Abstract
The common pathogen Streptococcus pyogenes colonizes the human skin and tonsils and can invade underlying tissues. This requires the adhesion of S. pyogenes to host surface receptors mediated through adhesins. The laminin-binding protein Lbp has been suggested as an adhesin, specific for the human extracellular matrix protein laminin. Sequence alignments, however, indicate a relationship between Lbp and a family of bacterial metal-binding receptors. To further analyze the role of Lbp in S. pyogenes and its potential role in pathogenicity, Lbp has been crystallized, and its structure has been solved at a resolution of 2.45 Å (R = 0.186; Rfree = 0.251). Lbp has the typical metal-binding receptor fold, comprising two globular (β/α)4 domains connected by a helical backbone. The two domains enclose the metal-binding site, which contains a zinc ion. The interaction of Lbp with laminin was further investigated and shown to be specific in vitro. Localization studies with antibodies specific for Lbp show that the protein is attached to the membrane. The data suggest that Lbp is primarily a zinc-binding protein, and we suggest that its interaction with laminin in vivo may be mediated via zinc bound to laminin.
The gram-positive bacterium Streptococcus pyogenes (group A Streptococcus) is a strictly human pathogen (9) that is responsible for a wide range of diseases. It colonizes the human skin, leading to erysipelas and pyoderma, and the tonsils, inducing strep throat or tonsillitis. Less often, it also invades underlying soft tissues, causing life-threatening conditions such as necrotizing fasciitis or streptococcal toxic shock syndrome (15). These host-pathogen interactions require the adhesion of S. pyogenes to dermal and epithelial cells, which is mediated by streptococcal adhesins that bind to human surface receptors. A number of potential adhesins have been described so far, e.g., hyaluronic acid (60), lipoteichoic acid (7), M protein (22), the pilus protein Cpa (35, 49, 54), or protein F1/SfbI (28).
An important human receptor for S. pyogenes is the extracellular matrix (ECM) protein laminin (67). Laminin is a large (900 kDa), highly glycosylated multidomain protein found in all human tissues. Adhesion to it is a starting point of tissue invasion for many pathogenic bacteria (59). Recently, the protein Lmb was identified as a potential streptococcal receptor for laminin in Streptococcus agalactiae (62). Later, a close homologue (98% sequence identity) was found in S. pyogenes and named Lbp or Lsp (23, 70). An S. agalactiae Δlmb mutant showed reduced adherence to laminin, while addition of exogenous recombinant Lmb inhibited adherence by the wild-type strain (62). Likewise, a Δlbp mutation abrogated laminin-binding for the S. pyogenes strains M1 and M49, and recombinant Lbp was shown to bind to laminin or interfere with bacterial adherence (23, 70, 71). Further work suggests that Lmb could also act as an invasin aiding S. agalactiae in the invasion of human brain microvascular endothelial cell lines (69).
Sequence alignments, however, suggest that Lbp is a member of the large group of bacterial metal-binding receptors (MBRs), which belong to the family of solute-binding receptors and have been referred to as cluster 9 of this family (21). MBRs encompass a vast number of conserved bacterial metal receptors in gram-negative and gram-positive bacteria. In the case of gram-positive bacteria, they are also named lipoprotein receptor antigen I (LraI) because of their attachment to the bacterial membrane by a lipid anchor (34). Structures of MBRs (6, 13, 40, 41, 43, 58, 64) have revealed an association with other solute-binding receptors such as the maltose-binding protein with which they share a structure of two (β/α)4 domains (63). Usually, MBRs are encoded in the bacterial genome in an operon together with a permease, an ATPase, and a regulator. It is assumed that the MBR scavenges metal and passes it on to the permease which, in complex with the ATPase, imports the ion into the bacterial cell (16). Lbp, however, appears to be an exception since it is not encoded together with a permease and an ATPase but, rather, with the histidine triad protein HtpA (23, 37).
Biological and sequence data combined raise the question of whether Lbp is a laminin-binding protein, a metal-binding receptor, or both. Therefore, we have solved the atomic structure of Lbp in order to elucidate its relation to MBRs and to gain insight into its potential role in laminin binding. We have also characterized the interaction between Lbp and laminin in vitro and the subcellular localization of Lbp in S. pyogenes. Here, we report the atomic structure of Lbp, which classifies it as a zinc-binding MBR, and suggest a possible mode of interaction with laminin that would reconcile both sets of data.
MATERIALS AND METHODS
Protein expression, purification, and crystallization.
The gene lmb (also called SPy_2007; NCBI gene identifier 901672) from S. pyogenes M1 strain SF370 was cloned, inserted into the Gateway expression vector, and transformed into Escherichia coli BL21(DE3) cells for heterologous overexpression as a His6-tagged protein, as described before (44). After purification by immobilized metal affinity chromatography, His6 tag removal, and size exclusion chromatography, the final purified protein comprised 286 amino acid residues, representing residues 5 to 287 of the mature protein, together with a 3-residue N-terminal extension (GSG) that remained after cleavage of the tag. Crystals of Lbp were grown in sitting drops by mixing equal volumes of protein solution (37 mg ml−1 Lbp and phosphate-buffered saline [PBS]-0.002% [wt/vol] NaN3) and reservoir solution (30% [wt/vol] polyethylene glycol 1500) at 291 K (44). Space group and unit cell dimensions are given in Table 1 .
TABLE 1.
Data collection and refinement statistics
| Parameter | Valuea |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.5418 |
| Space group | P21 |
| Unit cell dimension | |
| Axial lengths (Å) | a = 42.62, b = 92.16, c = 70.61 |
| Angles (°) | α = 90, β = 106.27, γ = 90 |
| Resolution range (Å) | 29.09-2.40 (2.53-2.40) |
| Total no. of observations | 150,310 (21627) |
| Unique reflections | 20,587 (3004) |
| Redundancy | 7.3 |
| Completeness (%) | 99.9 (100.0) |
| Mean I/σ(I) | 18.1 (2.6) |
| Rmerge (%)b | 9.8 (59.7) |
| Refinement statistics | |
| Resolution range (Å) | 27.98-2.45 (2.511-2.45) |
| Rwork (%)c | 18.63 (24.65) |
| Rfree (%)c | 25.07 (33.60) |
| No. of protein atoms | 3,981 |
| No. of water molecules | 79 |
| No. of zinc ions | 2 |
| RMS deviation from ideal geometry | |
| Bonds (Å) | 0.015 |
| Angles (°) | 1.560 |
| Wilson B factor (Å2) | 38.39 |
| Avg B factor (Å2) | |
| Main chain atoms | 39.12 |
| Side chain atoms | 39.54 |
| Zinc ions | 33.19 |
| Water molecules | 36.94 |
| Ramachandran plot—residues in:d | |
| Most favored regions (%) | 90.1 |
| Additional allowed regions (%) | 9.9 |
| Generously allowed regions (%) | 0 |
| Disallowed regions (%) | 0 |
Numbers in parentheses are values for the outermost shell.
Rmerge = Σhkl Σi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl).
Rwork and Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where Rfree was calculated over 5.1% of amplitudes that were chosen at random and not used in refinement.
Based on PROCHECK.
Data collection and structure solution.
