Abstract
Approximately one-third of the human population is asymptomatically colonized by Staphylococcus aureus. However, much of the global diversity within the carriage populations remains uncharacterized, and it is unclear to what degree the variation is geographically partitioned. We isolated 300 carriage isolates from 1,531 adults contemporaneously in four countries: France, Algeria, Moldova, and Cambodia. All strains were characterized by multilocus sequence typing. Six clonal complexes (CCs) were present in all four samples (CC30, -45, -121, -15, -5, and -8). Analyses based on the genotype frequencies revealed the French and Algerian samples to be most similar and the Cambodian sample to be most distinct. While this pattern is consistent with likely rates of human migration and geographic distance, stochastic clonal expansion also contributes to regional differences. Phylogenetic analysis revealed a highly divergent and uncharacterized genotype (ST1223) within Cambodia. This lineage is related to CC75, which has previously been observed only in remote aboriginal populations in northern Australia.
Although better known as an important human pathogen, Staphylococcus aureus is typically a commensal species and asymptomatically colonizes approximately one-third of the human population globally (18, 20, 29). This high carriage rate potentially represents a vast reservoir of as-yet-uncharacterized S. aureus diversity, an appreciation of which should shed light on the forces underpinning the diversification and dissemination of S. aureus. There are comparatively few studies examining spatial or temporal genotype distributions within carriage populations, and the extent of biogeographical structure is currently unclear, as is the level of discrimination which might be required to detect such structure.
Multilocus sequence typing (MLST) has proved to be very successful as an epidemiological tool in that it delimits S. aureus in to a small number of widespread and discrete clonal complexes (CCs) (6, 8). These can be readily identified as clusters of related genotypes which have diversified radially from “founder” genotypes (9), and because this organism is largely clonal (8), assignments of isolates to these groups is broadly robust to the many different typing methods employed (4, 10, 27). The high level of divergence between these lineages suggests that they are relatively ancient and temporally stable (7), and it is possible that isolated host populations may have been colonized by different S. aureus lineages in the past. However, any footprints of geographical partitioning are likely to have been compromised by high rates of migration in recent times, due largely to the advent of air travel.
Previous studies addressing the characterization of carried populations have tended to focus on samples from Western Europe or North America, and these have generally not provided strong evidence for geographical structuring. In a recent study using amplified fragment length polymorphism to compare the carried populations in Holland and North America, the authors noted considerable overlap between the samples, suggesting that they effectively constituted a single unstructured population (17). Similarly, independent MLST studies have revealed regional consistencies in Europe, such as the predominance of CC30 in the United Kingdom (8), Ireland (3), and Switzerland (25). Given the high rates of admixture within Europe and North America, the absence of obvious geographical structuring in the carried S. aureus population in these regions is perhaps not very surprising.
Although they are currently scarce, current data from carriage populations outside of Europe or North America point to greater geographical structuring. For example, a sample of carried S. aureus recovered from Bamako, Mali, has recently been characterized, constituting the first such study of an African population (23). Although many of the previously characterized CCs were also present in this sample, the authors noted a high frequency (∼25%) of a single genotype, ST152, which is phylogenetically divergent and noted very rarely in Europe. The high frequency of ST152 in this population raises the possibility that this genotype is endemic to the Malian population and possibly elsewhere in sub-Saharan Africa. This in turn hints at greater geographical partitioning on a global scale, although more representative samples are clearly required. To address this, we generated MLST data from contemporaneous carriage samples recovered from four countries representing three continents: France (Western Europe), Moldova (Eastern Europe), Algeria (North Africa), and Cambodia (Southeast Asia). To our knowledge, this is the first time such a study has been carried out on samples from Eastern Europe, North Africa, or Southeast Asia. These data were therefore generated to uncover diversity within the global carriage population but also to understand further the extent to which geographical distance and host migration can explain regional differences.
MATERIALS AND METHODS
Population and study design.
