Abstract
Exponentially growing populations of Bacillus subtilis contain two morphologically and functionally distinct cell types: motile individuals and nonmotile multicellular chains. Motility differentiation arises because RNA polymerase and the alternative sigma factor σD activate expression of flagellin in a subpopulation of cells. Here we demonstrate that the peptidoglycan-remodeling autolysins under σD control, LytC, LytD, and LytF, are expressed in the same subpopulation of cells that complete flagellar synthesis. Morphological heterogeneity is explained by the expression of LytF that is necessary and sufficient for cell separation. Moreover, LytC is required for motility but not at the level of cell separation or flagellum biosynthesis. Rather, LytC appears to be important for flagellar function, and motility was restored to a LytC mutant by mutation of either lonA, encoding the LonA protease, or a gene encoding a previously unannotated swarming motility inhibitor, SmiA. We conclude that heterogeneous activation of σD-dependent gene expression is sufficient to explain both the morphological heterogeneity and functional heterogeneity present in vegetative B. subtilis populations.
Growing populations of Bacillus subtilis are heterogeneous in cell morphology (17). Some members of these populations grow as single cells that are motile, whereas other cells in the populations grow in multicellular, nonmotile chains. The difference in motility between the two cell types is controlled at the level of gene transcription. The expression of the gene that encodes the flagellar filament (hag) is either ON or OFF and is under the control of RNA polymerase and the alternative sigma factor σD (9, 17, 29). Cells that are ON for flagellin expression complete flagellum assembly and constitute the motile subpopulation. Cells that are OFF for flagellin expression do not complete flagellum biosynthesis and constitute the nonmotile subpopulation.
The dimorphism of single cells and chaining cells in heterogeneous populations has not been resolved but is likely related to autolysin activity. Autolysins are bacterial enzymes that hydrolyze and remodel the peptidoglycan found in the bacterial cell wall (38, 42). B. subtilis is a gram-positive bacterium with a thick layer of peptidoglycan exterior to the cell membrane. During cell division, the cell wall grows inward by a process called septation to divide the mother into two identical daughters. The daughter cells subsequently remain joined by a shared layer of peptidoglycan, and autolysins are required for daughter cell separation (1, 10, 11, 25). Autolysins have also been implicated in cell wall turnover, sporulation, and motility (12, 33, 34, 39).
B. subtilis encodes at least 35 differentially regulated autolysins, but 3 of these autolysins in particular are strong candidates for participating in the single-cell-chain heterogeneity observed during exponential growth: LytC (an N-acetylmuramoyl-l-alanine amidase), LytD (an endo-β-N-acetylglucosaminidase), and LytF (a γ-d-glutamate meso-diaminopimelate muropeptidase) (20, 27, 26, 38). The genes that encode the LytD and LytF autolysins are under exclusive control of σD, and 70% of LytC expression is σD dependent (15, 20, 21, 26, 27). Furthermore, LytC, LytD, and LytF have been implicated in vegetative cell separation and motility (3, 31, 36). The simultaneous reduction in expression of the three autolysins is thought to account for the observation that a null mutant in which the gene that encodes σD is disrupted does not form separate cells and grows constitutively in chains (28).
Here we investigate the contribution of the σD-dependent LytC, LytD, and LytF autolysins to population heterogeneity in B. subtilis. We determine that the autolysins are expressed in the same subpopulation that expresses the flagellar filament. In an undomesticated strain background (B. subtilis 3610) in which the majority of cells in a population are ON for autolysin expression, we find that LytF is dedicated chiefly to cell separation, whereas LytC supports swarming motility. Thus, cells that are ON for σD express both the autolysin and flagellar genes and grow as separate motile cells. Cells that are OFF for σD do not express the autolysin or flagellar genes and grow as nonmotile chains. Finally, we demonstrate that the requirement of autolysins for motility is related to flagellar function rather than to flagellar biosynthesis or cell separation and that the motility defect could be genetically suppressed.
MATERIALS AND METHODS
Strains and growth conditions.
