Abstract
Pseudomonas aeruginosa exhibits swarming motility on semisolid surfaces (0.5 to 0.7% agar). Swarming is a more than just a form of locomotion and represents a complex adaptation resulting in changes in virulence gene expression and antibiotic resistance. In this study, we used a comprehensive P. aeruginosa PA14 transposon mutant library to investigate how the complex swarming adaptation process is regulated. A total of 233 P. aeruginosa PA14 transposon mutants were verified to have alterations in swarming motility. The swarming-associated genes functioned not only in flagellar or type IV pilus biosynthesis but also in processes as diverse as transport, secretion, and metabolism. Thirty-three swarming-deficient and two hyperswarming mutants had transposon insertions in transcriptional regulator genes, including genes encoding two-component sensors and response regulators; 27 of these insertions were newly identified. Of the 25 regulatory mutants whose swarming motility was highly impaired (79 to 97%), only 1 (a PA1458 mutant) had a major defect in swimming, suggesting that this regulator might influence flagellar synthesis or function. Twitching motility, which requires type IV pili, was strongly affected in only two regulatory mutants (pilH and PA2571 mutants) and was moderately affected in three other mutants (algR, ntrB, and nosR mutants). Microarray analyses were performed to compare the gene expression profile of a swarming-deficient PA3587 mutant to that of the wild-type PA14 strain under swarming conditions. PA3587 showed 63% homology to metR, which encodes a regulator of methionine biosynthesis in Escherichia coli. The observed dysregulation in the metR mutant of nine different genes required for swarming motility provided a possible explanation for the swarming-deficient phenotype of this mutant.
Pseudomonas aeruginosa, a ubiquitous gram-negative bacterium, is an important opportunistic human pathogen that causes eventually fatal chronic lung infections in cystic fibrosis (CF) patients (30). P. aeruginosa is also a major contributor to hospital-acquired bacterial infections, causing serious disease in immunocompromised patients with cancer and AIDS, as well as in patients with severe burns (60). Currently, the therapeutic options available to combat these robust infections are limited. In addition, there are now drug-resistant hospital strains of P. aeruginosa that cause untreatable infections (62). Infections with P. aeruginosa are extremely difficult to eradicate due to this bacterium's intrinsic resistance to a broad spectrum of antimicrobial agents and its repertoire of virulence factors (27, 60).
Motility is strongly associated with the pathogenesis of P. aeruginosa (4). Motility enables this bacterium to colonize different environments, attach to surfaces, and form biofilms (44). This bacterium is capable of twitching, swimming, swarming, and sliding motility. P. aeruginosa can exhibit twitching motility on solid surfaces or interfaces, which is mediated by type IV pili (52). P. aeruginosa also has a single polar flagellum that enables the bacterium to swim in an aqueous environment and in the presence of low agar concentrations (0.3%, wt/vol). P. aeruginosa is able to swarm on semisolid agar (viscous; 0.5 to 0.7% agar, wt/vol) under nitrogen-limited conditions and when certain amino acids are supplied as the sole nitrogen source (52). Most recently, this bacterium has also been observed to display sliding motility under the same conditions that allow swarming but in the absence of type IV pili and flagella (42).
Swarming is a social phenomenon that involves rapid coordinated movement of bacteria across a semisolid surface (33). Importantly, the conditions that promote swarming appear to mimic those in the mucous layers that overlay epithelial surfaces, such as those of the lung, a major site of infection by P. aeruginosa in patients with CF (30).
Swarmer cells are hyperflagellated and elongated compared to the normal single-flagellum and short vegetative cells (22). P. aeruginosa requires flagella and possibly type IV pili to swarm (33). Biosurfactants produced by the bacteria, such as rhamnolipids and 3-(3-hydroxyalkonoyloxy)alkanoic acids (HAAs), are also involved in swarming motility, as they aid in overcoming the surface tension between bacterial cells and their environment (10, 59). The dependence on these biosurfactants suggests that this bacterium's quorum-sensing systems, the las and rhl systems, play an important role in this type of surface propagation as these cell-to-cell signaling systems regulate the production of rhamnolipids and HAAs (60). In addition to the physical changes such as cell elongation, swarmer cells overexpress hundreds of virulence-related genes, including those encoding the type III secretion systems (46). Swarmer cells also exhibit elevated adaptive antibiotic resistance to ciprofloxacin, gentamicin, and polymyxin B (46). Thus, rather than merely being a form of locomotion, swarming is part of an alternative growth state for P. aeruginosa and is a complex adaptation process that occurs in response to specific environmental stimuli, resulting in changes in virulence gene expression, antibiotic resistance, and likely metabolism (46).
A recent screen of the 1,200 genes for which mutants are available in the P. aeruginosa PAO1 mini-Tn5-luxCDABE transposon mutant library (35) identified 36 mutants with altered swarming phenotypes (45). Moreover, a comparison of the gene expression profiles of swarmer cells and bacteria growing in the same medium under planktonic conditions showed that there was a >2-fold change in 7.5% of all P. aeruginosa genes. The 417 genes that were differentially regulated in swarmer cells included no less than 18 predicted or known transcriptional regulator genes. Furthermore, differentially regulated genes were found to function in processes as diverse as transport, secretion, metabolism, and motility, suggesting that the regulation of swarming behavior is complex (45).
To study the multifaceted regulation of swarming, we screened a P. aeruginosa strain PA14 transposon insertion mutant library (36) which covers about 75% of the predicted 5,962 PA14 genes to look for mutants with altered swarming phenotypes. As a result, 35 transcriptional regulators were found to be involved in swarming, including a variety of two-component sensors and response regulators. Interestingly, in contrast to the results of a small study described previously (45), an inverse relationship between the ability of mutants to swarm and their ability to form biofilms was observed in this study. The defect in one swarming-deficient mutant, a metR mutant, was confirmed by complementation in trans with the cloned gene. Microarray analysis of the metR mutant (PA3587) was performed and showed that there was differential regulation of 268 genes (≥2.0-fold change; P ≤ 0.05). Downregulation of the putative methionine synthase gene metH (PA1843) and the putative ABC transporter gene PA4223 might explain the swarming defect of the metR mutant, since the swarming of mutants with transposon insertions in these genes was impaired.
MATERIALS AND METHODS
Bacterial strains, primers, and growth conditions.
The bacteria used included P. aeruginosa PA14 (50), as well as the entire P. aeruginosa transposon mutant library (5,800 mutants) from Harvard University (36). Escherichia coli XL1-Blue (8) and plasmid pUCP18 (65) were used for cloning. The sequences of DNA primers (Alpha DNA, Montreal, Canada) used in these studies are available from us upon request. When required for plasmid or transposon selection or maintenance, gentamicin and carbenicillin were added to final concentrations of 15 and 500 μg/ml, respectively. Routine genetic manipulations were carried out using standard procedures (54).
Preliminary (broad) screen of the PA14 library for mutants defective in swarming motility.
To investigate genes involved in swarming motility in P. aeruginosa, the Harvard PA14 transposon insertion mutant library was screened for swarming defects as described elsewhere (45). Briefly, mutants were grown overnight in Luria-Bertani (LB) broth, and 1 μl of each culture was transferred using a custom 96-well pin device onto the surface of brain heart infusion agar plates containing 0.5% (wt/vol) Difco agar. Colonies were scored for differences in swarming motility compared to the wild type after 48 h of incubation at room temperature. Two independent experiments were performed with two replicates of each mutant.
Verification of the swarming-deficient mutants identified in the preliminary screen.
The mutants displaying swarming deficiencies identified in the preliminary screen were independently verified by performing swarming experiments on BM2 swarming agar plates (62 mM potassium phosphate buffer [pH 7], 2 mM MgSO4, 10 μM FeSO4, 0.4% [wt/vol] glucose, 0.1% [wt/vol] Casamino Acids, 0.5% [wt/vol] Difco agar) (46). One-microliter aliquots of mid-log-phase cultures grown in BM2 minimal medium [62 mM potassium phosphate buffer (pH 7), 7 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4, 0.4% (wt/vol) glucose] were inoculated onto the plates. Each experiment was carried out once with three replicates for each mutant. The verification experiments performed for transcriptional regulator or two-component sensor and response regulator mutants identified in the preliminary screen were performed three times, employing three to five replicates each time. All of the resulting dendritic colonies were analyzed by measuring the surface coverage on agar plates after 20 h of incubation at 37°C.