Between June 2005 and October 2006, asymptomatically carried S. aureus isolates were collected from four diverse countries contemporaneously and following the same sampling procedure. For each country, nasal swabs were obtained by an investigator trained in the coordinating center of the study, using a standard procedure. Nasal swabs were taken from patients (>15 to <80 years old) admitted to the hospital for emergency surgery within 8 h of the admission. Demographic data (age and sex) were collected prospectively for all patients admitted. The hospitals enrolled in the study were four tertiary care hospitals: Bichat-Claude-Bernard Hospital (956 beds) in Paris, France; Emergency Hospital (589 beds) in Chisinau, Moldova; Dr Tidjani Damerdji Hospital (850 beds) in Tlemcen, Algeria; and Calmette Hospital (270 beds) in Phnom Penh, Cambodia.
Isolate collection.
Nasal swabs were collected from both anterior nares of each patient, transported, and stored at 4°C for a maximum of 24 h until they were inoculated on mannitol salt agar plates (Oxoid, Basingstoke, United Kingdom) for S. aureus detection and transported frozen to the coordinating center of the study as described elsewhere (24). The plates were incubated at 37°C and examined for growth after 24 to 48 h. Isolates that produced yellow colonies were identified as S. aureus using the coagulase plasma test and triplex real-time PCR (22). Confirmed S. aureus isolates were stored at −80°C in the microbiology laboratory of Bichat-Claude-Bernard Hospital.
MLST.
Amplification and sequencing was performed as described previously (6), except that we used the tpi primers described elsewhere (2). For two isolates, we were unable to amplify aroE, and we redesigned the up and down primers for this gene as follows: aroE745-up, TTATCACCGTCGATGCATAGTGCA; aroE255-down, CGGAGTAGTATTTATCACAATATC. For Staphylococcus simiae, we were unable to amplify the glpF and gmk genes using the standard MLST primers, and we redesigned these primers as follows: glpF-down, ATTGGYAAWATHGCATGWGCRAT; glpF-up, TTYGGKGGTGGCGTTTGTGC; gmk-down, CGCGYTCTCKYTTYAARTGYTCWGC; gmk-up, TAATYGTTYTWTCAGGHCCWTCWGG. Allele and ST assignments were made by comparisons to the S. aureus MLST database (http://saureus.mlst.net), to which the final data were submitted.
16S rRNA gene sequencing.
For the two Cambodian strains in ST1223, a fragment of 1,330 bp of the 16S rRNA gene was amplified and sequenced using the primers described previously (21).
Data analysis.
The data were assigned to CCs using eBURST (www.eburst.mlst.net) (9). Heterozygosity (H) (the probability that two random isolates will be identical) was calculated for each sample in turn and for the pooled data set using Multilocus v1.3 (1). Cluster analysis of the frequency data was carried out based on Pearson distances and the average linkage method, as implemented in Minitab. To examine the extent of population differentiation between the samples, we used Multilocus v1.3 to calculate Wier's formulation for Wright's FST, θ (30). Phylogenetic analysis was carried out on the concatenated MLST data using the neighbor-joining algorithm (Kimura two-parameter distance estimation) as implemented in MEGA 4.0 (15).
Nucleotide sequence accession numbers.
The accession numbers for the aroE, glpF, gmk, pta, tpi, and yqiL sequences of S. simiae are FJ705815 to FJ705820, respectively.
RESULTS
Carriage samples.
Nasal swabs were taken from a total of 1,531 patients admitted to four hospitals: Bichat Claude Bernard Hospital (Paris, France) (n = 421), Emergency Hospital (Chisinau, Moldova) (n = 338), Dr Tidjani Damerdji Hospital (Tlemcen, Algeria) (n = 330), and Calmette Hospital (Phnom Penh, Cambodia) (n = 442). The mean age of all the patients was 38.6 years, and that when only the carriage-positive patients were considered was 37.3 years (Table 1). The mean ages of the carriers were very similar in France (41.1 years) and Algeria (41.8 years) but were significantly younger in Moldova (32.7 years) and Cambodia (31.6 years). In France, Algeria, and Cambodia the mean age of carriers was very similar to the mean age of all patients sampled. In Moldova carriers were on average younger (32.7 years) than expected from the whole patient sample from Moldova (mean, 39 years). There is no obvious explanation for this, as the same sampling regimen was used in all cases.
TABLE 1.