X-ray diffraction data to a resolution of 2.45 Å were collected in-house (Rigaku Micromax-007HF and Marresearch Mar345DTB instruments) at 100 K and processed as described previously (44). Details are given in Table 1. Molecular replacement calculations were performed using Phaser (48), with an ensemble of six search models of homologous proteins with 25 to 35% sequence identity. The ensemble consisted of the models for Treponema pallidum TroA (Protein Data Bank [PDB] codes 1TOA and 1K0F) (41, 42), Synechocystis sp. ZnuA (PDB 1PQ4) (6), Synechocystis sp. MntC (PDB 1XVL) (58), Bacillus subtilis YcdH (PDB 2O1E), and S. pyogenes MtsA (PDB 3HH8) (64). The initial model was refined by Phenix (1) to an Rwork/Rfree ratio of 40.9/47.4% using noncrystallographic symmetry, TLS (translation, libration, and screw-rotation) refinement, and simulated annealing. The model was further refined at a resolution of 2.45 Å through iterative cycles of manual building in COOT (24) and refinement in Phenix and REFMAC (50), using TLS. Noncrystallographic symmetry restraints were applied in the first refinement round, but after that the molecules were refined individually. Water molecules and zinc atoms were picked manually in COOT. The distances between the zinc atoms and surrounding atoms were restrained for refinement in Phenix after they had been characterized. Refinement statistics and model details are given in Table 1. Model quality was assessed with PROCHECK (39) and MolProbity (17). Protein structure comparisons were performed using secondary structure matching (SSM) (36). All figures were generated with PyMOL (18). Sequence and structural alignments were prepared using Clustal X, version 2.0.10 (38), and MAMMOTH-mult (46). Residue numbers for Lbp refer to the unprocessed full-length protein (NCBI accession number AAK34689).
Metal analysis.
Lbp solutions (50 to 100 μM) used for crystallization trials were analyzed by inductively coupled plasma-mass spectrometry (ICP-MS) (Hill Laboratories, Hamilton, New Zealand). X-ray fluorescence scans of crystals of Lbp were carried out at beamline PX-1 at the Australian Synchrotron, Victoria, Australia. Fluorescence within 300 eV of the Zinc Kα edge (9.6 keV) was measured by an Amptek XR100-CR. A peak at 9,666.91 eV was detected using the program CHOOCH (25).
Antiserum production.
Antiserum against Lbp was raised by the Hercus-Taieri Resource Unit, University of Otago, Dunedin, New Zealand. Briefly, a New Zealand White rabbit was bled at day 56 after intradermic injections of 300 μg of recombinant His6-Lbp mixed with incomplete Freund's adjuvant at days 0, 21, and 42. Antiserum was used directly, or polyclonal anti-Lbp antibodies were purified using recombinant Lbp coupled to CNBr-activated Sepharose 4B (GE Healthcare), according to the manufacturer's protocol.
Cell fractionation and Western blotting.
S. pyogenes serotype M1 (strain SF370) was grown in Todd-Hewitt broth with 0.2% yeast extract overnight at 310 K and harvested by centrifugation. S. pyogenes cells were fractionated based on a protocol adapted from Mora et al. (49). The cell pellet was resuspended in lysis buffer (100 mM KH2PO4, pH 6.2, 40% sucrose, 10 mM MgCl2) supplemented with Complete EDTA-free protease inhibitor cocktail tablets (Roche), 4 mg/ml lysozyme, and 100 U/ml mutanolysin (Sigma) and incubated at 310 K for 2 h 30 min with gentle rotation. Cells and debris were sedimented by centrifugation (15 min at 12,900 × g at 277 K); the supernatant containing proteins covalently attached to the cell wall such as the pili (49) was kept as the cell wall fraction. Cells were further lysed by ultrasonication on ice, and debris was removed by centrifugation (20 min at 5850 × g at 277 K). The cloudy supernatant was sedimented by ultracentrifugation (60 min at 180,000 × g at 277 K); the supernatant was kept as the cytosolic fraction, and the pellet was resuspended in 50 mM sodium phosphate (pH 8.0)-300 mM NaCl as the membrane fraction. Samples of each fraction and of recombinant Lbp were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto polyvinylidene difluoride membranes in duplicate. The membranes were blocked with TBST buffer (25 mM Tris-HCl, pH 7.4, 137 mM NaCl, 2.6 mM KCl, 0.1% Tween-20) and 5% milk powder and incubated with anti-Lbp antiserum or preimmune serum diluted 1:10,000 into TBST and 1% milk powder. Bound antibodies were detected using the Amersham ECL Western Blotting System (GE Healthcare) containing an anti-rabbit antibody conjugated with horseradish peroxidase and an LAS-3000 (Fujifilm) imaging system. The MultiGauge imaging program (Fujifilm) was used for quantitation of bands.
Immunoprecipitation of solubilized Lbp from S. pyogenes.
The S. pyogenes membrane fraction (prepared as above) was incubated with 1.1% (wt/vol) octyl-β-d-glucopyranoside at 277 K overnight and cleared by ultracentrifugation (60 min at 180,000 × g at 277 K). The supernatant was incubated for 60 min at room temperature with anti-Lbp polyclonal antibody (purified from rabbit anti-Lbp antiserum), which had been coupled to CNBr-activated Sepharose 4B (GE Healthcare) according to the manufacturer's protocol. Beads were harvested by centrifugation and washed five times with 10 mM Tris-HCl (pH 7.5)-500 mM NaCl. Bound protein was eluted with 100 mM glycine, pH 2.5, and separated by SDS-PAGE. A faint band was excised and submitted for liquid chromatography-tandem MS analysis at the University of Auckland Centre for Genomics and Proteomics.
Generation of a biotinylated form of Lbp.
The gene for Lbp was recloned using the primer 5′-GGC AGC GGC GCG TGC AAC CCC AAA CAG CCT ACG C-3′ and the Gateway Technology (Invitrogen) according to a protocol described previously (44). The codon shown in boldface codes for an additional cysteine. The protein was expressed in E. coli and purified by immobilized metal affinity chromatography and gel filtration the same way as Lbp (44), including the removal of the His6 tag, with the exception that 2 mM β-mercaptoethanol was added in the lysis and immobilized metal affinity chromatography steps. Cysteine-Lbp was biotinylated by the addition of sulfhydryl-specific biotin-maleimide (Sigma-Aldrich) to a concentration of 30 μg/ml and incubation for 2 h at room temperature under gentle rotation. Unbound biotin was removed by a further round of gel purification using PBS (pH 7.4)-0.002% (wt/vol) NaN3. Biotinylation of Lbp was tested by dot blot using streptavidin coupled to horseradish peroxidase (Invitrogen).
Dot blot assay for Lbp binding of ECM proteins.
Equal amounts of mouse laminin (Invitrogen), human fibronectin from plasma (Invitrogen), and collagen type I (Sigma-Aldrich) were spotted on a Hybond-ECL nitrocellulose membrane (GE Healthcare). Following blocking with TBST-3% bovine serum albumin (BSA) overnight at 277 K, the membrane was incubated with TBST-1% BSA-3.5 μg/ml biotinylated Lbp at room temperature for 2.5 h. After four washes with TBST, the membrane was incubated with TBST—1% BSA-0.5 μg/ml streptavidin coupled to horseradish peroxidase (Invitrogen). Streptavidin bound to Lbp was detected using the Amersham ECL Western Blotting System (GE Healthcare) and an LAS-3000 (Fujifilm) imaging system.
ELISA of the interaction between Lbp and laminin.