Growth curves.
P. aeruginosa PA14 mutants and the wild-type strain were grown overnight in BM2 swarming medium (62 mM potassium phosphate buffer, [pH 7], 2 mM MgSO4, 10 μM FeSO4, 0.4% [wt/vol] glucose, 0.1% [wt/vol] Casamino Acids). If necessary, cultures were diluted to obtain equal optical densities. Five-microliter portions of these cultures were added to 195 μl of fresh swarming medium in 96-well microtiter plates. The growth of these cultures at 37°C under shaking conditions was monitored with a TECAN Spectrofluor Plus by determining the absorbance at 620 nm every 20 min for 24 h. Two independent experiments were performed with three replicates for each mutant.
Swimming and twitching motility.
As P. aeruginosa PA14 exhibits very poor swimming motility on BM2 glucose swimming plates which are used for PAO1 (46), LB medium plates with 0.3% (wt/vol) agar were used instead. One-microliter aliquots of mid-log-phase cultures grown in LB broth were inoculated onto the plates. The diameters of the swimming zones were measured after incubation for 20 h at 37°C. Twitching motility was determined by measuring the diameters of the twitching zones after 24 h and 48 h of incubation of PA14 mutants on LB medium plates with 1% (wt/vol) agar as described previously (18). For both swimming and twitching assays, three independent experiments were performed with three replicates for each mutant.
Biofilm assays.
Biofilm formation was analyzed using an abiotic solid surface assay as described elsewhere (23). Dilutions (1/100) of overnight cultures were incubated in BM2 adjusted medium [62 mM potassium phosphate buffer (pH 7), 7 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4, 0.4% (wt/vol) glucose, 0.5% (wt/vol) Casamino Acids] in polystyrene microtiter plates (Falcon, United States) for 20 h at 37°C. Biofilms were stained with crystal violet, and the absorbance at 595 nm was measured using a microtiter plate reader (Bio-Tek Instruments Inc., United States).
Rhamnolipid production.
Rhamnolipid biosynthesis was analyzed by the agar plate method as described previously (16, 17, 19). Briefly, P. aeruginosa PA14 mutants were grown overnight in LB medium and spot inoculated onto agar plates containing iron-limited salt medium (0.7 g/liter KH2PO4, 0.9 g/liter NaHPO4, 2.0 g/liter NaNO3, 0.4 g/liter MgSO4 · H2O, 0.001 g/liter CaCl2 · 2H2O, 0.001 g/liter FeSO4 · 7H2O) supplemented with 1% (wt/vol) glucose, 0.5% (wt/vol) Casamino Acids, 0.02% (wt/vol) cetyltrimethylammonium bromide, 0.0005% (wt/vol) methylene blue, and 1.6% (wt/vol) agar. The plates were incubated for 37°C for 24 h and then at room temperature for an additional 24 h. Rhamnolipid production was determined by measuring the diameter of the dark blue halo that formed around a colony. Three independent experiments were performed for each mutant.
Phage PO4 sensitivity test.
Phage sensitivity assays were performed as previously described (7). Briefly, bacterial lawns were produced by adding 100 μl of an overnight P. aeruginosa LB broth culture to 3 ml of LB soft agar. The inoculated top agar was overlaid on an LB agar plate and allowed to solidify. Five microliters of a lysate of phage PO4 was spotted in duplicate onto lawns of P. aeruginosa wild-type and mutant strains. Zones of lysis (or clearing) on the plates were detected after incubation of the plates overnight at 37°C. Three independent experiments were performed for each mutant.
Mutant complementation.
The metR mutant P. aeruginosa PA14 ID46982 was complemented by amplifying the metR gene, including a 400-bp upstream region, from PA14 genomic DNA by PCR and cloning the gene into the pUCP18 vector (65) using BamHI and EcoRI restriction sites. The metR gene was cloned in the opposite direction to the lacZ promoter, ensuring gene expression from the native metR promoter rather than from the lacZ promoter. The hybrid plasmid pUCP-metR was transferred to the metR mutant by electroporation (13), yielding a metR+ strain.
DNA microarray experiment.
Two different independent microarray studies were performed, employing four or five independent assessments of transcriptional profiles. One microarray study involved harvesting RNA from the leading edges of dendritic swarm colonies of the metR mutant (PA14 ID46982) and wild-type strain PA14. As described previously (46), cells were resuspended in BM2 swarming medium supplemented with the RNAprotect reagent (Qiagen, Germany). Harvesting of cells, RNA isolation, cDNA synthesis, hybridization to P. aeruginosa PAO1 DNA microarray slides (aminosilane coated) from The Institute for Genomic Research Pathogenic Functional Genomics Resource Center, analysis of microarray slides using ArrayPipe (version 1.7), and quantitative reverse transcription-PCR (RT-qPCR) were performed as described previously (40). A second microarray experiment was performed to compare the gene expression profiles of the PA14 metR mutant and wild-type strain PA14 in liquid BM2 swarming medium. Briefly, five independent cultures of the metR mutant and wild-type strain PA14 were grown overnight in liquid BM2 swarming medium, diluted 1:100, and grown to mid-log phase (optical density at 600 nm, 0.4 to 0.5) before RNA isolation. Microarray studies were performed as described above, except that due to a manufacturing change by The Institute for Genomic Research epoxy-coated slides were used.
Microarray accession numbers.
ArrayExpress accession numbers are E-FPMI-19 and E-FPMI-20.
RESULTS
Identification of swarming-deficient mutants.
During a preliminary screen of the PA14 transposon insertion mutant library, 375 of approximately 5,600 mutants were identified as swarming deficient (data not shown). The phenotypes of these mutants were examined in more detail by performing three to five individual swarming experiments with each of the mutants on BM2 swarming agar plates (Fig. 1). As a result, 233 transposon insertion mutants from the PA14 mutant library were confirmed to have alterations in swarming motility (see Table S1 in the supplemental material). As expected from previous observations (33, 45), we found swarming-deficient mutants with transposon insertions in flagellum and type IV pilus biosynthesis and quorum-sensing genes (e.g., rhlR, rhlI, pqsD, and pqsH). Mutants with transposon insertions in genes belonging to a wide variety of functional classes other than the motility and quorum-sensing classes were also identified, and large numbers of these genes had functions in metabolism, transport of small molecules, adaptation, and protection. Moreover, 35 transcriptional regulators, including two-component sensors and response regulators, were identified as regulators involved in swarming motility (Table 1). To further probe the basis for the complex adaptive behavior in response to a viscous environment, we focused on regulation of swarming motility.
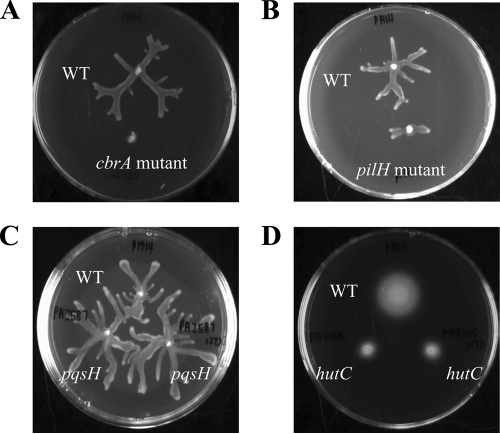
FIG. 1.
Selected P. aeruginosa PA14 transposon mutants displaying altered swarming and swimming motility compared to wild-type strain PA14 (WT). (A to C) Mutants displaying altered swarming motility on plates containing BM2 swarming medium with 0.5% (wt/vol) agar. (A) cbrA mutant displaying no swarming phenotype. (B) pilH mutant displaying a significantly reduced swarming phenotype. (C) pqsH mutants displaying a hyperswarming phenotype. (D) hutC mutants displaying a strong swimming motility defect on a plate containing LB medium with 0.3% (wt/vol) agar.
TABLE 1.