Patient characteristics
| Country (city) | Study period | No. of individuals | Prevalence of S. aureus carriers (%) | Mean age (yr)
|
Sex ratio (male/female)
|
||
|---|---|---|---|---|---|---|---|
| Carriers/noncarriers | All | Carriers/noncarriers | All | ||||
| France (Paris) | June to October 2005 | 421 | 19.2 | 41.1/44.4 | 43.8 | 3.26/1.64 | 1.84 |
| Republic of Moldova (Chisinau) | June to October 2005 | 338 | 25.1 | 32.7/41.2 | 39 | 0.85/0.85 | 0.85 |
| Algeria (Tlemcen) | June to October 2005 | 330 | 25.8 | 41.8/41.7 | 41.7 | 1.19/1.36 | 1.23 |
| Cambodia (Phnom Penh) | June to October 2006 | 442 | 11.1 | 31.6/31 | 31.1 | 1.45/2.6 | 2.43 |
| All | June 2005 to October 2006 | 1,531 | 19.6 | 37.3/38.9 | 38.6 | 1.48/1.53 | 1.52 |
Considering both carriers and noncarriers, a higher proportion of males was included in the Cambodian (male/female sex ratio, 2.43), French (sex ratio, 1.84), and Algerian (sex ratio, 1.23) samples, whereas a higher proportion of females was noted in the Moldovan sample (sex ratio, 0.85). Carriage rates were similar for France (19.2%), Moldova (25.1%), and Algeria (25.8%), but these rates were approximately twice as high as that observed in Cambodia (11.1%). We note no significant difference in the sex ratio between carriers and noncarriers in the samples from Algeria, Moldova, and Cambodia. For the French sample, however, we note that carriage is significantly more common in males (P = 0.014). This is in contrast with a previous study performed under the same conditions in Mali, which reported significantly lower carriage rates in males (23). Again, the reasons for this difference are unclear.
Comparisons of genotype frequency.
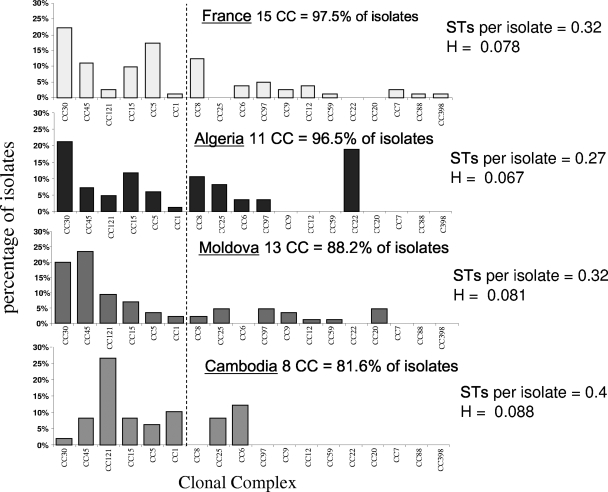
MLST defined 65 STs among the 300 isolates, which were grouped by eBURST into 18 CCs. A total of 30 novel (previously unrecorded) alleles were detected, 15 of which were in the Cambodian sample. The frequencies of the 18 CCs in the four samples are given in Fig. 1. Six CCs (CC30, CC45, CC121, CC15, CC5, and CC1) are present in each of the four countries. All of these except CC1 were also previously noted in the Malian population (23), thus confirming their dissemination throughout Europe, Asia, and Africa. CC30 is the most common complex in France (22%) and Algeria (21%) and the second most common in Moldova (20%). Our data are therefore consistent with previous reports suggesting that CC30 is stably maintained at a high frequency in the carriage population over a large area, including Europe and North America (3, 8, 17, 25), and the sample from Algeria reveals that this domain extends to North Africa.
FIG. 1.
Proportions all CCs identified within each of the four samples. eBURST analysis was used to subdivide the data into a total of 18 CCs, each named according to the ST of the assigned founder. Frequency profiles of these 18 CCs are given for the four countries. Six widely disseminated CCs (CC30, -45, -121, -15, -5, and -1) were observed in all four samples and are placed to the left of the dashed line. The diversity of each sample (given as STs per isolate and as H) is given.
