P. aeruginosa. In E. coli, both proteins are required for the addition of carboxymethylaminomethyl groups to the C5 carbon of uridine at position 34 of tRNAs that read codons ending with A or G in mixed-codon family boxes, including Glu, Gln, Lys, Leu, and Arg codons (4, 55). This modification enhances base-pairing specificity at the wobble position, allowing pairing with G and A, while restricting pairing with C and U. This prevents both misincorporation and +2 frameshifting (3, 4). It has been suggested that tRNA modification could function as a regulatory mechanism to adjust gene expression in response to nutrient deprivation through alteration of pools of cofactors required for the tRNA modification reactions (35).
In principle, GidA-mediated tRNA modification could affect the expression of all proteins in the cell. However, certain proteins will be more susceptible than others, if the respective genes contain a high proportion of codons that require decoding by tRNAs with U34 modifications. This includes many transcriptional regulators that contain clusters of positively charged Lys and Arg residues in their DNA-binding domains. Global, yet specific, regulation is consistent with previous studies. GidA controls virulence gene expression in A. hydrophila, P. syringae, S. flexneri, and S. pyogenes (5, 9, 21, 45) and fruiting-body development in M. xanthus (52), with little to no effect on growth. Posttranscriptional regulation has been shown in A. hydrophila, S. flexneri, and S. pyogenes, and involvement of a transcriptional regulator has been demonstrated in S. flexneri and S. pyogenes.
In this study, we have shown that GidA rather specifically controls RhlR expression in P. aeruginosa, also at the posttranscriptional level (see the model in Fig. 6). GidA affected RhlR protein but not transcript levels (Fig. 4), and introduction of rhlR into a gidA mutant partially restored rhl-dependent phenotypes (Fig. 5). Such phenotypic suppression is plausible, because overexpression of rhlR mRNA in a gidA mutant would result in a higher fraction of correctly translated RhlR, thereby compensating for the decreased level of functional protein translated from native mRNA. In S. flexneri and S. pyogenes, overexpression of the transcriptional regulators VirF and RopB, respectively, results in full suppression of gidA mutant phenotypes (5, 9). The fact that there was no full restoration to wild-type levels in P. aeruginosa suggests that GidA also affects rhl-dependent phenotypes independent of RhlR. It cannot be excluded, however, that a significant portion of the overexpressed RhlR, although soluble, is nonfunctional due to translation errors in the absence of GidA, resulting in only partial, rather than full, suppression. Despite the modest growth defect (Fig. 2), our results argue against a general gene expression defect in the gidA mutant: compared with the wild type, the gidA mutant did not show any deficiency in a las-specific phenotype (growth on adenosine as the sole carbon source), in LasR protein levels, and in acyl-HSL production. The observation that C4-HSL levels were not significantly altered in a gidA mutant is consistent with rhlI expression being primarily las dependent (7). While RhlR activates rhlI expression in the heterologous host E. coli (23), LasR rather than RhlR is the dominant regulator in P. aeruginosa (7).
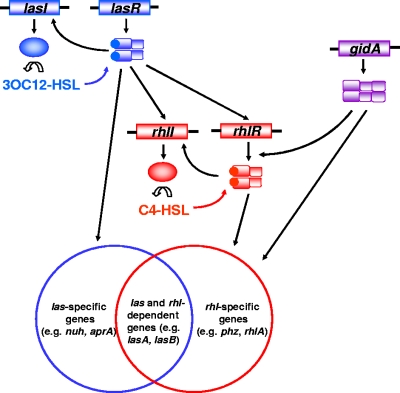
FIG. 6.
Model depicting the position of GidA in the QS circuitry. See the text for details.