Characteristics of transcriptional regulators involved in swarming
| Mutant | Product of gene | Polar effect possiblea | Change (fold) compared to wild-type strain PA14 (mean ± SD)
|
||
|---|---|---|---|---|---|
| Swarming | Swimming | Twitching | |||
| PA0037 (trpI)b | Transcriptional regulator TrpI | N | 0.03 ± 0.01c | 0.60 ± 0.02c | 0.98 ± 0.07 |
| PA0409 (pilH)b | Type IV pilus response regulator | Y | 0.17 ± 0.12c | 1.10 ± 0.03 | 0.03 ± 0.03c |
| PA0475b | Putative transcriptional regulator | N | 0.05 ± 0.02c | 0.59 ± 0.01c | 0.95 ± 0.05 |
| PA0479b | Putative transcriptional regulator | N | 0.04 ± 0.01c | 0.70 ± 0.02c | 0.94 ± 0.05 |
| PA0831 (oruR)b | Transcriptional regulator OruR | N | 0.05 ± 0.03c | 0.53 ± 0.05c | 0.89 ± 0.07 |
| PA0905 (rsmA) | Regulator of secondary metabolites | N | 0.12 ± 0.12c | 0.87 ± 0.01 | 0.78 ± 0.06 |
| PA0928 (gacS) | Sensor-response regulator hybrid | Y | 0.03 ± 0.01c | 0.68 ± 0.01c | 0.90 ± 0.10 |
| PA1180 (phoQ) | Two-component sensor PhoQ | N | 0.21 ± 0.22c | 0.87 ± 0.01 | 0.93 ± 0.10 |
| PA1196b | Putative transcriptional regulator | N | 0.04 ± 0.03c | 0.50 ± 0.06c | 0.87 ± 0.05 |
| PA1347b | Putative transcriptional regulator | N | 0.05 ± 0.03c | 0.89 ± 0.00 | 0.85 ± 0.03 |
| PA1422 (gbuR)b | Transcriptional regulator GbuR | N | 0.03 ± 0.01c | 0.77 ± 0.03 | 0.85 ± 0.04 |
| PA1431 (rsaL) | Regulatory protein RsaL | N | 0.40 ± 0.06c | 0.98 ± 0.02 | 0.95 ± 0.11 |
| PA1458b | Putative two-component sensor | N | 0.12 ± 0.03c | 0.09 ± 0.01c | 0.98 ± 0.08 |
| PA1544 (anr)b | Transcriptional regulator Anr | N | 0.66 ± 0.16c | 0.92 ± 0.01 | 0.74 ± 0.01c |
| PA1976b | Putative sensor histidine kinase | N | 0.37 ± 0.16c | 1.00 ± 0.01 | 1.03 ± 0.14 |
| PA2072b | Putative sensory box protein | N | 0.11 ± 0.03c | 0.50 ± 0.05c | 0.95 ± 0.06 |
| PA2332b | Putative AraC family regulator | N | 0.50 ± 0.07c | 0.99 ± 0.03 | 1.01 ± 0.04 |
| PA2571b | Putative histidine kinase | N | 0.09 ± 0.08c | 0.73 ± 0.07 | 0.10 ± 0.01c |
| PA2586 (gacA) | Response regulator GacA | Y | 2.28 ± 0.77c | 1.05 ± 0.03 | 0.94 ± 0.11 |
| PA2622 (cspD)b | Cold shock protein CspD | N | 1.72 ± 0.78c | 1.07 ± 0.03 | 0.89 ± 0.05 |
| PA2957b | Putative transcriptional regulator | N | 0.08 ± 0.05c | 0.56 ± 0.02c | 0.94 ± 0.09 |
| PA3391 (nosR)b | Regulatory protein for NO2 reductase | N | 0.09 ± 0.08c | 0.71 ± 0.30 | 0.60 ± 0.06c |
| PA3477 (rhlR) | Transcriptional regulator RhlR | N | 0.08 ± 0.02c | 1.05 ± 0.02 | 0.83 ± 0.06 |
| PA3587 (metR)b | Transcriptional regulator MetR | N | 0.18 ± 0.08c | 0.87 ± 0.02 | 0.75 ± 0.07 |
| PA3895b | Putative transcriptional regulator | N | 0.05 ± 0.03c | 0.54 ± 0.02c | 0.91 ± 0.11 |
| PA4315 (mvaT) | Transcriptional regulator MvaT | N | 0.55 ± 0.17c | 0.99 ± 0.02 | 0.87 ± 0.09 |
| PA4398b | Putative two-component response regulator | Y | 0.38 ± 0.12c | 0.96 ± 0.02 | 1.08 ± 0.08 |
| PA4725 (cbrA)b | Two-component sensor CbrA | N | 0.03 ± 0.01c | 0.97 ± 0.08 | 0.98 ± 0.05 |
| PA4778b | Putative transcriptional regulator | N | 0.04 ± 0.02c | 0.54 ± 0.01c | 0.85 ± 0.05 |
| PA4853 (fis)b | DNA-binding protein Fis | N | 0.07 ± 0.04c | 0.75 ± 0.06 | 0.99 ± 0.09 |
| PA5105 (hutC)b | Histidine utilization gene repressor | N | 0.09 ± 0.07c | 0.50 ± 0.04c | 0.97 ± 0.06 |
| PA5124 (ntrB)b | Two-component sensor NtrB | Y | 0.03 ± 0.01c | 0.90 ± 0.04 | 0.30 ± 0.17c |
| PA5125 (ntrC)b | Two-component response regulator NtrC | N | 0.12 ± 0.07c | 0.98 ± 0.01 | 1.00 ± 0.05 |
| PA5261 (algR) | Alginate biosynthesis regulator | N | 0.45 ± 0.18c | 0.99 ± 0.01 | 0.34 ± 0.03c |
| PA5536b | Putative C4-type zinc finger protein, DksA/TraR family | N | 0.49 ± 0.15c | 1.01 ± 0.01 | 1.00 ± 0.03 |
Possibility that a transposon insertion affects expression of downstream genes. Y, yes; N, no.
Regulatory mutant identified as swarming deficient in this study.
The phenotype was statistically significantly altered (P < 0.05, Student's t test), with a >25% change compared to P. aeruginosa wild-type strain PA14.
Closer study of the mutants with transposon insertions in transcriptional regulator genes confirmed that 33 mutants exhibited reduced swarming and 2 mutants (gacA and cspD mutants) exhibited hyperswarming (Table 1). To our knowledge, only the swarming phenotypes of mutants with mutations in gacS (PA0928) (6), rhlR (PA3477) (33), rsmA (PA0905) (29), phoQ (PA1180) (6), rsaL (PA1431) (61), algR (PA5261) (45), mvaT (PA4315) (20), and gacA (PA2586) (31) have been described previously. Therefore, 27 swarming-deficient transcriptional regulators, 10 of which were two-component sensors or response regulators, were newly identified in this study (Table 1). In summary, we confirmed that 27 mutants (22 new mutants) showed either no swarming or minimal swarming, 6 mutants (4 new mutants) showed reduced (34 to 62% reduced) swarming, and 2 mutants (1 new mutant) had a hyperswarming phenotype.
To test whether swarming deficiency may have resulted from a growth defect, all mutants were tested to determine their growth behavior in liquid swarming medium. Most of the mutants grew like P. aeruginosa wild-type strain PA14 (data not shown). Mutants with transposon insertions in the cbrA (PA4725), gacS (PA0928), fis (PA4853), and PA4778 genes showed significant reductions in the growth rate in this medium (data not shown); however, it seems unlikely that these defects, which reduced the growth rate up to twofold, could entirely explain the 93 to 97% decrease in swarming motility observed for these mutants.
The possibility that polar effects on downstream genes, rather than the genes in which the insertions occurred, might be responsible for the observed swarming phenotype of the transposon mutants was examined (see Table S1 in the supplemental material). In particular, the swarming abilities were examined for available mutants with transposon insertions in genes that were part of a predicted or known operon and downstream of the mutations of the 35 regulatory mutants, based on the operon prediction tool at www.pseudomonas.com (67); as the prediction tool is currently available only for the PAO1 genome, PA14 genes that were not orthologs of genes in the PAO1 genome could not be predicted. As a result, the swarming phenotypes of 5 of the 35 regulatory mutants (pilH, gacA, gacS, ntrB, and PA4398 mutants) might have been affected by the altered expression of downstream genes, but the remainder of the regulatory mutants were either mutants with orphan genes or mutants for which transposon insertions in downstream genes did not result in swarming defects (Table 1).
Characterization of swarming-deficient regulators.