For enzyme-linked immunosorbent assay (ELISA) analysis of binding of Lbp to immobilized laminin, mouse laminin (Invitrogen) was diluted to 10 μg/ml in carbonate buffer, pH 9.6, and coated on Greiner PS 96-well microplates (50 μl/well) overnight at 277 K; 1% BSA (fraction V; Sigma-Aldrich) in carbonate buffer (pH 9.6; 50 μl/well) was used as a control. The following day, wells were washed with 100 μl/well TBST and blocked with 100 μl/well TBST-3% BSA for 2 h at room temperature. Increasing concentrations of recombinant Lbp (0.5 μg/ml to 1 mg/ml [purified as described in reference 44]; 50 μl/well) diluted into TBST-1% BSA were incubated for 2 h at room temperature. Wells were washed four times with 100 μl/well TBST and incubated with purified rabbit polyclonal anti-Lbp antibodies (4 μg/ml in TBST-1% BSA; 50 μl/well) for 1 h. After four washes (100 μl/well TBST), wells were incubated with donkey anti-rabbit antibody coupled with horseradish peroxidase (GE Healthcare) diluted 1:1,000 in TBST-1% BSA for 1 h. SigmaFast o-phenylenediamine dihydrochloride (Sigma-Aldrich) and an EnVision plate reader (Perkin-Elmer) were used for quantification. Absorption was read at 490 nm and 650 nm, and values for 650 nm were subtracted from those for 490 nm. All experiments were performed in quadruplicate and comprised controls for unspecific binding (BSA) and for the antibodies. Background absorption was subtracted from all values. A binding curve was generated using SigmaPlot, version 10.0 (Systat Software), and a Kd (dissociation constant) was calculated using a one-site binding hyperbola and the following equation: Y = (Bmax X)/ (Kd + X), where X is the concentration of Lbp, Y is binding (absorption units), and Bmax is maximum binding (as determined by nonlinear regression).
For ELISA analysis of the binding of laminin to immobilized Lbp, recombinant Lbp was diluted in carbonate buffer, pH 9.6, to 1 μg/ml, and 50 μl/well was coated on Costar 96-well plates (Corning) overnight at 277 K. Wells were washed with 100 μl of TBST and blocked with 100 μl of TBST-1% BSA for 2 h at room temperature. Increasing concentrations (0 to 50 μg/ml) of mouse laminin (Invitrogen) diluted in TBST-1% BSA (50 μl/well) were incubated for 2 h at room temperature. After four washes (100 μl/well), bound laminin was detected using a rabbit anti-laminin antibody (Sigma-Aldrich), diluted 1:1,000 in TBST-1% BSA, and the donkey anti-rabbit antibody as described above. All experiments were performed in quadruplicate and included controls for unspecific binding (BSA) and for the antibodies. Background absorption was subtracted from all values.
For competition experiments, ELISA analysis of the binding of laminin to immobilized Lbp was performed as above. However, increasing concentrations of mouse laminin diluted in TBST-1% BSA were concomitantly incubated with a constant concentration of recombinant Lbp (1 mg/ml) on the plates, coated with Lbp, and blocked with BSA.
Protein structure accession number.
The PDB accession number for Lbp is 3GI1.
RESULTS
Three-dimensional structure of Lbp.
The structure of Lbp was solved by molecular replacement using an ensemble of search models and refined at a resolution of 2.45 Å (Table 1). There are two molecules of Lbp, designated A and B, per asymmetric unit (AU) in the crystal, which superimpose extremely closely, with a root mean square (RMS) difference in Cα atom positions of 0.41 Å over the whole model. Therefore, only molecule A will be discussed. The first 9 residues from the N terminus of the crystallized construct are disordered, with no visible electron density, and although there is weak electron density for the loops from residue Q60 to G64 (Q60-G64), G123-T137, and E229-E231, the significance of this observation is not unambiguous. The final model comprises residues T30-I59, I65-K122, L138-P228, and P232-K306 for each of the two molecules in the AU.
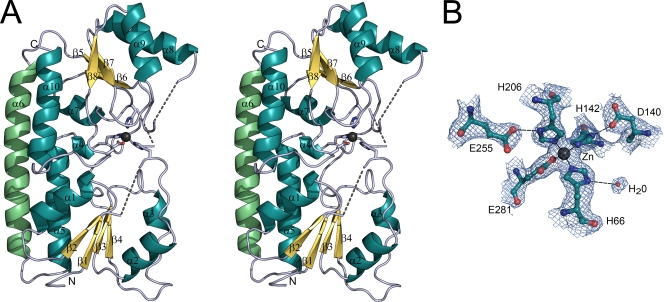
The fold is typical of a bacterial MBR (cluster 9 of solute-binding receptors), comprising two globular domains linked by a long backbone α-helix (Fig. 1A). The two domains are structurally similar and can be superimposed with an RMS difference in Cα positions of 3.11 Å (79 aligned residues). Each domain consists of a central β-sheet of four parallel β-strands surrounded by four α-helices [a (β/α)4 domain] although one helix is kinked. One helix of each domain packs tightly to the linker helix to form a rigid backbone (Fig. 1A).
FIG. 1.
Three-dimensional structure of Lbp. (A) The fold of Lbp is drawn as a ribbon diagram in stereoview. Residues of the metal-binding site are represented as stick models, with oxygen and nitrogen atoms shown in red and blue, respectively. The zinc ion is shown as a black sphere. Dashed lines indicate the connectivity for loops which were not built in the model. (B) The final 2Fobs − Fc (where Fobs is the observed and Fc is the calculated structure factor amplitude) electron density map around the zinc-binding site of Lbp is shown contoured at 1σ. Residues are drawn in ball-and-stick representation with carbon, oxygen, and nitrogen atoms shown in cyan, red, and blue, respectively. The zinc ion is depicted as a black sphere, and a water molecule is shown as a red sphere. Metal-ligand bonds are indicated by thin solid lines, and probable hydrogen bonds are shown by broken lines.
The metal-binding site of Lbp.
The two globular domains enclose a zinc ion in their interface. The metal site was detected by a strong peak in the anomalous Fourier electron density map, and the bound ion was identified as zinc by an X-ray fluorescence scan. ICP-MS quantitative analysis showed that Lbp solutions used for crystallization contained an Lbp/Zn ratio of ca. 1:1 (Table 2). In our structural model, the zinc ion is coordinated by the side chains of three histidines and one glutamate in a slightly distorted tetrahedral geometry (Fig. 1B). The histidines interact with the zinc ion through their Nɛ2 atoms with metal-ligand distances of 2.1 Å (H66, H142, and H206) (Fig. 1B). The glutamate E281 binds it in a strictly syn-monodentate manner through Oɛ2 at 2.0 Å. It is possible, however, that there is some very weak long-range interaction between zinc and E281 through the second carboxylate oxygen Oɛ1, since this atom does not otherwise interact with the surrounding structure. The Zn-O distance is 3.1 Å, however, and any interaction must be very weak. Hydrogen bonds from the side chains of residues D140 and E255 stabilize H142 (2.8 Å) and H206 (2.6 Å), respectively. A water molecule forms a hydrogen bond with H66 (3.2 Å) in chain A but not in chain B.
TABLE 2.
ICP-MS quantification of zinc concentrations in samples of purified recombinant Lbp solutions
| Sample no. | Concn in sample (μM)
|
Ratio of [Zn]/[Lbp] | |
|---|---|---|---|
| Lbp | Zn | ||
| 1 | 88 | 90 | 1.02:1 |
| 2 | 88 | 83 | 0.94:1 |
| 3 | 102 | 96 | 0.94:1 |
| Avg | 93 | 90 | 0.97:1 |
Structural comparison between Lpb and MBR homologues.