B. subtilis strains were grown in Luria-Bertani (LB) broth (10 g tryptone per liter, 5 g yeast extract per liter, 5 g NaCl per liter) or on LB agar plates containing 1.5% Bacto agar at 37°C. When appropriate, antibiotics were included at the following concentrations: 10 μg/ml tetracycline, 100 μg/ml spectinomycin, 5 μg/ml chloramphenicol, 5 μg/ml kanamycin, and 1 μg/ml erythromycin plus 25 μg/ml lincomycin (mls). Isopropyl β-d-thiogalactopyranoside (IPTG) (Sigma) was added to the medium at the concentration indicated below when appropriate.
Swarm expansion assay.
Cells were grown to mid-log phase at 37°C in LB broth and resuspended at an optical density at 600 nm (OD600) of 10 in 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) containing 0.5% India ink (Higgins). Plates containing freshly prepared LB agar fortified with 0.7% Bacto agar (25 ml/plate) were dried for 20 min in a laminar flow hood, centrally inoculated with 10 μl of the cell suspension, dried for another 10 min, and incubated at 37°C. The India ink marked the origin of a colony, and the swarm radius was measured relative to the origin. For consistency, an axis was drawn on the back of each plate, and the swarm radius was measured along this axis (17).
Microscopy.
Fluorescence microscopy was performed with a Nikon 80i microscope with an Excite 120 metal halide lamp. Green fluorescent protein (GFP) and Alexa Fluor 488 C5 maleimide fluorescent signals were viewed using a phase-contrast objective (Nikon Plan Apo 100X) and were visualized using a C-FL HYQ fluorescein isothiocyanate filter cube (excitation filter wavelength, 460 to 500 nm; barrier filter wavelength, 515 to 550 nm). FM4-64 was visualized with a C-FL HYQ Texas Red filter cube (excitation filter wavelength, 532 to 587 nm; barrier filter wavelength, >590 nm). Images were captured with a Photometrics Coolsnap HQ2 camera in black and white, false colored, and superimposed using Metamorph image software.
For fluorescent microscopy of flagella, 0.5 ml of a broth culture was harvested at an OD600 of 0.5 and washed once in 1.0 ml of T-Base buffer [15 mM (NH4)2SO4, 80 mM K2HPO4, 44 mM KH2PO4, 3.4 mM sodium citrate, 3.0 mM MgSO4·6H2O]. The suspension was pelleted, resuspended in 50 μl of T-Base buffer containing 5 μg/ml Alexa Fluor 488 C5 maleimide (Molecular Probes), and incubated for 5 min at room temperature. Cells were then washed twice with 500 μl T-Base buffer. Membranes were stained by resuspension in 50 μl of T-Base buffer containing 5 μg/ml FM4-64 (Molecular Probes) and incubated for 10 min at room temperature. Three microliters of the suspension was placed on a microscope slide and immobilized with a poly-l-lysine-treated coverslip (4).
For GFP microscopy, cells were grown overnight at 22°C in LB broth to an OD600 of 0.5, and 1 ml was pelleted and resuspended in 0.1 ml PBS. Membranes were stained by resuspension in 50 μl of PBS containing 5 μg/ml FM4-64 and incubated for 10 min at room temperature. Samples were observed by spotting 3 μl of the suspension on a glass microscope slide and were immobilized with a poly-l-lysine-treated glass coverslip. Images were captured with Metamorph software.
Strain construction.
All constructs were first introduced into domesticated strain PY79 by natural competence and then transferred to the 3610 background using SPP1-mediated generalized phage transduction (46). All strains used in this study are listed in Table 1. All plasmids used in this study are listed in Table S1 in the supplemental material. All primers used in this study are listed in Table S2 in the supplemental material.
TABLE 1.