Swarming-deficient mutants with transposon insertions in transcriptional regulator genes were characterized further to investigate how these genes influence swarming behavior. One way in which swarming may be regulated is by modulating flagellum biosynthesis or function, as flagella have been shown to be essential for swarming motility in P. aeruginosa. As swimming motility depends entirely on a functional flagellum, we tested swarming-defective mutants to determine their abilities to swim in 0.3% agar.
Only one mutant with a mutation in the putative two-component sensor gene PA1458 had a major defect in swimming motility. Another 10 mutants (including the PA0037/trpI, PA0475, PA0831/oruR, PA1196, PA2072, PA3895, PA4778, and PA5105/hutC mutants) had significant (P ≤ 0.05, Student's t test) but modest (40 to 50%) swimming defects (Table 1) that are unlikely per se to explain the much more extreme swarming defect. Also, it was observed that most mutants that were completely unable to swarm also showed at least mild swimming defects (range, 10 to 50% inhibition). Thus, it seems that the swarming defects were generally much more extreme than the swimming defects and that the loss of flagellar function could not fully explain the substantial swarming alterations observed (>92% for 19 of the regulatory mutants). Mutants that showed more moderate deficiencies in swarming (40 to 60%) exhibited minor (anr and PA4398 mutants) or no (rsaL, mvaT, PA1976, algR, PA5536, and PA2332 mutants) swimming defects. Particularly in the case of the cbrA, ntrB, ntrC, rhlR, and PA1347 mutants, absolutely no association was observed between swarming and swimming, since the swarming of these mutants was strongly impaired but their swimming motility was (almost) normal.
In addition to flagella, type IV pili are known to be important for swarming motility, although the dependence on type IV pili has been reported to be different in different strains. For PAO1, knockout mutants with mutations in pilus synthesis genes were found to be swarming deficient (33, 45). In contrast, in one study, specific type IV pilus synthesis genes were shown to be dispensable for this type of migration in P. aeruginosa PA14 (58). In our screen of the PA14 mutant library for swarming-deficient mutants, we identified 10 genes that were known to be involved in type IV pilus biogenesis and twitching motility (a type of motility mediated by type IV pili). This indicates that type IV pili are involved in the swarming motility of P. aeruginosa strain PA14 under our experimental conditions. The twitching motility of swarming-deficient regulators was largely unaffected, except for strong effects (P ≤ 0.05, Student's t test) for two mutants with transposon insertions in pilH and PA2571 and intermediate effects for algR, ntrB, and nosR mutants compared to the wild type (Table 1).
To further investigate whether the deficiencies in twitching observed in the pilH, PA2571, algR, ntrB, and nosR mutants were related to the inability of these mutants to generate functional type IV pili, the sensitivity to pilus-dependent bacteriophage PO4 was assessed for all 35 regulatory mutants (5). As expected, the pilH, PA2571, algR, ntrB, and nosR mutants showed resistance to phage PO4 infection, while the rest of the regulatory mutants showed zones of lysis where phage PO4 was spotted.
Another factor known to be involved in the swarming of P. aeruginosa is rhamnolipid production. Rhamnolipids are synthesized and excreted into the external medium by P. aeruginosa to aid in overcoming the surface tension between bacterial cells and their environment and thus facilitate movement of the bacteria across semiviscous surfaces. Previous studies showed that mutants with defects in genes involved in the rhamnolipid biosynthesis pathway displayed major defects in swarming motility (e.g., rhlA) or showed an altered swarming pattern (e.g., rhlB) (12). To test whether the swarming-deficient phenotype of the regulatory mutants was related to a lack of rhamnolipid biosynthesis, agar plate assays were performed. On the agar plates inoculated with the rhlR, rsaL, and anr regulatory mutants, a lack of visible haloes around the colonies was observed, indicating that the mutants were not able to excrete rhamnolipid. The PA1347 and PA4398 mutants consistently had larger and smaller halos, respectively, around their colonies, while for the remaining 30 regulatory mutants the formation of a blue halo around a colony was the same as that for the wild-type strain (data not shown).
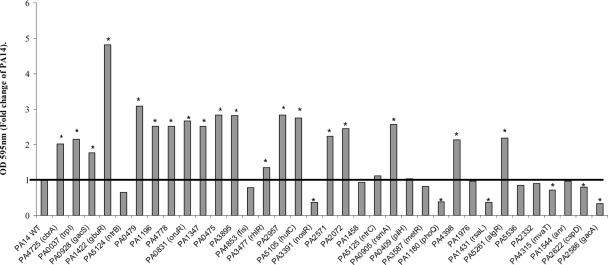
A characteristic of P. aeruginosa that has been associated with swarming motility is its ability to form biofilms. Intercellular signaling through quorum-sensing systems is thought to play a role in both types of surface-associated behavior, although the contribution of quorum sensing to biofilms seems to depend on environmental conditions (for a review, see reference 32). Simple biofilm formation was measured in 96-well microtiter plates after 20 h of incubation as described above. All of the mutants except the PA1458, PA1976, PA5536, PA2332, fis, ntrB, ntrC, pilH, cspD, and anr mutants showed significant (P ≤ 0.05, Student's t test) changes in biofilm formation (Fig. 2). Mutants displaying severe swarming defects generally showed biofilm overproduction. In contrast, mutants with more moderate deficiencies in swarming or hyperswarmers showed normal or decreased biofilm formation, and only the phoQ, rsaL, gacA, and nosR mutants had major deficiencies in biofilm formation. Thus, while swarming motility and biofilm formation seem to be related, the data seem to imply that there is an inverse relationship between these processes.
FIG. 2.
Biofilm formation by mutants with altered swarming motility. Mutants were sorted on the basis of increasing swarming motility. Bacteria were incubated in adjusted BM2 medium in polystyrene microtiter plates for 20 h at 37°C, and biofilms were stained with crystal violet. The data are representative of three independent experiments with eight replicates for each mutant and are expressed as the fold change in the optical density at 595 nm (OD 595nm) of the mutant compared to the wild-type. Asterisks indicate mutants that showed a statistically significant change in biofilm production (P ≤ 0.05, Student's t test). WT, wild type.
Regulation of swarming by the transcriptional regulator MetR.
Of the 35 transcriptional regulator mutants whose swarming behavior was altered, one interesting mutant, the PA3587 mutant, whose transcriptional regulator shows 63% homology to the transcriptional regulator MetR from E. coli (39), was studied in more detail. There have been no reports on metR in P. aeruginosa, but various functions of MetR in E. coli have been described, including the regulation of methionine biosynthesis through transcriptional activation of the metH and metE genes and probably the metA gene (9). In our assays, the P. aeruginosa metR mutant exhibited a severe defect in swarming (82% decrease compared to the wild type) but displayed only slight deficiencies in swimming and twitching (only 13 and 25% decreases compared to the wild type, respectively) and normal growth in swarming medium. The strong selective swarming motility defect and the fact that a corresponding MetR regulator in E. coli regulates processes other than flagellum or type IV pilus biosynthesis or function encouraged us to analyze this regulator in more detail.
The metR gene was cloned onto pUCP18 to obtain pUCP-metR. As shown in Fig. 3, complementation of the metR mutant with pUCP-metR completely restored the swarming phenotype to the levels observed for the wild-type strain. To determine the conditions under which this gene is expressed in wild-type PA14 cells, RNA was extracted from wild-type cultures growing in LB or swarming medium at the mid-log, early stationary, and late stationary phases of growth. Semiquantitative PCR showed that the expression of the metR gene was similar under all conditions (data not shown). To analyze the regulon of metR, two sets of DNA microarray experiments were performed to compare the transcriptome of the metR mutant to that of wild-type P. aeruginosa PA14 cells that were either grown for 20 h at 37°C on swarming medium plates or, to test for medium effects, grown to mid-log phase in liquid swarming medium. All of the genes with a change in expression of ≥2-fold compared to the wild type with a P value of ≤0.05 are shown in Table S2 in the supplemental material. Comparison of the gene expression profiles of the metR mutant and the wild type showed that far more genes (268 genes) were dysregulated under semiviscous swarming conditions than under liquid swarming conditions, under which only 49 dysregulated genes were observed, indicating that MetR is selectively employed as a regulator under semiviscous swarming conditions. Indeed, only eight genes were dysregulated under swarming conditions and in liquid swarming medium, and all except two of these genes were more highly dysregulated under swarming conditions; it is noteworthy that the number of overlapping genes increased when a lower cutoff (1.5-fold) was used for the liquid medium arrays. It is also noteworthy that the results for at least one gene in each functional category listed in Table 2 were confirmed by RT-qPCR (data not shown).