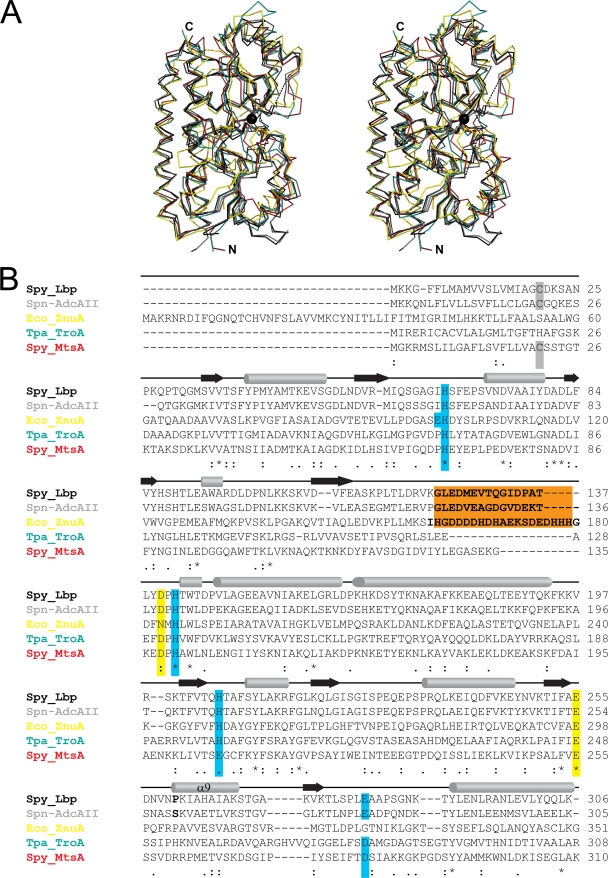
The overall fold of Lbp is very similar to that of other MBRs such as Synechocystis sp. MntC (PDB code 1XVL; RMS difference, 1.63 Å) (58) and ZnuA (1PQ4; RMS difference, 1.7 Å) (6), S. pyogenes MtsA (3HH8; RMS difference, 1.72 Å) (64), T. pallidum TroA (1TOA; RMS difference, 1.73 Å) (41), Streptococcus pneumoniae PsaA (1PSZ; RMS difference, 1.82 Å) (40), B. subtilis YcdH (2O1E; RMS difference, 1.88 Å), and E. coli ZnuA (2OSV; RMS difference, 1.97 Å) (43) (Fig. 2A). During the preparation of the manuscript, the structure of a close homologue of Lbp, AdcAII from S. pneumoniae, was published (45) (PDB code 3CX3). Lbp and AdcAII display a sequence identity of 63%, and their structures are nearly identical (RMS difference, 0.98 Å). Only helix 9 (residues 257 to 270) shows a slight displacement between the two structures. It is possible that Lbp residue P260 (Fig. 2B) at the start of helix α9 induces this conformational shift, but it remains unclear whether this shift is the result of different crystal packing or is a true structural divergence. Other changes include tyrosine Y302, which protrudes from the surface of Lbp but is replaced by alanine (A301) in AdcAII, and variations in the positions of some proline residues (Fig. 2B). The metal-binding sites are completely superimposable between Lbp and AdcAII. In both cases, H66 (Lbp) and H71 (AdcAII) form a hydrogen bond with a water molecule in only one of the two chains present in the AU.
FIG. 2.
Structural alignment of Lbp with other bacterial MBRs. (A) Stereo view of the Cα trace of Lbp (black), S. pneumoniae AdcAII (PDB code 3CX3; gray), E. coli ZnuA (2OSV; yellow), T. pallidum TroA (1TOA; green), and S. pyogenes MtsA (3HH8; red). The zinc ion in Lbp is indicated by a black sphere. Dashed lines indicate the connectivity for loops which were not built in Lbp. The structures were aligned using MAMMOTH-mult (46). (B) Sequence alignment of Lbp (UniProt accession number Q99XV3) with S. pneumoniae AdcAII (B2DT57), E. coli ZnuA (B3HHJ6), T. pallidum TroA (P96116), and S. pyogenes MtsA (P0A4G4). Turquoise boxes indicate residues of the primary shell of the metal-binding site; yellow boxes indicate secondary shell residues. An extended loop characteristic of Zn receptors is highlighted with a brown box. Residues set in bold indicate the unmodeled part of this loop in the structures of Lbp and ZnuA. The membrane-anchored cysteines of the mature proteins from gram-positive bacteria are marked with a gray box. Secondary structure is indicated by arrows (β-strands) or cylinders (α-helices). Sequences were aligned using Clustal W, version 2.0.10 (38). The alignment was modified based on the optimal structural superposition.
Significance of disordered loops.
Loops which are disordered in the structure of Lbp are either disordered in AdcAII (Lbp residues G123-T137) or have very high B factors (Q60-G64 and E229-E231). This is consistent with observations from other MBR structures where loops around the metal-binding site are often disordered. In the structures of the zinc-binding MBRs Synechocystis ZnuA (6), E. coli ZnuA (43), and B. subtilis YcdH (PDB code 2O1E), there is a long loop consisting primarily of histidines and charged residues that remains unmodeled. The locations of the start and end points of these disordered histidine-rich loops correspond exactly to those of the unordered Lbp (G123-T137) and AdcAII loops (Fig. 2B). Although the Lbp and AdcII loops are not histidine rich and do not display a particularly high density of charged residues, it is likely that the mobility displayed by this loop reflects a common functionality among MBRs, possibly an involvement in complex formation with permeases during metal ion transfer. Differences in amino acid sequence in this loop may reflect a divergence in function. Thus, the zinc-binding MBRs of Lbp and ZnuA have large mobile loops, whereas structures of the manganese- or iron-binding MBRs of S. pneumoniae PsaA (40), Synechocystis MntC (58), and S. pyogenes MtsA (64) display only a very short loop at a position homologous to Lbp G123-T137 (Fig. 2B). The same is true for T. pallidum TroA (41), which is most likely a manganese-receptor (19) (Fig. 2B).
Relationship between structure and metal-binding specificity.
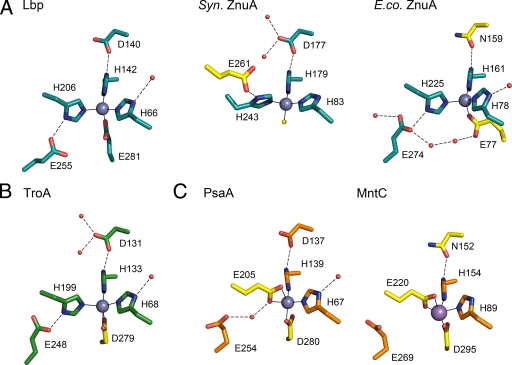
The majority of bacterial MBRs characterized so far are highly specific receptors for either zinc or manganese ions (14). Exceptions are T. pallidum TroA, which displays almost equal affinities for zinc and manganese (19, 29), and S. pyogenes MtsA, which appears to prefer iron over manganese (64). An overview of the metal-binding site architecture of known MBR structures is given in Fig. 3. Although there is a high degree of conservation between the metal-binding sites of MBRs, there is some structural variation. Lbp residues H66 and H142 are completely conserved in all the MBRs (Fig. 2B and Fig. 3). Aspartate D140 of Lbp is highly conserved, and the only substitution is an asparagine in E. coli ZnuA (43) and Synechocystis MntC (58). The third histidine, H206 of Lbp, is conserved in the group of zinc receptors and TroA but is replaced by a glutamate at a homologous position in the manganese receptors PsaA (40) and MntC (58) and in the iron receptor MtsA (64). The greatest variation is found at the position of the fourth residue of the Lbp zinc-binding site, which can be absent as in Synechocystis ZnuA (6); replaced by an aspartate as in TroA (41), the manganese receptors PsaA (40) and MntC (58), or the iron receptor MtsA (64); or replaced by a nonhomologous glutamate originating from a different domain, as in E. coli ZnuA (43). The last of these indicates that the seemingly similar metal-binding sites of Lbp and E. coli ZnuA, both with three histidines and one glutamate, are not entirely homologous but are the result of convergent evolution, possibly from an ancestral MBR comparable to Synechocystis ZnuA.