There is no evidence for a high frequency of CC30 in the Cambodian sample, where this lineage accounted for only 2% of the isolates. This is similar to the frequency found previously in Mali, where CC30 accounted for only 4.5% of the isolates (23). Thus, the high frequency of this genotype does not extend as far south as sub-Saharan Africa or as far east as Southeast Asia. The Cambodian sample does reveal the presence of a single dominant complex, however; CC121 accounts for 26.5% of these isolates. This contrasts with the frequency of CC121 in the three other samples, where this complex accounts for <10% of the isolates. There are other noteworthy differences between the Cambodian sample and the other three samples. CC1 is consistently rare in France, Algeria, and Moldova (∼2.5%) but accounts for >10% of the Cambodian sample. The Cambodian sample is also the most diverse (in terms of heterozygosity and STs per isolate) (Fig. 1) and contains the highest number of novel alleles (which likely reflects the biases in the current database). We also note that the overall frequency of carriage in Cambodia is approximately half of that observed in the other countries.
These trends point to the distinctiveness of the Cambodian sample, consistent with its relative geographical isolation from the other samples. To examine this, we first used a conservative multivariate clustering approach based only on the frequencies of the six CCs found in all four samples. The French and Algerian samples clustered, and the Cambodian sample was the most distinct; this pattern is robust to different linkage and distance methods (not shown). This approach does not take into account the full range of allelic diversity in the data, as it discounts those CCs (and singletons) which are not present in all samples and combines related genotypes into a single CC. We examined population differentiation between the samples further by calculating Weir's θ, an unbiased estimator of population differentiation (30). The values of θ for each pair of samples support the view that French and Algerian samples are the least differentiated (i.e., most similar) (θ = 0.023), while the Cambodian sample is the most distinct (θ = 0.061) from all other samples. The Moldovan sample gave intermediate values of 0.037 and 0.042 compared to the French and Algerian samples, respectively.
In broad terms these analysis suggest that differences in local S. aureus populations are consistent with geographical distance and migration of the host. The Cambodian sample is most distinct (consistent with geographical isolation), whereas the French and Algerian samples are most similar (consistent with frequent migration). However, a closer inspection of the data reveals a number of anomalies. For example, CC45 accounts for 5 to 10% of the samples from France, Cambodia, and Algeria but 23.5% of the Moldovan sample. Similarly CC22 accounts for >18% of the Algerian sample but is absent in all the other samples. Unlike ST152, the predominant genotype in Mali (23), CC45 and -22 are known to be widespread, and therefore these high frequencies probably do not reflect endemism resulting from geographical isolation or limited gene flow. These observations might instead be explained by a combination of host movement and the stochastic expansion of newly introduced strains.
Although we made every effort to design the study so as to draw meaningful comparisons between the different samples, the possibility of sampling artifacts due to age or gender should be considered. For example, the similarities between the French and Algerian samples may reflect the similar ages of the carriers in these two countries (mean ages, 41.1 and 41.7 years, respectively), whereas the mean ages of the carriers in Moldova and Cambodia are approximately 10 years lower. However, this could not explain the differences between the Cambodian and Moldovan samples. Similarly, it is possible that the distinctiveness of the Cambodian sample is due to a higher proportion of males in the original patient sample, although we consider it very unlikely that this relatively small bias toward males could explain the paucity of CC30 and abundance of CC121 in Cambodia.
Nucleotide diversity and phylogenetic analysis.