FIG. 3.
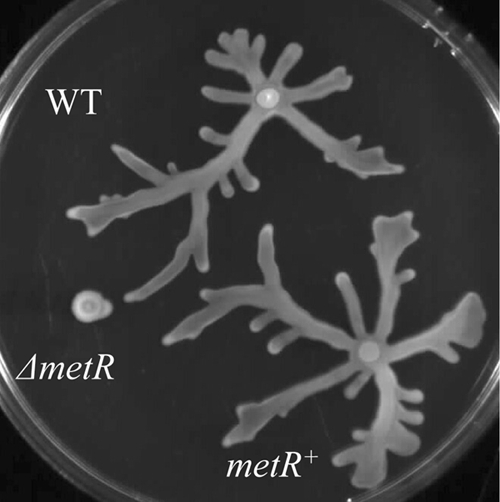
Complementation of the metR mutant. The defect in swarming motility of the metR transposon insertion mutant (ΔmetR) was restored to a level similar to that of P. aeruginosa wild-type strain PA14 (WT) by transformation of this mutant with a plasmid containing the metR gene, pUCP-metR.
TABLE 2.
Selected P. aeruginosa genes that were dysregulated in the metR mutant compared to wild-type strain PA14 under semiviscous and liquid swarming conditions
| Gene | Designation | Function | Change (fold)a | Pa |
|---|---|---|---|---|
| Regulators | ||||
| PA0248 | Probable transcriptional regulator | −2.72 | 0.03 | |
| PA0610 | prtN | Transcriptional regulator PrtN | 2.84 | 0.00 |
| PA0707 | toxR | Transcriptional regulator ToxR | −5.38 | 0.00 |
| PA0906 | Probable transcriptional regulator | 2.92 | 0.03 | |
| PA0929 | Two-component response regulator | −3.33 | 0.02 | |
| PA1637 | kdpE | Two-component response regulator KdpE | −2.37 | 0.04 |
| PA1705 | pcrG | Regulator in type III secretion | −2.19 | 0.01 |
| PA2056 | Probable transcriptional regulator | 2.16 | 0.02 | |
| PA2082 | Probable transcriptional regulator | −3.72 | 0.02 | |
| PA2337 | mtlR | Transcriptional regulator MtlR | 2.13 | 0.00 |
| PA2383 | Probable transcriptional regulator | −2.16 | 0.02 | |
| PA2577 | Probable transcriptional regulator | 2.57 | 0.00 | |
| PA2665 | Anaerobic nitric oxide reductase transcription regulator | 3.73 | 0.02 | |
| PA2846 | Probable transcriptional regulator | −2.60 | 0.03 | |
| PA2917 | Probable transcriptional regulator | −2.68 | 0.03 | |
| PA3206 | Probable two-component sensor | 2.92 | 0.00 | |
| PA3583 | glpR | Glycerol-3-phosphate regulon repressor | −2.27 | 0.00 |
| PA3830 | Probable transcriptional regulator | −3.00 | 0.00 | |
| PA5125 | ntrC | Two-component response regulator NtrC | 2.22 | 0.00 |
| PA5380 | Probable transcriptional regulator | 2.40 | 0.02 | |
| Type III secretion system and effectors | ||||
| PA0044 | exoT | Exoenzyme T | −2.08 | 0.01 |
| PA1695 | pscP | Translocation protein in type III secretion | −3.85 | 0.01 |
| PA1696 | pscO | Translocation protein in type III secretion | 21.12 | 0.00 |
| PA1699 | Conserved hypothetical protein in type III secretion | −3.57 | 0.02 | |
| PA1705 | pcrG | Regulator in type III secretion | −2.19 | 0.01 |
| PA1716 | pscC | Type III secretion outer membrane protein PscC precursor | −2.11 (2.42) | 0.00 (0.00) |
| PA1717 | pscD | Type III export protein PscD | −2.80 | 0.01 |
| PA1718 | pscE | Type III export protein PscE | −7.61 | 0.01 |
| PA1719 | pscF | Type III export protein PscF | −2.19 | 0.00 |
| PA1721 | pscH | Type III export protein PscH | −3.50 | 0.00 |
| PA1724 | pscK | Type III export protein PscK | −2.32 | 0.00 |
| Pyoveridine-related genes | ||||
| PA2385 | pvdQ | PvdQ | −18.74 | 0.00 |
| PA2386 | pvdA | l-Ornithine N5-oxygenase | −9.60 | 0.00 |
| PA2392 | pvdP | PvdP | −31.94 | 0.00 |
| PA2394 | pvdN | PvdN | −15.56 | 0.00 |
| PA2395 | pvdO | PvdO | −7.55 | 0.00 |
| PA2396 | pvdF | Pyoverdine synthetase F | −5.07 | 0.00 |
| PA2397 | pvdE | Pyoverdine biosynthesis protein PvdE | −7.66 | 0.00 |
| PA2399 | pvdD | Pyoverdine synthetase D | −5.28 | 0.02 |
| PA2400 | pvdJ | PvdJ | −10.38 | 0.00 |
| PA2402 | Probable nonribosomal peptide synthetase | −13.28 | 0.00 | |
| PA2413 | pvdH | Diaminobutyrate-2-oxoglutarate aminotransferase | −7.53 | 0.00 |
| PA2424 | pvdL | PvdL | −15.24 | 0.00 |
| PA2425 | pvdG | PvdG | −8.19 | 0.00 |
| Pyochelin-related genes | ||||
| PA4219 | Hypothetical protein | −2.79 | 0.00 | |
| PA4220 | Conserved hypothetical protein | −2.82 | 0.01 | |
| PA4221 | fptA | Fe(III)-pyochelin outer membrane receptor precursor | −6.97 | 0.01 |
| PA4222 | Probable ATP-binding component of ABC transporter | −4.06 (−3.54) | 0.00 (0.00) | |
| PA4223 | Probable ATP-binding component of ABC transporter | −3.85 (−2.77) | 0.01 (0.00) | |
| PA4224 | pchG | Pyochelin biosynthetic protein PchG | −4.67 | 0.00 |
| PA4225 | pchF | Pyochelin synthetase | −5.06 | 0.00 |
| PA4226 | pchE | Dihydroaeruginoic acid synthetase | −3.15 | 0.00 |
| PA4229 | pchC | Pyochelin biosynthetic protein PchC | −2.27 | 0.00 |
| PA4230 | pchB | Salicylate biosynthesis protein PchB | −2.83 (−2.93) | 0.00 (0.00) |
| PA4231 | pchA | Salicylate biosynthesis isochorismate synthase | −3.45 | 0.00 |
| Bacteriophage Pf1 genes | ||||
| PA0717 | Hypothetical protein of bacteriophage Pf1 | 342.12 (10.08) | 0.00 (0.00) | |
| PA0718 | Hypothetical protein of bacteriophage Pf1 | 271.30 (5.74) | 0.00 (0.00) | |
| PA0719 | Hypothetical protein of bacteriophage Pf1 | 109.98 | 0.00 | |
| PA0720 | Helix-destabilizing protein of bacteriophage Pf1 | 325.95 (24.11) | 0.00 (0.00) | |
| PA0721 | Hypothetical protein of bacteriophage Pf1 | 375.78 | 0.00 | |
| PA0725 | Hypothetical protein of bacteriophage Pf1 | 7.01 (3.69) | 0.00 (0.00) | |
| PA0726 | Hypothetical protein of bacteriophage Pf1 | 47.72 | 0.00 | |
| PA0727 | Hypothetical protein from bacteriophage Pf1 | 11.96 | 0.00 |
The values not in parentheses are data for semiviscous swarming conditions, and the values in parentheses are data for liquid swarming conditions.