FIG. 3.
Comparison of the metal-binding site of Lbp with sites of other bacterial MBRs. Shown are the relevant residues of the primary and secondary shell of the binding site for receptors for zinc (A) and manganese (C) and for those with dual affinities for zinc and manganese (B). This categorization is based on biological data (see text for details); the metal present in the PsaA structure (C) has not been conclusively identified. Residues are shown in stick format, with oxygen and nitrogen atoms shown in red and blue, respectively. The bound metal is shown as a sphere (gray for zinc and pink for manganese), and waters are shown as small red spheres. Metal-ligand bonds are indicated as thin full lines, and hydrogen bonds are shown as broken lines. Residue numbers refer to those of the corresponding PDB file. Residues and important water molecules that distinguish a structure from Lbp are highlighted in yellow. Structures shown are S. pyogenes Lbp (PDB code 3GI1), Synechocystis ZnuA (1PQ4), E. coli ZnuA (2OSV), T. pallidum TroA (1TOA), S. pneumoniae PsaA (1PSZ), and Synechocystis MntC (1XVL).
Laminin-binding of Lbp.
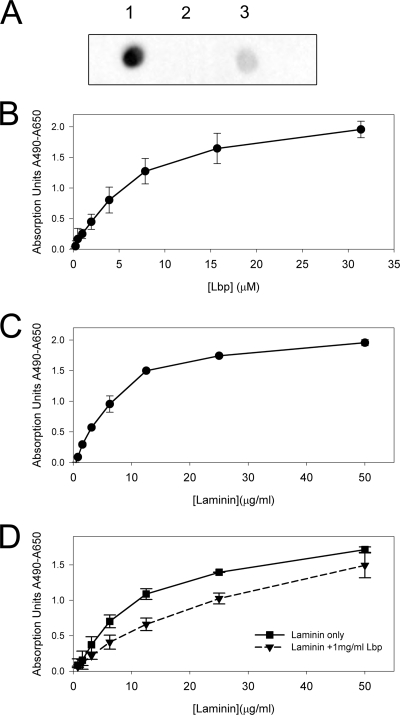
Using ELISA and a dot blot assay, we were able to confirm previous reports (23, 70, 71) that Lbp interacts with laminin (Fig. 4A) but not with fibronectin or collagen. The ELISA data for binding of Lbp to laminin (Fig. 4B) could be fitted to a one-binding-site hyperbola, and the resulting Kd was in the lower micromolar range. Our data correspond to those obtained for the binding to laminin of a truncated construct of the human laminin receptor LamR, which has a similar Kd in the lower micromolar range (31). Furthermore, we were able to inhibit binding of laminin to immobilized Lbp with an excess of Lbp in solution (Fig. 4D). This suggests a specific mode of binding between Lbp and laminin.
FIG. 4.
Analysis of the interaction between Lbp and laminin. (A) Dot blot assay for the binding of biotinylated Lbp to laminin (1), collagen type I (2), and fibronectin (3). (B) ELISA analysis of the binding of different concentrations of Lbp to immobilized laminin. (C) ELISA analysis of the binding of different concentrations of laminin to immobilized Lbp. (D) ELISA analysis of the binding of different concentrations of laminin to immobilized Lbp with and without the presence of an excess of Lbp in solution. In panels B to D, all values are the averages of repeat experiments performed in triplicate or quadruplicate, with the background absorption subtracted. Error bars indicate standard deviations. See Materials and Methods for details.
Subcellular localization of Lbp in S. pyogenes and binding to laminin.
An important facet of the question of whether Lbp is a laminin-binding protein involved in adhesion is its surface exposure. The Lbp sequence displays the motif IAGCD (Fig. 2B), which is similar to the classical lipidation signal LA(A/G) C(S/G) (66), suggesting that Lbp is anchored to the membrane. As noted by Lawrence et al. (40), MBRs are also anchored to the cell membrane and are of a maximal length of 60 Å (in the case of Lbp); therefore, they are unlikely to protrude from a cell wall of an average thickness of 150 to 300 Å (10). However, there is no current experimental evidence that conclusively shows the cellular location of Lbp, and it could be argued that potential adhesins such as Lbp are not anchored to the membrane after all but to the cell wall.
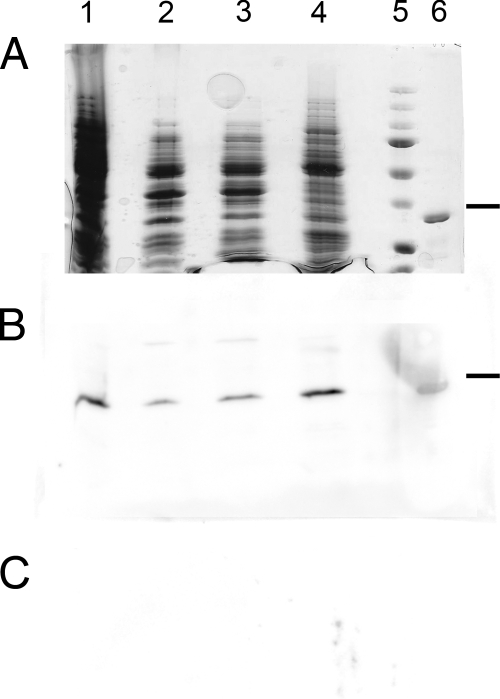
To test this hypothesis, we fractionated S. pyogenes serotype M1 (strain SF370) cells into cell wall, cytosolic, and membrane fractions. Using rabbit antiserum raised against recombinant Lbp, we detected the highest concentration of Lbp in the membrane fraction (Fig. 5) in amounts equal to that of the whole-cell fraction. Small amounts, ca. 20 to 30% compared to the whole-cell fraction, were detected in the cytosolic and cell wall fractions (Fig. 5). Whereas the former is expected, as Lbp is transcribed in the cytosol, we believe that the latter is due to Lbp contamination from the cytosol in the process of the fractionation. Our protocol, based on Mora et al. (49), for removing the cell wall uses mutanolysin to produce protoblasts that are naturally sensitive to mechanical damage and can easily release their cytosolic content (61). Nevertheless, we cannot exclude the possibility that there could be a small proportion of Lbp that is associated with the cell wall. The majority, however, is located to the membrane fraction (Fig. 5). Moreover, using purified polyclonal anti-Lbp antibodies, we could immunoprecipitate a protein from the S. pyogenes membrane fraction after solubilization through the detergent octyl-β-d-glucopyranoside. MS analysis identified this protein as Lbp. Thus, we are able to confirm that Lbp is a membrane-anchored protein.