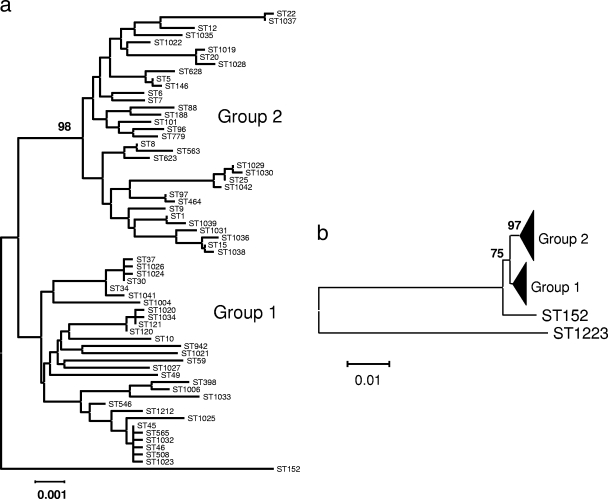
In order to examine the phylogeny and diversity of the combined samples, we constructed a neighbor-joining tree based on the concatenated MLST alleles of all unique STs (Fig. 2). Sixty-three of the 65 STs (97%) fell into the two major clades noted in previous studies (groups 1 and 2) (Fig. 2a), and the delineation of group 1 is well supported by bootstrap resampling (97%). We did not detect any obvious clustering within each of the major clades corresponding to geographical source and thus can provide no convincing evidence for the geographic restriction of particular phylogenetic clusters.
FIG. 2.
(a) Neighbor-joining tree reconstructed from the concatenated sequences of each unique ST observed in the pooled data set. The Kimura two-parameter distance measure was used as implemented in MEGA 4.0. The analysis resolved the STs in the two main groups identified previously; group 2 is supported by a bootstrap score of 98. Two STs did not cluster within the two major groups. One of these, ST1223, is omitted for the sake of clarity, while ST152 (corresponding to a single French isolate) is approximately equidistant from the two main groups. The use of ST152 as an outgroup confirms that group 1 is basal to group 2. (b) Neighbor-joining tree showing the divergent position of ST1223. Groups 1 and 2 are collapsed for the sake of clarity, and ST1223 is ∼10% diverged from these groups. Bootstrap scores for the group 2 node and the ancestral node of groups 1 and 2 are given.
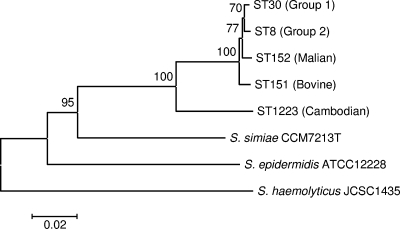
Two STs, ST152 and ST1223, did not cluster with the other genotypes. ST152 was represented by a single isolate from the French sample and is approximately equidistant from the two major clades. ST1223 is far more divergent from the other STs (Fig. 2b), and differs from ST8 (taken as a typical ST) at an average of 9.6% of nucleotide sites over all seven loci (ranging from 5.8% at gmk to 15.8% at aroE). We confirmed that these coagulase-positive strains were S. aureus on the basis of their 16S rRNA gene sequences (not shown), and phylogenetic analysis reveals them to be more similar to S. aureus than to S. simiae, which is the most closely related coagulase-negative species (19) (Fig. 3). Four other STs in the MLST database, ST75, ST258, ST883, and ST850, are closely related to ST1223. These strains were recovered from aboriginal communities in northern Australia (16).
FIG. 3.
Neighbor-joining tree of selected S. aureus genotypes and closely related coagulase-negative staphylococcal species, based on the concatenated sequences of six MLST genes. arcC is excluded due to difficulties in amplifying this gene in S. simiae. This analysis confirms that ST1223 is more closely related to other S. aureus genotypes than to the nearest coagulase-negative species and clusters with S. aureus with a bootstrap support of 100%. This is consistent with the taxonomic status of ST1223 as S. aureus.
DISCUSSION
We present MLST data from 300 isolates recovered contemporaneously from 1,531 patients in four countries. These data confirm the global dissemination of six-well documented clonal lineages: CC30, -45, -121, -15, -5, and -8. In particular, we note that the frequency of CC30 is consistent at ∼20 to 25% throughout Europe (West and East) and North Africa. A similar frequency was also recently reported from a carriage study in Switzerland (25), and CC30 accounted for 33% of isolates from a carriage study in the United Kingdom (8). It is also notable that ST30 has, since the start of 2008, been reported to be predominant in samples of community-acquired methicillin (meticillin)-resistant S. aureus from Alaska (5), Greece (12), Turkey (13), Kuwait (28), and Hong Kong (11). This points to the ecological success and transmissibility of this CC and suggests that it is not specifically adapted to populations of European descent. It is, however, rarely observed at frequencies of higher than ∼30% in carriage samples. In fact, curiously, evidence from the current study and from previous studies suggests that 30% appears to be the approximate maximum frequency for any single CC within carriage samples, although two lineages may be detected at a similarly high frequency (with the Moldovan sample being a good example). Presumably this reflects competition between lineages, which effectively caps the frequency of any single genotype within a given sample.