Strains useda
| Strain | Genotype |
|---|---|
| PY79 | Lab strain, swrAPY79sfp0 |
| 3610 | Wild type |
| DS108 | ΔlytD::mls |
| DS323 | ΔsigD::tetb |
| DS1122 | srfAC::Tn10 |
| DS1447 | ΔlytF::tet |
| DS1499 | ΔlytD::mls ΔlytF::tet |
| DS1895 | [PY79] swrAPY79amyE::Phag-hag(T209C) spec |
| DS1916 | amyE::Phag-hag(T209C) specc |
| DS2856 | thrC::PlytF-CFP mls lacA::Phag-YFP tet |
| DS3032 | [PY79] swrAPY79thrC::PlytF-CFP mls lacA::Phag-YFP tet |
| DS3159 | ΔsigD::tet amyE::Phag-hag(T209C) spec |
| DS3437 | ΔswrA::kan amyE::Physpank-lytF spec lacA::Phag-YFP tet |
| DS3519 | ΔsigD::tet amyE::Physpank-lytC spec |
| DS3520 | ΔsigD::tet amyE::Physpank-lytF spec |
| DS3521 | ΔsigD::tet amyE::Physpank-lytD spec |
| DS3819 | ΔlytC::kan |
| DS3820 | ΔlytC::kan ΔlytF::tet |
| DS3821 | ΔlytC::kan ΔlytD::mls |
| DS3822 | ΔlytC::kan ΔlytD::mls ΔlytF::tet |
| DS3823 | ΔlytC::kan ΔlytD::mls ΔlytF::tet amyE::Phag-GFP cat |
| DS3824 | ΔlytC::kan ΔlytD::mls ΔlytF::tet amyE::Phag-hag(T209C) spec |
| DS3894 | sigD::Tn10 spec thrC::PlytF-CFP mls lacA::Phag-YFP tet |
| DS3961 | ΔlytC::cat ΔlytD::spec |
| DS4066 | ΔlytC::cat ΔlytD::spec pMARa kan mls |
| DS4071 | ΔlytC::cat ΔlytD::spec smiAΩTnYLB kan (TAAAAGCGTAT) |
| DS4186 | ΔlytC::cat ΔlytD::spec lonAΩTnYLB kan (TATTAATAAAA) |
| DS4187 | ΔlytC::cat ΔlytD::spec smiAΩTnYLB kan (TAGATCCGATC) |
| DS4190 | ΔlytC::cat ΔlytD::spec smiAΩTnYLB kan (TAATAAGCTGA) |
| DS4191 | ΔlytC::cat ΔlytD::spec lonAΩTnYLB kan (TAGGTCCGTCT) |
| DS4195 | ΔlytC::cat ΔlytD::spec lonAΩTnYLB kan (TAGTTGCGGAT) |
| DS4196 | ΔlytC::cat ΔlytD::spec lonAΩTnYLB kan (TAATAAACCTC) |
| DS4987 | ΔsmiA |
| DS4988 | ΔlytC::cat ΔlytD::spec ΔsmiA |
| DS4995 | ΔlytC::cat ΔlytD::spec ΔsmiA thrC::PsmiA-smiA mls |
| DS4996 | ΔlytC::cat ΔlytD::spec ΔsmiA thrC::PfliD-smiA mls |
| DS5036 | ΔlytC::cat ΔlytD::spec ΔsmiA thrC::Physpank-smiA mls |
| DS5109 | ΔsmiA lytC::kan lytD::mls |
| DS5125 | ΔsmiA lytC::kan lytF::tet |
| DS5126 | ΔsmiA lytC::kan lytD::mls lytF::tet |
| DS5127 | ΔsmiA lytD::mls lytF::tet |
| DS5268 | srfAC::Tn10 spec lytD::mls |
| DS5269 | srfAC::Tn10 spec lytC::kan lytF::tet |
| DS5270 | srfAC::Tn10 spec lytF::tet |
| DS5271 | srfAC::Tn10 spec lytC::kan lytF::tet |
| DS5272 | srfAC::Tn10 spec lytC::kan |
| DS5273 | srfAC::Tn10 spec lytC::kan lytD::mls lytF::tet |
| DS5274 | srfAC::Tn10 spec lytD::mls lytF::tet |
| DS5286 | ΔlonA |
| DS5300 | ΔlonA lytC::cat lytD::spec |
| DS5302 | ΔlonA lytC::kan lytF::tet |
| DS5303 | ΔlonA lytD::mls lytF::tet |
| DS5318 | ΔlonA lytC::cat lytD::spec thrC::PlonA-lonA mls |
| DS5351 | ΔlonA lytC::kan lytD::mls |
| DS5352 | ΔlonA lytC::kan lytD::mls lytF::tet |
(i) ΔlytC::kan.