Under both conditions, it was found that disruption of the metR gene resulted in downregulation of the metH (methionine synthase) gene. The metH gene was downregulated 1.5- and 2.9-fold under liquid swarming and semiviscous swarming conditions, respectively, and these values were confirmed by RT-qPCR. Furthermore, the metH mutant was completely defective in both swarming and swimming motility when it was tested using the appropriate motility assay. Significant twofold downregulation was also observed in the metR mutant for the metF (5,10-methylenetetrahydrofolate reductase) gene, but only under semiviscous swarming conditions; this change was confirmed by RT-qPCR. The swarming motility of the metF mutant was not assayed as this mutant did not grow in BM2 minimal medium. Interestingly, the gene adjacent to metR, PA3586, which encodes a putative hydrolase and is transcribed divergently from metR, was highly upregulated (17.3-fold) under semiviscous swarming conditions and was upregulated less under liquid swarming medium conditions (2-fold). PA3586 was, however, not required for swarming.
A large number of virulence-related genes were downregulated in the metR mutant under semiviscous swarming conditions. These virulence-related genes include 11 genes involved in type III secretion, as well as the effector gene exoT and 11 and 13 genes involved in the pyochelin and pyoverdine siderophore biosynthesis pathways, respectively. In contrast, only three prospective pyochelin-related virulence genes were downregulated at least twofold in the metR mutant in liquid swarming medium. With the exception of the PA4223 mutant, which displayed a 64% decrease in swarming motility, no swarming defects compared to the wild type were observed for the other mutants with transposon insertions in the virulence-related genes (data not shown).
Strong upregulation in the metR mutant (Table 2) was observed for several genes in the filamentous bacteriophage Pf1 cluster, and this upregulation was greater under semiviscous swarming conditions than in liquid medium. Bacteriophage Pf1 has been suggested to be important in CF infections, and studies have shown that Pf1 genes are upregulated during biofilm development (49, 64). An increase in expression as great as 325-fold compared to wild type was noted for the helix-destabilizing protein (PA0720) of the phage. Confirmation of the expression levels of the PA0720 gene by RT-qPCR showed even stronger upregulation, 1,721-fold. However, these genes do not appear to be required for swarming motility as transposon insertions in them did not affect the swarming motility of the mutants compared to the wild type.
Only two transcriptional regulator genes and two sensor kinase genes were observed to be differentially expressed (least twofold) in the metR mutant in liquid swarming medium (data not shown). In contrast, 20 transcriptional regulators and two-component system sensor kinases or response regulators were dysregulated in the metR mutant under semiviscous conditions (Table 2). None of the regulators identified using liquid swarming medium were among the dysregulated regulators identified using semiviscous swarming medium. This result suggests that the viscosity of the bacterium's environment may cause the metR transcriptional regulator to exert different downstream effects by inducing or repressing different regulators. Interestingly, one of the dysregulated regulators was NtrC, which was required for swarming motility, but metR negatively regulated NtrC.
Comparison of the genes that were positively regulated by MetR (i.e., that exhibited reduced expression in the metR mutant) with the genes required for swarming motility revealed PA1843 (metH), PA2399 (pvdD), PA2413 (pvdH), PA2891, PA4144, and PA4223. Conversely, PA0633, PA0848, and PA5125 (ntrC) were negatively regulated by metR and also were required for swarming motility. These genes are proposed to be the genes that mediate the effects of MetR on swarming motility.
DISCUSSION
In this study, 233 mutants from the P. aeruginosa strain PA14 transposon insertion mutant library were verified to have alterations in swarming motility. The large number of genes (4% of the PA14 genome) found to be involved in this form of motility and the variety of functions performed by these genes support the previous finding that swarming is a complex adaptive behavior.
A classification of all 233 swarming-associated genes according to their predicted functions (www.pseudomonas.com) is shown in Fig. S1A in the supplemental material. When these genes were compared with the functional composition of the entire PA14 genome (see Fig. S1B in the supplemental material), they were found to be in almost all of the functional categories for PA14 except the “noncoding RNA gene” and “related to phage, transposon, or plasmid” categories. Interestingly, a significant proportion of the swarming-associated genes (12%) were regulator genes (i.e., transcriptional regulator or two-component system genes). As well, a comparison of the functional composition of the swarming-associated genes with that of the PA14 genome revealed that 6% of all known or putative regulator genes in the PA14 genome are involved in swarming (see Fig. S1C in the supplemental material). The relatively large number of regulators involved in this form of motility supports the hypothesis that swarming may be highly regulated. As well, our study revealed that the regulatory system for swarming overlaps with other complex adaptations, such as biofilm and quorum sensing. A greater effort to study the functions of genes in the PA14 genome is important as nearly 30% of the swarming-associated genes identified are currently labeled hypothetical genes.
In this study, we confirmed that 35 P. aeruginosa PA14 mutants showed altered swarming motility and had transposon insertions in genes encoding transcriptional regulators. Twenty-seven of these mutants were newly identified in this study. There are hints that some of these regulators might be involved in part in controlling flagellum biosynthesis or function, but only a single mutant showed a strong swimming motility defect. Thus, the great disparity in the extents of the defects of all mutants except the PA1458 mutant leads to the conclusion that other factors must be important in determining swarming deficiency in most mutants. Similarly, only five mutants (pilH, PA2571, algR, nosR, and ntrB mutants) had substantial or moderate deficiencies in twitching motility and displayed resistance to phage PO4, indicating that most regulators are not obligately involved in type IV pilus functioning.
With the exception of the rhlR, rsaL, anr, PA4398, and PA1347 mutants, all the regulatory mutants showed the same formation of a dark blue halo around a colony as the wild-type strain, indicating that there was efficient production of the biosurfactant. RhlR is a well-studied transcriptional regulator that plays a central role in the quorum-sensing response. Upon interaction with its quorum-sensing molecule, N-butyryl-l-homoserine lactone, RhlR influences the expression of a variety of quorum-sensing-regulated genes, including the rhamnolipid biosynthesis genes (11). Evidently, a mutation in the rhlI gene, which is involved in the biosynthesis of N-butyryl-l-homoserine lactone, leads to the inability of the rhlI mutant to swarm (see Table S1 in the supplemental material). The rsaL gene is located between the lasR and lasI genes, and its expression is directly activated by LasRI. In turn, the RsaL protein can specifically represses the transcription of lasI, which is involved in the synthesis of the quorum-sensing signal molecule N-3-oxo-dodecanoyl-l-homoserine lactone (51). We are unable to decipher why this regulator decreases rhamnolipid synthesis, as observed in this study. A study by Caiazza et al. suggested that HAAs (rhamnolipid precursors) are the minimal surfactants required for swarming in P. aeruginosa, and although rhamnolipids are not required for swarming motility, they modulate the swarming pattern (11). Therefore, it is also important to study the mechanism by which rsaL, anr, PA1347, and PA4398 influence the generation of HAAs.
While it was reported that mutations in O-antigen or lipopolysaccharide (LPS) synthesis eliminate the swarming motility of Salmonella enterica serovar Typhimurium (12), in our study, transposon insertions in O-antigen and LPS synthesis genes in P. aeruginosa did not seem to affect the ability of strains to swarm (see Table S1 in the supplemental material). It is possible that the production of secreted biosurfactants, rhamnolipids and HAAs, make P. aeruginosa less dependent on LPS and O antigen to improve the surface “wettability” that is required for swarming colony expansion. In contrast, LPS and O antigen play an important role in the swarming motility of Salmonella strains, as these bacteria do not excrete biosurfactants and most likely utilize their outer surface as a biosurfactant (12).
For some of the swarming-deficient mutants, previous studies provided greater insight into how the corresponding regulators might affect swarming. The gbuR mutant was unable to swarm in our assay. Expression of the gbuR gene is normally induced by the alternative sigma factor RpoS (56). Interestingly, RpoS is involved in the control of expression of a large number of quorum-sensing-regulated genes. More specifically, many of the genes that are controlled by RpoS are also under the control of rhlR and lasR, which encode two quorum-sensing regulators that are important for swarming (56).
A strong swarming deficiency was also observed for the PA1458 mutant. The PA1458 gene is predicted to be involved in chemotaxis. PA1458 in P. aeruginosa is homologous to cheA in Pseudomonas putida and E. coli. Upon chemoattractant recognition at the cell surface by a methyl-accepting chemotaxis protein, CheA is autophosphorylated. CheY is then phosphorylated by CheA and interacts with switch proteins in the flagellar motor. Disruption of the cheA gene in P. putida resulted in a nonswarming, nonchemotactic mutant that was unable to change the direction of flagellar rotation but was still able to swim (21). We found an even stronger effect on flagellar function in the P. aeruginosa PA1458 mutant, as this mutant displayed no swimming and almost no swarming motility. Another indication that PA1458 is involved in flagellar functioning is the observation that PA1458 expression depends on FleQ, the key regulator of flagellar biosynthesis (14).