The current study expands the known range of CC30 predominance to North Africa but also confirms that this pattern is not global. CC30 accounts for only 2% of isolates in Cambodia, whereas CC121 accounts for 26.5% of these isolates. Although it is possible that this pattern extends throughout Southeast Asia and China, a recent study of pediatric carriage suggests that CC121 predominance does not reach as far north as Korea, where CC30 again accounts for ∼30% of the carriage population (14). However, serious difficulties can arise from comparing studies varying in sampling design, and the magnitudes of the biases relating to the age and gender characteristics of the samples remain unclear.
Cluster analysis and pairwise measures of population differentiation both point to the French and Algerian samples being the most similar and the Cambodian sample being the most distinct in terms of genotype frequency. While this pattern is consistent with high rates of migration between France and Algeria and the geographical separation of Cambodia, other aspects of the data are more difficult to explain. Stochastic clonal expansion, due to rapid rates of local transmission, is likely to have introduced considerable noise. There also remain substantial gaps in our knowledge of the global diversity. For example, it is currently unknown how far the range of CC30 predominance extends moving east out of Europe and into Asia. Although MLST provides broad clues as to the diversification and spread of this species, detailed dissection of long-term historical trends from short-term stochastic effects and increased migration will ultimately require far larger data sets. Ideally such studies would generate higher-resolution data from comparable samples over time.
In addition to considering genotype frequencies, we also examined the nucleotide sequence variation within the samples through a phylogenetic analysis. This identified the two major clades previously noted in the S. aureus population, but we note little evidence of a general association between phylogeny and geographic source.
However, the phylogenetic analysis revealed the presence of a highly atypical genotype, ST1223, in the Cambodian sample. This genotype exhibits an average of ∼10% nucleotide divergence from typical S. aureus yet remains more closely related to S. aureus than to S. simiae, which is the most closely related coagulase-negative species. This atypical genotype has previously been observed in northern Australia, and thus it is possible that it is more prevalent in Southeast Asia and Australia than in other parts of the world. We report considerable difficulty in PCR amplifying and sequencing the MLST genes for ST1223 using the standard MLST primers, and we succeeded in this only by designing a new primer for aroE (see Materials and Methods). It is possible that such difficulties may have resulted in this genotype being discarded in previous studies. A recent study of carried S. aureus strains from Indonesia reported three diverged strains which were not characterized by MLST owing to difficulties in PCR amplifying aroE, thus providing anecdotal evidence for the presence of this lineage in Indonesia (26).
In conclusion, we have compared four geographically diverse samples of carried S. aureus in order to provide evidence as to the biogeographical structuring and diversity of this species. The stable predominance of CC30 throughout Europe and North Africa and the distinctiveness of the Cambodian sample point to the possibility that long-term geographical differences may not have been entirely eroded by recent admixture and stochastic clonal expansion. However, larger data sets, particularly from Asia and Africa, are required to explore regional differences more closely. Finally, our survey has revealed the presence of a highly diverged genotype in Cambodia, ST1223. This lineage is related to CC75 and may represent an ancestral lineage intermediate between coagulase-negative species and typical S. aureus. If so, more extensive sequence characterization may reveal important clues as to how S. aureus acquired such high virulence potential from relatively benign forebears.
Acknowledgments
We are grateful to all the depositors to the S. aureus MLST database. We are grateful to Nadine Richard and Patricia Lawson-Body for technical assistance and to Sabine Couriol and Marie-Jeanne Julliard for secretarial work. We thank Ivo Sedlacek and Dana Novakova (Czech Collection of Microorganisms, Faculty of Science, Masaryk University Bmo, 602 00 Bmo, Czech Republic) for providing the Staphylococcus simiae reference strain used in this study.