The ΔlytC::kan insertion-deletion allele was generated by long flanking homology PCR (using primer pairs 1185/1186 and 1187/1188), and DNA containing a kanamycin drug resistance gene (pDG780) was used as a template for marker replacement (13, 43). The ΔlytC::cat insertion-deletion allele was generated by long flanking homology PCR with the same primer combinations, and DNA containing a chloramphenicol drug resistance gene (pAC225) was used as a template for marker replacement (pAC225 was a generous gift from Amy Camp and Rich Losick, Harvard University).
(ii) ΔlytD::mls.
The ΔlytD::mls insertion-deletion allele was generated by long flanking homology PCR (using primer pairs 61/62 and 63/64), and DNA containing a erythromycin drug resistance gene (pDG646) was used as a template for marker replacement (13, 43). The ΔlytD::spec insertion-deletion allele was generated by long flanking homology PCR with the same primer combinations, and DNA containing a spectinomycin drug resistance gene (pAH54) was used as a as a template for marker replacement (pAH54 was a generous gift from Amy Camp and Rich Losick, Harvard University).
(iii) ΔlytF::tet.
The ΔlytF::tet insertion-deletion allele was generated by long flanking homology PCR (using primer pairs 494/495 and 496/497), and DNA containing a tetracycline drug resistance gene (pDG1515) was used as a template for marker replacement (13, 43).
(iv) ΔsmiA.
To generate the ΔsmiA in-frame markerless deletion construct pKB117, the region upstream of smiA was PCR amplified using the 1555/1556 primer pair and digested with EcoRI and SalI, and the region downstream of smiA was PCR amplified using the 1557/1558 primer pair and digested with SalI and BamHI. The two fragments were then simultaneously ligated into the EcoRI and BamHI sites of pMiniMAD, which carries a temperature-sensitive origin of replication and an erythromycin resistance cassette, to generate pKB117 (32). Plasmid pKB117 was introduced into PY79 by single-crossover integration by transformation at the restrictive temperature for plasmid replication (37°C) using mls resistance for selection. The integrated plasmid was then transduced into strain DS3961. To evict the plasmid, the strain was incubated in 3 ml LB broth at a temperature permissive for plasmid replication (22°C) for 14 h, diluted 30-fold in fresh LB broth, and incubated at 22°C for another 8 h. Dilution and outgrowth were repeated two more times. Cells were then serially diluted and plated on LB agar at 37°C. Individual colonies were patched on LB agar plates and LB agar plates containing mls to identify mls-sensitive colonies that had evicted the plasmid. Chromosomal DNA from colonies in which the plasmid had been excised was purified and screened by PCR using primers 1555 and 1558 to determine which isolate had retained the ΔsmiA allele.
(v) ΔlonA.
To generate the ΔlonA in-frame markerless deletion construct pDP307, the region upstream of lonA was PCR amplified using the 1609/1610 primer pair and digested with BamHI and XhoI, and the region downstream of lonA was PCR amplified using the 1611/1612 primer pair and digested with XhoI and EcoRI. The two fragments were then simultaneously ligated into the EcoRI and BamHI sites of pMiniMAD (32). pDP307 was integrated and excised as described above for ΔsmiA. Chromosomal DNA from colonies in which the plasmid had been excised was purified and screened by PCR using primers 1609 and 1612 to determine which isolate had retained the ΔlonA allele.
(vi) Inducible lyt constructs.
To generate the inducible amyE::Physpank-lytC spec construct pKB67, a PCR product containing lytC was amplified from B. subtilis 3610 chromosomal DNA using the 1059/1060 primer pair, digested with SalI and SphI, and cloned into the SalI and SphI sites of pDR111 containing a spectinomycin resistance cassette, a polylinker downstream of the Physpank promoter, and the gene encoding the LacI repressor between the arms of the amyE gene (2).
To generate the inducible amyE::Physpank-lytD spec construct pKB68, a PCR product containing lytD was amplified from B. subtilis 3610 chromosomal DNA using the 1061/1062 primer pair, digested with SalI and SphI, and cloned into the SalI and SphI sites of pDR111.
To generate the inducible amyE::Physpank-lytF spec construct pKB69, a PCR product containing lytF was amplified from B. subtilis 3610 chromosomal DNA using the 1063/1064 primer pair, digested with SalI and SphI, and cloned into the SalI and SphI sites of pDR111.