Interestingly, the coding regions of LasR and RhlR both contain roughly equal numbers of codons (17 and 18, respectively) that require decoding by GidA/MnmE-modified tRNAs, which appears to be inconsistent with the observation that GidA affected RhlR but not LasR protein levels. However, the frequency of two-family box triplet codons alone does not determine the impact on protein function. Although none of the affected amino acid residues are highly conserved, the contribution of individual, less conserved residues to protein stability and activity may well be different in LasR and RhlR. Both proteins are only 33% identical and function quite differently. While LasR requires its acyl-HSL ligand for proper folding (43), RhlR does not (26). In addition, frameshifts, which tend to be much more deleterious than misincorporations, are also strongly dependent on the base composition surrounding the respective codons, although the precise mechanism is not clear (4). It is therefore plausible that GidA-mediated tRNA modification could have a selective effect on RhlR but not LasR protein levels and consequently could affect rhl-dependent but not las-dependent phenotypes.
Further experiments will be needed to elucidate the precise molecular mechanism of GidA function in P. aeruginosa. Taking our results together, we have identified an additional regulatory component of the QS network in P. aeruginosa that affects rhl-dependent gene expression through posttranscriptional control of RhlR.
Supplementary Material
Acknowledgments
We thank Nicole Liberati and Frederick Ausubel, Department of Molecular Biology, Massachusetts General Hospital, for the nonredundant set of P. aeruginosa PA14 transposon insertion mutants.
This work was supported by startup funds from Oregon State University (to M.S.), by NIH grant AI079454 (to M.S.) and by an HHMI/URISC scholarship (to T.R.G.).
Footnotes
Published ahead of print on 10 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1793928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjarnsholt, T., and M. Givskov. 2007. The role of quorum sensing in the pathogenicity of the cunning aggressor Pseudomonas aeruginosa. Anal. Bioanal. Chem. 387409-414. [DOI] [PubMed] [Google Scholar]
- 3.Bjork, G. R., and G. Hagervall. July 2005, posting date. Chapter 4.6.2, Transfer modification. In R. Curtiss III, A. Bock, J. L. Ingrahan, J. B. Kaper, S. Maloy, F. C. Neidhardt, M. M. Riley, C. L. Squires, and B. L. Wanner (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington DC. http://www.ecosal.org.
- 4.Bregeon, D., V. Colot, M. Radman, and F. Taddei. 2001. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 152295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, K. H., and M. G. Caparon. 2008. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect. Immun. 763176-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Kievit, T., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 1812175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Kievit, T. R., Y. Kakai, J. K. Register, E. C. Pesci, and B. H. Iglewski. 2002. Role of the Pseudomonas aeruginosa las and rhl quorum-sensing systems in rhlI regulation. FEMS Microbiol. Lett. 212101-106. [DOI] [PubMed] [Google Scholar]
- 8.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Camara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 1842576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durand, J. M., B. Dagberg, B. E. Uhlin, and G. R. Bjork. 2000. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 35924-935. [DOI] [PubMed] [Google Scholar]
- 10.Essar, D. W., L. Eberly, A. Hadero, and I. P. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girard, G., and G. V. Bloemberg. 2008. Central role of quorum sensing in regulating the production of pathogenicity factors in Pseudomonas aeruginosa. Future Microbiol. 397-106. [DOI] [PubMed] [Google Scholar]
- 12.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 223803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heurlier, K., V. Denervaud, M. Haenni, L. Guy, V. Krishnapillai, and D. Haas. 2005. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 1874875-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heurlier, K., F. Williams, S. Heeb, C. Dormond, G. Pessi, D. Singer, M. Camara, P. Williams, and D. Haas. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1862936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 2877-86. [DOI] [PubMed] [Google Scholar]
- 16.Joanny, G., J. Le Derout, D. Brechemier-Baey, V. Labas, J. Vinh, P. Regnier, and E. Hajnsdorf. 2007. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucleic Acids Res. 352494-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jude, F., T. Kohler, P. Branny, K. Perron, M. P. Mayer, R. Comte, and C. van Delden. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J. Bacteriol. 1853558-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juhas, M., L. Eberl, and B. Tummler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7459-471. [DOI] [PubMed] [Google Scholar]