This work was supported in part by grant 5710AND90 from the Institut de Médecine et Epidémiologie Africaines (IMEA-Fondation MBA) (Mali) and by contracts 05 MDU 666 for Algeria and COCOP 0209-MOL-413-014 for Moldova.
Footnotes
Published ahead of print on 10 July 2009.
REFERENCES
- 1.Agapow, P.-M., and A. Burt. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1101-102. [Google Scholar]
- 2.Armand-Lefevre, L., R. Ruimy, and A. Andremont. 2005. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11711-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collery, M. M., D. S. Smyth, J. M. Twohig, A. C. Shore, D. C. Coleman, and C. J. Smyth. 2008. Molecular typing of nasal carriage isolates of Staphylococcus aureus from an Irish university student population based on toxin gene PCR, agr locus types and multiple locus, variable number tandem repeat analysis. J. Med. Microbiol. 57348-358. [DOI] [PubMed] [Google Scholar]
- 4.Conceicao, T., M. Aires de Sousa, and H. de Lencastre. 2009. Staphylococcal interspersed repeat unit typing of Staphylococcus aureus: evaluation of a new multilocus variable-number tandem-repeat analysis typing method. J. Clin. Microbiol. 471300-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David, M. Z., K. M. Rudolph, T. W. Hennessy, S. Boyle-Vavra, and R. S. Daum. 2008. Molecular epidemiology of methicillin-resistant Staphylococcus aureus, rural southwestern Alaska. Emerg. Infect. Dis. 141693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feil, E. J. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2483-495. [DOI] [PubMed] [Google Scholar]
- 8.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 1853307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, T. A., R. Sampath, L. B. Blyn, R. Ranken, C. Ivy, R. Melton, H. Matthews, N. White, F. Li, V. Harpin, D. J. Ecker, L. K. McDougal, B. Limbago, T. Ross, D. M. Wolk, V. Wysocki, and K. C. Carroll. 2009. Rapid molecular genotyping and clonal complex assignment of Staphylococcus aureus isolates by PCR coupled to electrospray ionization-mass spectrometry. J. Clin. Microbiol. 471733-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, P. L., S. K. Chuang, Y. F. Choi, R. A. Lee, A. C. Lit, T. K. Ng, T. L. Que, K. C. Shek, H. K. Tong, C. W. Tse, W. K. Tung, and R. W. Yung. 2008. Community-associated methicillin-resistant and methicillin-sensitive Staphylococcus aureus: skin and soft tissue infections in Hong Kong. Diagn. Microbiol. Infect. Dis. 61245-250. [DOI] [PubMed] [Google Scholar]
- 12.Karapsias, S., E. T. Piperaki, I. Spiliopoulou, G. Katsanis, and A. Tseleni-Kotsovili. 2008. Methicillin-resistant Staphylococcus aureus nasal carriage among healthy employees of the Hellenic Air Force. Eur. Surveill. 13:9-11. [DOI] [PubMed]
- 13.Kilic, A., G. Mert, Z. Senses, O. Bedir, H. Aydogan, A. C. Basustaoglu, and P. C. Appelbaum. 2008. Molecular characterization of methicillin-resistant Staphylococcus aureus nasal isolates from Turkey. Antonie van Leeuwenhoek 94615-619. [DOI] [PubMed] [Google Scholar]
- 14.Ko, K. S., J. Y. Lee, J. Y. Baek, K. R. Peck, J. Y. Rhee, K. T. Kwon, S. T. Heo, K. M. Ahn, and J. H. Song. 2008. Characterization of Staphylococcus aureus nasal carriage from children attending an outpatient clinic in Seoul, Korea. Microb. Drug Resist. 1437-44. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald, M., A. Dougall, D. Holt, F. Huygens, F. Oppedisano, P. M. Giffard, J. Inman-Bamber, A. J. Stephens, R. Towers, J. R. Carapetis, and B. J. Currie. 2006. Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote aboriginal communities. J. Clin. Microbiol. 443720-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melles, D. C., F. C. Tenover, M. J. Kuehnert, H. Witsenboer, J. K. Peeters, H. A. Verbrugh, and A. van Belkum. 2008. Overlapping population structures of nasal isolates of Staphylococcus aureus from healthy Dutch and American individuals. J. Clin. Microbiol. 46235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nulens, E., I. Gould, F. MacKenzie, A. Deplano, B. Cookson, E. Alp, E. Bouza, and A. Voss. 2005. Staphylococcus aureus carriage among participants at the 13th European Congress of Clinical Microbiology and Infectious Diseases. Eur. J. Clin. Microbiol. Infect. Dis. 24145-148. [DOI] [PubMed] [Google Scholar]