To generate the inducible thrC::Physpank-smiA mls construct pAP9, a PCR product containing smiA was amplified from B. subtilis 3610 chromosomal DNA using the 1366/1367 primer pair, digested with HindIII and NheI, and cloned into the HindIII and NheI sites of pDP150 containing an mls resistance cassette, a polylinker downstream of the Physpank promoter, and the gene encoding the LacI repressor between the arms of the thrC gene (18).
(vii) Complementation constructs.
To generate the thrC::PlonA-lonA mls complementation construct pDP291, the PlonA-lonA region was PCR amplified from B. subtilis 3610 chromosomal DNA using the 1360/1361 primer pair and digested with EcoRI and BamHI. The digested fragment was then cloned into the EcoRI and BamHI sites of pDG1664 containing an erythromycin resistance cassette (mls) and a polylinker between the arms of the thrC gene (14).
To generate the thrC::PsmiA-smiA mls complementation construct pKB104, the PsmiA-smiA region was PCR amplified from B. subtilis 3610 chromosomal DNA using the 1444/1508 primer pair and digested with EcoRI and HindIII. The digested fragment was then cloned into the EcoRI and HindIII sites of pDG1664 containing an erythromycin resistance cassette (mls) and a polylinker between the arms of the thrC gene (14).
To generate the thrC::PfliD-smiA mls complementation construct pKB105, the PfliD region was PCR amplified from B. subtilis 3610 chromosomal DNA using the 1509/1510 primer pair and digested with BamHI and HindIII, whereas the smiA region was PCR amplified from B. subtilis 3610 chromosomal DNA using the 1444/1508 primer pair and digested with HindIII and EcoRI. Both fragments were simultaneously ligated into the BamHI and EcoRI sites of pDG1664.
(viii) Cytological reporters.
To generate the lacA::Phag-YFP reporter pLC20, the Phag promoter region was PCR amplified from B. subtilis 3610 chromosomal DNA using the 380/381 primer pair and digested with EcoRI and NheI. The yellow fluorescent protein (YFP) gene with an artificial optimized ribosome binding site was PCR amplified from plasmid pKL187 (23) using the 562/563 primer pair and digested with NheI and BamHI. The two fragments were then simultaneously ligated into the EcoRI and BamHI sites of plasmid pNC018, which carries a tetracycline resistance marker and a polylinker between two arms of the lacA gene (a gift from Natalie Campo and David Rudner, Harvard Medical School).
To generate the thrC::PlytF-CFP reporter pDP224, the PlytF promoter region was PCR amplified from B. subtilis 3610 chromosomal DNA using the 917/918 primer pair and digested with EcoRI and SphI. The cyan fluorescent protein (CFP) gene, codon optimized for B. subtilis, was PCR amplified from plasmid pDR200 (a gift from David Rudner, Harvard Medical School) using the 633/634 primer pair and digested with SphI and BamHI. The two fragments were then simultaneously ligated into the EcoRI and BamHI sites of plasmid pDG1664, which carries an erythromycin resistance marker and a polylinker between two arms of the thrC gene (14).
Transposon mutagenesis.
The temperature-sensitive vector pMarA carrying TnYLB was introduced into strain DS3961 by SPP1-mediated generalized transduction to generate DS4066 (22). Next, a transposon library was generated by inoculating DS4066 into 3 ml LB broth containing kanamycin and incubating the culture at 22°C in a roller drum for 14 h. The culture was then serially diluted and spread on prewarmed LB agar plates fortified with 1.5% agar and kanamycin and incubated overnight at 42°C. The colonies were pooled and spotted in the center of a 0.7% swarm agar plate. Most of the cells remained nonmotile, but rare motile flares emerged from the central colony and were clonally isolated. Cells from the motile flares likely contained transposons linked to the mutation of interest. To confirm that the transposon was linked to the suppressor mutation, a lysate was generated for the suppressor mutant and the transposon was transduced into the parent strain lacking the suppressor. Transposon insertion sites were identified by partially degenerate touchdown PCR using primer 766, hybrid degenerate primer 749, 250 ng of purified chromosomal DNA, and Phusion polymerase (New England Biolabs) (24).