- 19.Kay, E., B. Humair, V. Denervaud, K. Riedel, S. Spahr, L. Eberl, C. Valverde, and D. Haas. 2006. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 1886026-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessler, E., M. Safrin, J. C. Olson, and D. E. Ohman. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 2687503-7508. [PubMed] [Google Scholar]
- 21.Kinscherf, T. G., and D. K. Willis. 2002. Global regulation by gidA in Pseudomonas syringae. J. Bacteriol. 1842281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohler, T., C. van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 1835213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 211137-1146. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. H., Y. Lequette, and E. P. Greenberg. 2006. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59602-609. [DOI] [PubMed] [Google Scholar]
- 25.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 1032833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina, G., K. Juarez, B. Valderrama, and G. Soberon-Chavez. 2003. Mechanism of Pseudomonas aeruginosa RhlR transcriptional regulation of the rhlAB promoter. J. Bacteriol. 1855976-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer, S., A. Scrima, W. Versees, and A. Wittinghofer. 2008. Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate. J. Mol. Biol. 380532-547. [DOI] [PubMed] [Google Scholar]
- 28.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227197-203. [DOI] [PubMed] [Google Scholar]
- 29.Ochsner, U. A., A. Fiechter, and J. Reiser. 1994. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 26919787-19795. [PubMed] [Google Scholar]
- 30.Ochsner, U. A., A. K. Koch, A. Fiechter, and J. Reiser. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1762044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohfuji, K., N. Sato, N. Hamada-Sato, T. Kobayashi, C. Imada, H. Okuma, and E. Watanabe. 2004. Construction of a glucose sensor based on a screen-printed electrode and a novel mediator pyocyanin from Pseudomonas aeruginosa. Biosens. Bioelectron. 191237-1244. [DOI] [PubMed] [Google Scholar]
- 32.Passador, L., K. D. Tucker, K. R. Guertin, M. P. Journet, A. S. Kende, and B. H. Iglewski. 1996. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J. Bacteriol. 1785995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 921490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1795756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persson, B. C. 1993. Modification of tRNA as a regulatory device. Mol. Microbiol. 81011-1016. [DOI] [PubMed] [Google Scholar]
- 36.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1793127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rumbaugh, K. P., J. A. Griswold, and A. N. Hamood. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 21721-1731. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Sandoz, K., S. Mitzimberg, and M. Schuster. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. USA 10415876-15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuster, M., and E. P. Greenberg. 2007. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics 8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuster, M., and E. P. Greenberg. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 29673-81. [DOI] [PubMed] [Google Scholar]
- 42.Schuster, M., C. P. Lohstroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 1852066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 10115833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 15815-22. [DOI] [PubMed] [Google Scholar]
- 45.Sha, J., E. V. Kozlova, A. A. Fadl, J. P. Olano, C. W. Houston, J. W. Peterson, and A. K. Chopra. 2004. Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect. Immun. 721084-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 656-60. [DOI] [PubMed] [Google Scholar]
- 47.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 962408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, L. S., J. S. Webb, S. A. Rice, and S. Kjelleberg. 2003. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220187-195. [DOI] [PubMed] [Google Scholar]
- 49.Venturi, V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30274-291. [DOI] [PubMed] [Google Scholar]
- 50.von Meyenburg, K., B. B. Jorgensen, J. Nielsen, and F. G. Hansen. 1982. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Mol. Gen. Genet. 188240-248. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 1852080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, D. J., R. Merod, B. Thomasson, and P. L. Hartzell. 2001. GidA is an FAD-binding protein involved in development of Myxococcus xanthus. Mol. Microbiol. 42503-517. [DOI] [PubMed] [Google Scholar]
- 53.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 9613904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winzer, K., C. Falconer, N. C. Garber, S. P. Diggle, M. Camara, and P. Williams. 2000. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 1826401-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yim, L., I. Moukadiri, G. R. Bjork, and M. E. Armengod. 2006. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 345892-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.