An interesting trend was found for the relationship between swarming and biofilm formation. Both of these surface-associated behaviors depend on quorum sensing, flagella, and type IV pili (32, 43). For some P. aeruginosa mutants, defects in swarming coincide with deficiencies in biofilm formation (1, 61). The results obtained here, however, support the idea that there is an inverse relationship between the regulation of swarming and biofilm formation, which has also been proposed in other studies (10, 41). Most of the nonswarming mutants with transposon insertions in transcriptional regulator genes showed strong increases in biofilm formation. In contrast, the hyperswarming gacA mutant is deficient in biofilm formation (47). Importantly, swarming has been proposed to influence the early stages of biofilm formation, with actively swarming cells forming flat biofilms and swarming-deficient cells forming structured biofilms (57). As we observed an increase in the total biofilm mass in our solid-surface biofilm assays for most mutants with strong swarming defects, this may be explained by less-motile cells forming structured biofilms (15, 34).
To gain insight into how genes putatively involved in metabolism influence the complex swarming behavior, we took a closer look at the metR mutant by analyzing its gene expression profile. The function of metR in P. aeruginosa has not been described, but as mentioned above, the metR regulator gene of E. coli, which is 62% similar, has been studied in some detail. In E. coli the MetR protein activates expression of the metH and metE genes and probably metA, all of which are involved in methionine synthesis. Also, metR has been shown to downregulate its own transcription and to be repressed by MetJ (9, 24). The methionine synthase genes metH (PA1843) and metE (PA1927) are present in P. aeruginosa and are 79% and 74% similar to their E. coli counterparts, respectively. In addition to methionine synthesis, metR regulates the activation of transcription of glyA, which encodes a serine hydroxymethyltransferase that catalyzes the interconversion of serine, glycine, and one-carbon units. This reaction is the major source of one-carbon units in the cell and is used in a variety of metabolic processes, including methionine biosynthesis (37, 48). In addition, a role for metR in biofilm formation in E. coli has been suggested (28).
In E. coli, the last step of methionine synthesis can be catalyzed by either MetH or MetE synthase (25). In P. aeruginosa, there is no experimental evidence that MetH and MetE act as methionine synthases, but these proteins have been predicted to have the same function as their counterparts in E. coli, based on similarities in their protein domains. We hypothesized that the swarming-deficient phenotype of the metR mutant of P. aeruginosa might have been caused by reduced intracellular methionine levels, due to impairment of methionine biosynthesis. This idea was supported by the finding that metH was downregulated in the metR mutant and by the finding that a metH mutant was completely unable to swarm. However, metE and metX were not found to be differentially regulated in the microarray experiment, and mutants with mutations in these genes swarmed at levels comparable to the wild-type level. MetX has been predicted to catalyze an essential step earlier in the pathway of biosynthesis of this amino acid (2). The normal swarming behavior of metE and metX mutants indicates that defects in methionine biosynthesis do not per se impair swarming. Rather, the effect on swarming is specific to metH and metR and is likely to be caused by functions of these genes other than methionine biosynthesis. Moreover, in exploratory studies, addition of high levels (200 μg/ml) of methionine or homocysteine to swarm plates did not restore swarming in either the metR or metH mutant. As well, the metR mutant is not auxotrophic for methionine since it grows as well as the wild type in BM2 minimal medium, which lacks methionine. When the data are taken together, while the downregulation of metH expression in the metR mutant can partially explain the swarming-defective phenotype of this mutant, it seems unlikely that low methionine levels caused the impaired swarming motility in our assays, and this indicates that metH probably functions in processes other than methionine synthesis in P. aeruginosa.
Besides metH, 13 genes located in a cluster involved in pyochelin biosynthesis (PA4218 to PA4231) were found to be downregulated in the swarming-deficient metR mutant under semiviscous swarming conditions. Interestingly, Overhage et al. found increased expression of most genes in this cluster in a microarray comparing the gene expression of swarming cells to the gene expression of cells growing under planktonic conditions (46). The swarming motility of a PA4223 mutant was found to be impaired, whereas other mutants with mutations in this cluster of genes (fptA, PA4222 [this study], pchF, and pchE [46]) had no swarming defects. This indicates that although nine genes in this cluster are downregulated in the metR mutant, only PA4223 is likely to contribute to the swarming-deficient phenotype of this mutant. Although PA4223 is located in a gene cluster related to pyochelin synthesis, studies show that it does not have a function in this process. The PA4223 protein resembles an ABC transporter with an export function. This transporter does not export pyochelin as pyochelin levels were not affected by mutations in PA4223 (53). The molecule(s) that is exported via this transporter is currently unknown. An interesting role for PA4223 was suggested recently, as transposon insertions in this gene were found to cause a strong decrease in bacterial virulence against the amoeba Dictyostelium. This phenotype was explained by the observation that PA4223 is involved in induction of the type III secretion system (3). Overall, the downregulation of virulence-related genes in the metR mutant under semiviscous swarming conditions suggests that the MetR transcriptional regulator may play an important role in regulating these virulence-associated genes. It will be of great interest to investigate MetR in greater detail to learn how this regulator contributes to the pathogenesis of the bacterium, including its role in the process of iron acquisition in the host and possibly colonization and dissemination.
Another interesting observation resulting from the microarray analysis of the metR mutant was the very strong upregulation of nine genes from the bacteriophage Pf1 gene cluster (PA0717 to PA0728). Remarkably, a very similar pattern of expression of phage genes was found in P. aeruginosa biofilms, leading to 100- to 1,000-fold increases in Pf1 phages in the environment of the biofilm compared to planktonic cell cultures (66). Phage induction in biofilms was associated with bacterial cell lysis and dispersion of cells. It was hypothesized that cell lysis may contribute to creating voids within biofilm microcolonies, thereby facilitating dispersal of a subpopulation of surviving cells (63). In contrast, “sticky” P. aeruginosa colony variants, characterized by autoaggregation in liquid culture and hyperadherence to solid surfaces, displayed the opposite pattern of gene expression, with a strong decrease in phage gene transcription. The similarity between the patterns of phage gene expression in biofilms and in the metR mutant is striking, but the relationship between these two phenotypes is unclear. Besides upregulation of the bacteriophage gene cluster, there was very little overlap between the gene expression patterns of biofilm cells (66) and those of the metR mutant (Table 2) grown under planktonic conditions. Also, no common transcriptional regulators were found in the two microarrays. As metR was not downregulated in biofilm cells, no direct effect of metR on phage gene expression is expected in this situation.
By performing preliminary screening of the P. aeruginosa PA14 transposon insertion mutant library, we found that approximately 4% of the genes for which mutants are present in this library are involved in swarming behavior. The complexity of this form of motility is reflected by the high number of regulators that were found to be involved in swarming behavior. Seventeen regulators and 10 two-component sensors and response regulators were newly identified in this study. To further understand how swarming behavior is regulated, it is important to study the hierarchy of the transcriptional regulators involved.
Supplementary Material
Acknowledgments
We thank Hanne C. Winther-Larsen (University of Oslo, Oslo, Norway) for providing bacteriophage PO4.