FIG. 5.
Subcellular localization of Lbp within S. pyogenes serotype M1 (strain SF370). SDS-PAGE gel (A) and Western immunoblots of S. pyogenes fractions using rabbit anti-Lbp antiserum (B) or preimmune serum (C). Lane 1, S. pyogenes whole cells; lane 2, cell wall fraction prepared using mutanolysin; lane 3, cytosolic fraction (soluble phase) after ultrasonication and ultracentrifugation; lane 4, membrane fraction after ultracentrifugation; lane 5, protein mass standards (37-kDa band is indicated with a dash); lane 6, recombinant Lbp.
DISCUSSION
Our results throw some light on the conflicting functional annotations for Lbp. As discussed below, both the crystal structure and the genomic context of the lbp gene strongly support the assignment to Lbp of a role in zinc acquisition and homeostasis. On the other hand, our functional studies also confirm the ability of Lbp to bind laminin in vitro, consistent with its originally proposed role as an adhesin.
The crystal structure shows that Lbp has the typical fold of an MBR, comprising two (β/α)4 domains and a helical backbone. Furthermore, we have shown that Lbp binds zinc and that its metal-binding site is consistent with that of other zinc-binding MBRs such as Synechocystis ZnuA (6) and E. coli ZnuA (43). At the same time, the genomic context of the lbp gene is intriguingly different from the genes of canonical MBRs, which are found in operons that also include genes for a cognate permease, an ATPase, and a regulator (14). Lbp, in contrast, has no known associated permease or ATPase but is, instead, encoded in an operon with a member of the family of streptococcal histidine triad proteins, HtpA (23, 37). The same organization is found for the Lbp homologues Lmb in S. agalactiae (62) and AdcAII in S. pneumoniae (45). HtpA, too, has been shown to bind zinc in vitro (37) and contains five histidine triad motifs (HXXHXH) such as have been shown to take part in zinc binding in the structure of an S. pneumoniae homologue, PhtA (56). Although the function of HtpA or other histidine triad proteins is still speculative, the cotranscription of the two zinc-binding proteins Lbp and HtpA suggests that the Lpb/HtpA operon is involved in bacterial zinc homeostasis.
Further evidence supporting such a role is that the Lbp/HtpA operon includes an AdcR promoter site in both S. pyogenes and S. pneumoniae (53). AdcR is part of the recognized MBR operon AdcABCR, which has been identified as a zinc-acquisition system in S. pneumoniae (20, 21). It consists of the regulator AdcR, the MBR AdcA, the permease AdcB, and the ATPase AdcC (20, 21). There are homologues of high sequence identity (60 to 80%) to all four of these proteins in S. pyogenes although AdcA is isolated from the operon (based on the genome of S. pyogenes serotype M1) (26). The transcriptional activity of AdcR has been shown to be regulated by zinc in S. pneumoniae (51), and it is striking that both the AdcABCR and Lbp/HtpA systems, together with a second histidine triad protein, Slr (11), are all regulated by the equivalent AdcR regulator in S. pyogenes. This evidence points to the regulation of Lbp expression by zinc, and we conclude that Lbp is indeed involved in zinc homeostasis in some way even though a potential permease/ATPase specific for Lbp has not been identified.
Based on sequence comparisons and biochemical data, S. pyogenes expresses three MBRs: Lbp (23, 70), MtsA, (32, 33, 64), and the yet uncharacterized AdcA, which is a close homologue to S. pneumoniae AdcA (20, 21). MtsA is a receptor for manganese and iron (32, 33, 64, 65), whereas Lbp and AdcA are both probable zinc receptors. AdcA differs from Lbp (and its S. pneumoniae homologue AdcAII) in that it has a histidine-rich loop similar to that in E. coli ZnuA (Lbp residues G123-T137) and, significantly, an additional 22-kDa C-terminal domain, YodA, which contains additional zinc-binding sites (45). The histidine-rich loop of AdcA may bind zinc ions as in Synechocystis ZnuA (72) and E. coli ZnuA (73). As Lbp has a similar loop in the same position, but without histidines, we speculate that these loops could give rise to different functions in Lbp and AdcA. Whether this implies that they associate with different permeases or that they have different roles in zinc homeostasis is unknown.
In apparent conflict with its role in binding zinc, Lbp has also been reported to act as an adhesin by binding to laminin (23, 62, 70). Laminin is a prominent component of the ECM in animals, and Lbp transcription has been shown to be highly upregulated during infection of cutaneous tissues in mouse models (11). Mutational studies (23, 62), together with our own binding data, confirm a possible role for Lbp in binding of streptococci to laminin (67). How might these activities be reconciled?
It is known that laminin binds zinc (2) and that zinc can enhance binding of laminin to entactin (nidogen), collagen type IV (2), and a laminin-binding protein of the human pathogen Leishmania (5). A crystal structure of the laminin-entactin complex reveals that metal ions (cadmium, in this case) are involved in the interaction, albeit in a nonessential manner (PDB code 1NPE) (68). By analogy, we suggest that the interaction between Lbp and laminin could be zinc mediated. This would explain how an MBR could have the additional role of binding to proteins of the ECM.
The question remains as to whether the laminin-binding activity shown here in vitro also applies in vivo. Previous studies have suggested adhesive roles for a number of MBRs from streptococci, but these were often contradicted later, as in the case of Streptococcus parasanguis FimA (12, 27, 52) or Streptococcus gordonii Sca (3, 30). Discussion is still ongoing with regard to S. pneumoniae PsaA, where evidence points to an involvement in adhesion: a knockout mutant of the S. pneumoniae MBR PsaA displayed impaired adherence to A549 cells (pneumocytes) (8), and an anti-PsaA antibody inhibited adhesion to nasopharyngeal cells (57). The human receptor E-cadherin has been implicated in the latter interaction (4). Nevertheless, the actual role of PsaA in pneumococcal adhesion has been contested as its dimensions appear too small to protrude from the cell wall and thus to be able to take part in adhesion directly (40). Similarly, although our data imply a specific interaction between Lbp and laminin, we have also shown that Lbp is membrane bound. Lbp has dimensions similar to those of PsaA, and in the light of recent results showing that many gram-positive bacteria have a pseudo-periplasmic space of about 15 nm between cell membrane and cell wall, in addition to a cell wall proper of 20 nm (47, 74), we have to assume that Lbp extends into only the streptococcal periplasmic space. We cannot exclude the possibility, however, that Lbp could become exposed when cell deformation occurs during cell-cell interactions and that it could then engage with laminin.
Our structure of Lbp and recent biological data strongly suggest that Lbp is an MBR and plays a role in zinc homeostasis. We believe that this is the primary function of Lbp. Nevertheless, the in vitro data do suggest that Lbp could act as an adhesin as well, perhaps in a metal-mediated manner. This is similar to the case of PsaA from S. pneumoniae (20), which is a proven MBR but seems also to have adhesive function (55).
Acknowledgments
We thank Fiona Clow for help with the S. pyogenes culture, Martin Middleditch for help with MS, and Neil Paterson and Julian Adams for help with the X-ray fluorescence scan.