- 19.Pantucek, R., I. Sedlacek, P. Petras, D. Koukalova, P. Svec, V. Stetina, M. Vancanneyt, L. Chrastinova, J. Vokurkova, V. Ruzickova, J. Doskar, J. Swings, and V. Hajek. 2005. Staphylococcus simiae sp. nov., isolated from South American squirrel monkeys. Int. J. Syst. Evol. Microbiol. 551953-1958. [DOI] [PubMed] [Google Scholar]
- 20.Peacock, S. J., I. de Silva, and F. D. Lowy. 2001. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 9605-610. [DOI] [PubMed] [Google Scholar]
- 21.Ruimy, R., V. Breittmayer, P. Elbaze, B. Lafay, O. Boussemart, M. Gauthier, and R. Christen. 1994. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int. J. Syst. Bacteriol. 44416-426. [DOI] [PubMed] [Google Scholar]
- 22.Ruimy, R., M. Dos-Santos, L. Raskine, F. Bert, R. Masson, S. Elbaz, C. Bonnal, J. C. Lucet, A. Lefort, B. Fantin, M. Wolff, M. Hornstein, and A. Andremont. 2008. Accuracy and potential usefulness of triplex real-time PCR for improving antibiotic treatment of patients with blood cultures showing clustered gram-positive cocci on direct smears. J. Clin. Microbiol. 462045-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruimy, R., A. Maiga, L. Armand-Lefevre, I. Maiga, A. Diallo, A. K. Koumare, K. Ouattara, S. Soumare, K. Gaillard, J. C. Lucet, A. Andremont, and E. J. Feil. 2008. The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J. Bacteriol. 1903962-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruppe, E., F. Barbier, Y. Mesli, A. Maiga, R. Cojocaru, M. Benkhalfat, S. Benchouk, H. Hassaine, I. Maiga, A. Diallo, A. K. Koumare, K. Ouattara, S. Soumare, J. B. Dufourcq, C. Nareth, J. L. Sarthou, A. Andremont, and R. Ruimy. 2009. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 53442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakwinska, O., G. Kuhn, C. Balmelli, P. Francioli, M. Giddey, V. Perreten, A. Riesen, F. Zysset, D. S. Blanc, and P. Moreillon. 2009. Genetic diversity and ecological success of Staphylococcus aureus strains colonizing humans. Appl. Environ. Microbiol. 75175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severin, J. A., E. S. Lestari, K. Kuntaman, D. C. Melles, M. Pastink, J. K. Peeters, S. V. Snijders, U. Hadi, D. O. Duerink, A. van Belkum, and H. A. Verbrugh. 2008. Unusually high prevalence of Panton-Valentine leukocidin genes among methicillin-sensitive Staphylococcus aureus strains carried in the Indonesian population. J. Clin. Microbiol. 461989-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tewitt, R., V. Kanhai, and W. B. van Leeuwen. 2009. Comparison of the DiversiLab system™, pulsed-field gel electrophoresis and multi-locus sequence typing for the characterisation of epidemic reference MRSA strains. J. Microbiol. Methods 77130-133. [DOI] [PubMed]
- 28.Udo, E. E., F. G. O'Brien, N. Al-Sweih, B. Noronha, B. Matthew, and W. B. Grubb. 2008. Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J. Clin. Microbiol. 463514-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Belkum, A. 2006. Staphylococcal colonization and infection: homeostasis versus disbalance of human (innate) immunity and bacterial virulence. Curr. Opin. Infect. Dis. 19339-344. [DOI] [PubMed] [Google Scholar]
- 30.Weir, B. S., and W. G. Hill. 2002. Estimating F-statistics. Annu. Rev. Genet. 36721-750. [DOI] [PubMed] [Google Scholar]