This research was funded by the Canadian Institutes of Health Research. R.E.W.H. holds a Canada Research Chair. I.W. was supported by the Juergen Manchot Foundation. J.O. was supported by a fellowship from the Canadian Cystic Fibrosis Foundation. A.T.Y.Y. received studentships from the Canadian Cystic Fibrosis Foundation and the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 10 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abbas, A., C. Adams, N. Scully, J. Glennon, and F. O'Gara. 2007. A role for TonB1 in biofilm formation and quorum sensing in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 274269-278. [DOI] [PubMed] [Google Scholar]
- 2.Alaminos, M., and J. L. Ramos. 2001. The methionine biosynthetic pathway from homoserine in Pseudomonas putida involves the metW, metX, metZ, metH and metE gene products. Arch. Microbiol. 176151-154. [DOI] [PubMed] [Google Scholar]
- 3.Alibaud, L., T. Köhler, A. Coudray, C. Prigent-Combaret, E. Bergeret, J. Perrin, M. Benghezal, C. Reimmann, Y. Gauthier, C. van Delden, I. Attree, M. O. Fauvarque, and P. Cosson. 2008. Pseudomonas aeruginosa virulence genes identified in a Dictyostelium host model. Cell. Microbiol. 10729-740. [DOI] [PubMed] [Google Scholar]
- 4.Arora, S. K., A. N. Neely, B. Blair, S. Lory, and R. Ramphal. 2005. Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 734395-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley, D. E. 1973. Basic characterization of a Pseudomonas aeruginosa pilus-dependent bacteriophage with a long noncontractile tail. J. Virol. 121139-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinkman, F. S., E. L. Macfarlane, P. Warrener, and R. E. W. Hancock. 2001. Evolutionary relationships among virulence-associated histidine kinases. Infect. Immun. 695207-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzik, J. M., W. A. Rosche, A. Rietsch, and G. A. O'Toole. 2004. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J. Bacteriol. 1863270-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullock, W. O., J. M. Fernandez, and J. M. Stuart. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5376-379. [Google Scholar]
- 9.Cai, X. Y., M. E. Maxon, B. Redfield. R. Glass, N. Brot, and H. Weissbach. 1989. Methionine synthesis in Escherichia coli: effect of the MetR protein on metE and metH expression. Proc. Natl. Acad. Sci. USA 864407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caiazza, N. C., J. H. Merritt, K. M. Brothers, and G. A. O'Toole. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1893603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caiazza, N. C., R. M. Shanks, and G. A. O'Toole. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 1877351-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, B. G., L. Turner, and H. C. Berg. 2007. The wetting agent required for swarming in Salmonella enterica serovar Typhimurium is not a surfactant. J. Bacteriol. 1898750-8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64391-397. [DOI] [PubMed] [Google Scholar]
- 14.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. [DOI] [PubMed] [Google Scholar]
- 15.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1851027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deziel, E., G. Paguette, R. Villemur, F. Lepine, and J. G. Bisaillon. 1996. Biosurfactant production by a soil Pseudomonas strain growing on polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 621908-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deziel, E., F. Lepine, D. Dennie, D. Boismenu, O. A. Manner, and R. Villemur. 1999. Liquid chromatography/mass spectrometry analysis of mixtures or rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta 1440244-252. [DOI] [PubMed] [Google Scholar]
- 18.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 1831195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 1492005-2013. [DOI] [PubMed] [Google Scholar]
- 20.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Cámara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of n-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 1842576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ditty, J. L., A. C. Grimm, and C. S. Harwood. 1998. Identification of a chemotaxis gene region from Pseudomonas putida. FEMS Microbiol. Lett. 159267-273. [DOI] [PubMed] [Google Scholar]
- 22.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2630-635. [DOI] [PubMed] [Google Scholar]
- 23.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51675-690. [DOI] [PubMed] [Google Scholar]
- 24.Fritsch, P. S., M. L. Urbanowski, and G. V. Stauffer. 2000. Role of the RNA polymerase alpha subunits in MetR-dependent activation of metE and metH: important residues in the C-terminal domain and orientation requirements within RNA polymerase. J. Bacteriol. 1825539-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez, J. C., R. V. Banerjee, S. Huang, J. S. Sumner, and R. G. Matthews. 1992. Comparison of cobalamin-independent and cobalamin-dependent methionine synthases from Escherichia coli: two solutions to the same chemical problem. Biochemistry 316045-6056. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Hancock, R. E. W., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updates 32622-2630. [DOI] [PubMed] [Google Scholar]
- 28.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heurlier, K., F. Williams, S. Heeb, C. Dormond, G. Pessi, D. Singer, M. Cámara, P. Williams, and D. Haas. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1862936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchison, M. L., and J. R. Govan. 1999. Pathogenicity of microbes associated with cystic fibrosis. Microbes Infect. 11005-1014. [DOI] [PubMed] [Google Scholar]
- 31.Kay, E., B. Humair, V. Dénervaud, K. Riedel, S. Spahr, L. Eberl, C. Valverde, and D. Haas. 2006. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 1886026-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirisits, M. J., and M. R. Parsek. 2006. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell. Microbiol. 81841-1849. [DOI] [PubMed] [Google Scholar]
- 33.Köhler, T., L. K. Curty, F. Barja, C. Van Delden, and J. C. Pechère. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 1825990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lequette, Y., and E. P. Greenberg. 2005. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 18737-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewenza, S., R. K. Falsafi, G. Winsor, W. J. Gooderham, J. B. McPhee, F. S. Brinkman, and R. E. Hancock. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 1032833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenz, E., and G. V. Stauffer. 1995. Characterization of the MetR binding sites for the glyA gene of Escherichia coli. J. Bacteriol. 1774113-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Maxon, M. E., B. Redfield, X. Y. Cai, R. Shoeman, K. Fujita, W. Fisher, G. Stauffer, H. Weissbach, and N. Brot. 1989. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc. Natl. Acad. Sci. USA 8685-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McPhee, J. B., M. Bains, G. Winsor, S. Lewenza, A. Kwasnicka, M. D. Brazas, F. S. Brinkman, and R. E. Hancock. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 1883995-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and G. A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 1898154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray, T. S., and B. I. Kazmierczak. 2008. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J. Bacteriol. 1902700-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 44.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 45.Overhage, J., S. Lewenza, A. K. Marr, and R. E. W. Hancock. 2007. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 1892164-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overhage, J., M. Bains, M. D. Brazas, and R. E. W. Hancock. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 1902671-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkins, M. D., H. Ceri, and D. G. Storey. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 401215-1226. [DOI] [PubMed] [Google Scholar]
- 48.Plamann, M. D., and G. V. Stauffer. 1989. Regulation of the Escherichia coli glyA gene by the metR gene product and homocysteine. J. Bacteriol. 1714958-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Platt, M. D., M. J. Schurr, K. Sauer, G. Vazquez, I. Kukavica-Ibrulj, E. Potvin, R. C. Levesque, A. Fedynak, F. S. Brinkman, J. Schurr, S. H. Hwang, G. W. Lau, P. A. Limbach, J. J. Rowe, M. A. Lieberman, N. Barraud, J. Webb, S. Kjelleberg, D. F. Hunt, and D. J. Hassett. 2008. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J. Bacteriol. 1902739-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 2681899-1902. [DOI] [PubMed] [Google Scholar]
- 51.Rampioni, G., F. Polticelli, I. Bertani, K. Righetti, V. Venturi, E. Zennaro, and L. Leoni. 2007. The Pseudomonas quorum-sensing regulator RsaL belongs to the tetrahelical superclass of H-T-H proteins. J. Bacteriol. 1891922-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 974885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimmann, C., H. M. Patel, L. Serino, M. Barone, C. T. Walsh, and D. Haas. 2001. Essential PchG-dependent reduction in pyochelin biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 183813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Plainview, NY.
- 55.Reference deleted.
- 56.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51973-985. [DOI] [PubMed] [Google Scholar]
- 57.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 621264-1277. [DOI] [PubMed] [Google Scholar]
- 58.Toutain, C. M., M. E. Zegans, and G. A. O'Toole. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tremblay, J., A. P. Richardson, F. Lépine, and E. Déziel. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 92622-2630. [DOI] [PubMed] [Google Scholar]
- 60.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner, V. E., L. L. Li, V. M. Isabella, and B. H. Iglewski. 2007. Analysis of the hierarchy of quorum-sensing regulation in Pseudomonas aeruginosa. Anal. Bioanal. Chem. 387469-479. [DOI] [PubMed] [Google Scholar]
- 62.Wang, C. J., J. S. Jerng, K. Y. Chen, L. N. Lee, C. J. Yu, P. R. Hsueh, and P. C. Yang. 2006. Pandrug-resistant Pseudomonas aeruginosa among hospitalised patients: clinical features, risk factors and outcomes. Clin. Microbiol. Infect. 1263-68. [DOI] [PubMed] [Google Scholar]
- 63.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1854585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Webb, J. S., M. Lau, and S. Kjelleberg. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1868066-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 14881-86. [DOI] [PubMed] [Google Scholar]
- 66.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413860-864. [DOI] [PubMed] [Google Scholar]
- 67.Winsor, G. L., T. Van Rossum, R. Lo, B. Khaira, M. D. Whiteside, R. E. W. Hancock, and F. S. Brinkman. 2009. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.