C.L. was supported by a Ph.D. scholarship of the German Academic Exchange Service (DAAD), a New Zealand International Doctoral Research Scholarship (Education New Zealand), and a University of Auckland Plus Scholarship. This work was supported by the Health Research Council of New Zealand and by the Maurice Wilkins Centre for Molecular Biodiscovery.
Footnotes
Published ahead of print on 17 July 2009.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. Phenix: building new software for automated crystallographic structure determination. Acta Crystallogr. D 581948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Ancsin, J. B., and R. Kisilevsky. 1996. Laminin interactions important for basement membrane assembly are promoted by zinc and implicate laminin zinc finger-like sequences. J. Biol. Chem. 2716845-6851. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1993. Cloning of the Streptococcus gordonii PK488 gene, encoding an adhesin which mediates coaggregation with Actinomyces naeslundii PK606. Infect. Immun. 61981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderton, J. M., G. Rajam, S. Romero-Steiner, S. Summer, A. P. Kowalczyk, G. M. Carlone, J. S. Sampson, and E. W. Ades. 2007. E-cadherin is a receptor for the common protein pneumococcal surface adhesin A (PsaA) of Streptococcus pneumoniae. Microb. Pathog. 42225-236. [DOI] [PubMed] [Google Scholar]
- 5.Bandyopadhyay, K., S. Karmakar, A. Ghosh, and P. K. Das. 2002. High affinity binding between laminin and laminin binding protein of Leishmania is stimulated by zinc and may involve laminin zinc-finger like sequences. Eur. J. Biochem. 2691622-1629. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee, S., B. Wei, M. Bhattacharyya-Pakrasi, H. B. Pakrasi, and T. J. Smith. 2003. Structural determinants of metal specificity in the zinc transport protein ZnuA from Synechocystis 6803. J. Mol. Biol. 3331061-1069. [DOI] [PubMed] [Google Scholar]
- 7.Beachey, E. H., and I. Ofek. 1976. Epithelial cell binding of group A streptococci by lipoteichoic acid on fimbriae denuded of M protein. J. Exp. Med. 143759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry, A. M., and J. C. Paton. 1996. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 645255-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessen, D. E. 2009. Population biology of the human restricted pathogen, Streptococcus pyogenes. Infect. Genet. Evol. 9581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beveridge, T. J., and V. R. F. Matias. 2006. Ultrastructure of gram-positive cell walls, p. 849. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 11.Brenot, A., B. F. Weston, and M. G. Caparon. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol. Microbiol. 631185-1196. [DOI] [PubMed] [Google Scholar]
- 12.Burnette-Curley, D., V. Wells, H. Viscount, C. L. Munro, J. C. Fenno, P. Fives-Taylor, and F. L. Macrina. 1995. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect. Immun. 634669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra, B. R., M. Yogavel, and A. Sharma. 2007. Structural analysis of ABC-family periplasmic zinc binding protein provides new insights into mechanism of ligand uptake and release. J. Mol. Biol. 367970-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claverys, J. P. 2001. A new family of high-affinity ABC manganese and zinc permeases. Res. Microbiol. 152231-243. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson, A. L., E. Dassa, C. Orelle, and J. Chen. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72317-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 19.Desrosiers, D. C., Y. C. Sun, A. A. Zaidi, C. H. Eggers, D. L. Cox, and J. D. Radolf. 2007. The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles. Mol. Microbiol. 65137-152. [DOI] [PubMed] [Google Scholar]
- 20.Dintilhac, A., G. Alloing, C. Granadel, and J. P. Claverys. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25727-739. [DOI] [PubMed] [Google Scholar]
- 21.Dintilhac, A., and J. P. Claverys. 1997. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res. Microbiol. 148119-131. [DOI] [PubMed] [Google Scholar]
- 22.Ellen, R. P., and R. J. Gibbons. 1972. M protein-associated adherence of Streptococcus pyogenes to epithelial surfaces: prerequisite for virulence. Infect. Immun. 5826-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsner, A., B. Kreikemeyer, A. Braun-Kiewnick, B. Spellerberg, B. A. Buttaro, and A. Podbielski. 2002. Involvement of Lsp, a member of the LraI-lipoprotein family in Streptococcus pyogenes, in eukaryotic cell adhesion and internalization. Infect. Immun. 704859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 25.Evans, G., and R. F. Pettifer. 2001. CHOOCH: a program for deriving anomalous-scattering factors from X-ray fluorescence spectra. J. Appl. Crystallogr. 3482-86. [Google Scholar]
- 26.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, L. H. Shing, L. S. Ping, Y. Qian, J. H. Gui, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 984658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Froeliger, E. H., and P. Fives-Taylor. 2000. Streptococcus parasanguis FimA does not contribute to adherence to SHA. J. Dent. Res. 79337. [Google Scholar]
- 28.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 896172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazlett, K. R. O., F. Rusnak, D. G. Kehres, S. W. Bearden, C. J. La Vake, M. E. La Vake, M. E. Maguire, R. D. Perry, and J. D. Radolf. 2003. The Treponema pallidum tro operon encodes a multiple metal transporter, a zinc-dependent transcriptional repressor, and a semi-autonomously expressed phosphoglycerate mutase. J. Biol. Chem. 27820687-20694. [DOI] [PubMed] [Google Scholar]
- 30.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38140-153. [DOI] [PubMed] [Google Scholar]
- 31.Jamieson, K. V., J. Wu, S. R. Hubbard, and D. Meruelo. 2008. Crystal structure of the human laminin receptor precursor. J. Biol. Chem. 2833002-3005. [DOI] [PubMed] [Google Scholar]
- 32.Janulczyk, R., J. Pallon, and L. Björck. 1999. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol. Microbiol. 34596-606. [DOI] [PubMed] [Google Scholar]
- 33.Janulczyk, R., S. Ricci, and L. Björck. 2003. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect. Immun. 712656-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkinson, H. F. 1994. Cell surface protein receptors in oral streptococci. FEMS Microbiol. Lett. 121133-140. [DOI] [PubMed] [Google Scholar]
- 35.Kreikemeyer, B., M. Nakata, S. Oehmcke, C. Gschwendtner, J. Normann, and A. Podbielski. 2005. Streptococcus pyogenes collagen type I-binding Cpa surface protein: Expression profile, binding characteristics, biological functions, and potential clinical impact. J. Biol. Chem. 28033228-33239. [DOI] [PubMed] [Google Scholar]
- 36.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 602256-2268. [DOI] [PubMed] [Google Scholar]
- 37.Kunitomo, E., Y. Terao, S. Okamoto, T. Rikimaru, S. Hamada, and S. Kawabata. 2008. Molecular and biological characterization of histidine triad protein in group A streptococci. Microbes Infect. 10414-423. [DOI] [PubMed] [Google Scholar]
- 38.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26283-291. [Google Scholar]
- 40.Lawrence, M. C., P. A. Pilling, V. C. Epa, A. M. Berry, A. D. Ogunniyi, and J. C. Paton. 1998. The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure 61553-1561. [DOI] [PubMed] [Google Scholar]
- 41.Lee, Y. H., R. K. Deka, M. V. Norgard, J. D. Radolf, and C. A. Hasemann. 1999. Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat. Struct. Biol. 6628-633. [DOI] [PubMed] [Google Scholar]
- 42.Lee, Y. H., M. R. Dorwart, K. R. O. Hazlett, R. K. Deka, M. V. Norgard, J. D. Radolf, and C. A. Hasemann. 2002. The crystal structure of Zn(II)-free Treponema pallidum TroA, a periplasmic metal-binding protein, reveals a closed conformation. J. Bacteriol. 1842300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, H., and G. Jogl. 2007. Crystal structure of the zinc-binding transport protein ZnuA from Escherichia coli reveals an unexpected variation in metal coordination. J. Mol. Biol. 3681358-1366. [DOI] [PubMed] [Google Scholar]
- 44.Linke, C., T. T. Caradoc-Davies, T. Proft, and E. N. Baker. 2008. Purification, crystallization and preliminary crystallographic analysis of Streptococcus pyogenes laminin-binding protein Lbp. Acta Crystallogr. F 64141-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loisel, E., L. Jacquamet, L. Serre, C. Bauvois, J. L. Ferrer, T. Vernet, A. M. Di Guilmi, and C. Durmort. 2008. AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: biochemical and structural analysis. J. Mol. Biol. 381594-606. [DOI] [PubMed] [Google Scholar]
- 46.Lupyan, D., A. Leo-Macias, and A. R. Ortiz. 2005. A new progressive-iterative algorithm for multiple structure alignment. Bioinformatics 213255-3263. [DOI] [PubMed] [Google Scholar]
- 47.Matias, V. R. F., and T. J. Beveridge. 2006. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 1881011-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. O. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA 10215641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 51.Ogunniyi, A. D., M. Grabowicz, L. K. Mahdi, J. Cook, D. L. Gordon, T. A. Sadlon, and J. C. Paton. 2009. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 23731-738. [DOI] [PubMed] [Google Scholar]
- 52.Oligino, L., and P. Fives-Taylor. 1993. Overexpression and purification of a fimbria-associated adhesin of Streptococcus parasanguis. Infect. Immun. 611016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of bacterial zinc regulons: Enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 1009912-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Podbielski, A., M. Woischnik, B. A. B. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 311051-1064. [DOI] [PubMed] [Google Scholar]
- 55.Rajam, G., J. M. Anderton, G. M. Carlone, J. S. Sampson, and E. W. Ades. 2008. Pneumococcal surface adhesin A (PsaA): a review. Crit. Rev. Microbiol. 34131-142. [DOI] [PubMed] [Google Scholar]
- 56.Riboldi-Tunnicliffe, A., N. W. Isaacs, and T. J. Mitchell. 2005. 1.2 Å Crystal structure of the S. pneumoniae PhtA histidine triad domain a novel zinc binding fold. FEBS Lett. 5795353-5360. [DOI] [PubMed] [Google Scholar]
- 57.Romero-Steiner, S., T. Pilishvili, J. S. Sampson, S. E. Johnson, A. Stinson, G. M. Carlone, and E. W. Ades. 2003. Inhibition of pneumococcal adherence to human nasopharyngeal epithelial cells by anti-PsaA antibodies. Clin. Diagn. Lab. Immunol. 10246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rukhman, V., R. Anati, M. Melamed-Frank, and N. Adir. 2005. The MntC crystal structure suggests that import of Mn2+ in cyanobacteria is redox controlled. J. Mol. Biol. 348961-969. [DOI] [PubMed] [Google Scholar]
- 59.Schéele, S., A. Nyström, M. Durbeej, J. F. Talts, M. Ekblom, and P. Ekblom. 2007. Laminin isoforms in development and disease. J. Mol. Med. 85825-836. [DOI] [PubMed] [Google Scholar]
- 60.Schrager, H. M., S. Alberti, C. Cywes, G. J. Dougherty, and M. R. Wessels. 1998. Hyaluronic acid capsule modulates M protein-mediated adherence and acts as a ligand for attachment of group A streptococcus to CD44 on human keratinocytes. J. Clin. Investig. 1011708-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siegel, J. L., S. F. Hurst, and E. S. Liberman. 1981. Mutanolysin-induced spheroplasts of Streptococcus mutans are true protoplasts. Infect. Immun. 31808-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, N. Schnitzler, R. Lütticken, and A. Podbielski. 1999. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spurlino, J. C., G. Y. Lu, and F. A. Quiocho. 1991. The 2.3-Å resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J. Biol. Chem. 2665202-5219. [DOI] [PubMed] [Google Scholar]
- 64.Sun, X., H. M. Baker, R. Ge, H. Sun, Q.-Y. He, and E. N. Baker. 22 May 2009, posting date. Crystal structure and metal binding properties of the lipoprotein MtsA, responsible for iron transport in Streptococcus pyogenes. Biochemistry. doi: 10.1021/bi900552c. [DOI] [PubMed]
- 65.Sun, X., G. Ruiguang, C. Jen-Fu, S. Hongzhe, and H. Qing-Yu. 2008. Lipoprotein MtsA of MtsABC in Streptococcus pyogenes primarily binds ferrous ion with bicarbonate as a synergistic anion. FEBS Lett. 5821351-1354. [DOI] [PubMed] [Google Scholar]
- 66.Sutcliffe, I. C., and D. J. Harrington. 2002. Pattern searches for the identification of putative lipoprotein genes in gram-positive bacterial genomes. Microbiology 1482065-2077. [DOI] [PubMed] [Google Scholar]
- 67.Switalski, L. M., P. Speziale, M. Höök, T. Wadström, and R. Timpl. 1984. Binding of Streptococcus pyogenes to laminin. J. Biol. Chem. 2593734-3738. [PubMed] [Google Scholar]
- 68.Takagi, J., Y. Yang, J. H. Liu, J. H. Wang, and T. A. Springer. 2003. Complex between nidogen and laminin fragments, reveals a paradigmatic β-propeller interface. Nature 424969-974. [DOI] [PubMed] [Google Scholar]
- 69.Tenenbaum, T., B. Spellerberg, R. Adam, M. Vogel, K. S. Kim, and H. Schroten. 2007. Streptococcus agalactiae invasion of human brain microvascular endothelial cells is promoted by the laminin-binding protein Lmb. Microbes Infect. 9714-720. [DOI] [PubMed] [Google Scholar]
- 70.Terao, Y., S. Kawabata, E. Kunitomo, I. Nakagawa, and S. Hamada. 2002. Novel laminin-binding protein of Streptococcus pyogenes, Lbp, is involved in adhesion to epithelial cells. Infect. Immun. 70993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wahid, R. M., M. Yoshinaga, J. Nishi, N. Maeno, J. Sarantuya, T. Ohkawa, A. M. Jalil, K. Kobayashi, and K. Miyata. 2005. Immune response to a laminin-binding protein (Lmb) in group A streptococcal infection. Pediatr. Int. 47196-202. [DOI] [PubMed] [Google Scholar]
- 72.Wei, B., A. M. Randich, M. Bhattacharyya-Pakrasi, H. B. Pakrasi, and T. J. Smith. 2007. Possible regulatory role for the histidine-rich loop in the zinc transport protein, ZnuA. Biochemistry 468734-8743. [DOI] [PubMed] [Google Scholar]
- 73.Yatsunyk, L. A., J. A. Easton, L. R. Kim, S. A. Sugarbaker, B. Bennett, R. M. Breece, I. I. Vorontsov, D. L. Tierney, M. W. Crowder, and A. C. Rosenzweig. 2008. Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. J. Biol. Inorg. Chem. 13271-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuber, B., M. Haenni, T. Ribeiro, K. Minnig, F. Lopes, P. Moreillon, and J. Dubochet. 2006. Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J. Bacteriol. 1886652-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]