RESULTS
Autolysin expression is heterogeneous.
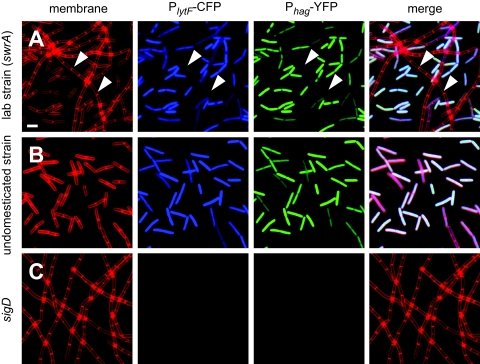
Transcription of the genes encoding the vegetative autolysins LytC, LytD, and LytF is under the control of RNA polymerase and the alternative sigma factor σD (28). It has recently been demonstrated that the expression of at least one σD-dependent gene, hag (which encodes flagellin), is heterogeneous in growing populations (18). To determine whether σD-dependent autolysin gene expression was also heterogeneous, a laboratory strain was doubly labeled with two compatible fluorescent reporters. One reporter fused expression of CFP to the promoter of the LytF autolysin (PlytF) and was integrated at the ectopic thrC locus (thrC::PlytF-CFP). The other reporter fused expression of YFP to the promoter of flagellin (Phag) and was integrated at the ectopic lacA locus (lacA::Phag-YFP). Observation of exponentially growing cells by fluorescence microscopy revealed that expression of lytF was heterogeneous in the population; it was ON in single cells and OFF in long chains (Fig. 1A). The same subpopulation of cells that expressed lytF also expressed flagellin (Fig. 1A). Neither reporter was expressed in a cell in which sigD, the gene encoding the σD sigma factor, was mutated (Fig. 1C). We conclude that heterogeneous expression is not flagellin specific, that the expression of lytF is also heterogeneous in a population, and that σD coordinately activates both motility and autolysin genes in the same subpopulation of cells.
FIG. 1.
The LytF autolysin and flagellin are expressed in the same subpopulation of cells. Fluorescence microscopy images were obtained for cells of laboratory strain PY79 (DS3032) (A), wild-type undomesticated strain 3610 (DS2856) (B), and a sigD null mutant (DS3894) (C) that were grown to mid-log phase (OD600, 0.5) and stained to reveal membranes (FM4-64; false color, red). Each strain also contained the promoter of the lytF (autolysin) gene fused to CFP expression (PlytF-CFP; false color, blue) and the promoter of the hag (flagellin) gene fused to YFP expression (Phag-YFP; false color, green). Arrowheads indicate the location of a chain. Scale bar = 2 μm.
Flagellin expression in the majority of cells in an exponentially growing population of laboratory strains (like strain PY79) is OFF because of a loss-of-function frameshift mutation in the swrA gene (5, 16, 18, 19). In contrast, the ancestral undomesticated strain B. subtilis 3610 synthesizes functional SwrA, and flagellin expression is ON in the majority of cells during exponential growth (18, 47) (Fig. 1B). As observed for flagellin expression, a larger population of the undomesticated strain than of the laboratory strain expressed the PlytF promoter (Fig. 1B). From this we conclude that the proportion of the population that expresses the σD-dependent autolysin genes is greater in undomesticated strains than in laboratory strains.
LytF is necessary and sufficient for cell separation.
LytC, LytD, and LytF have been implicated in cell separation in laboratory strains (3, 31). We now realize that laboratory strains grow primarily as chains because a majority of the cells in populations fail to express the autolysin genes, and this may obscure cell separation defects of autolysin mutants (Fig. 1A). We therefore investigated the roles of LytC, LytD, and LytF in chain separation in an undomesticated strain, the majority of cells of which express the three autolysins and grow as separate individuals (Fig. 2). Cells in which lytC or lytD was mutated grew predominantly as individuals, but cells in which lytF was mutated grew predominantly as chains (Fig. 2). Double mutation of the lyt genes had additive phenotypic effects (Fig. 2). Simultaneous mutation of lytC, lytD, and lytF was required to produce a strain that grew constitutively as long chains and resembled a strain in which sigD was mutated (Fig. 2). From this we conclude that all of the σD-dependent autolysins contribute to cell separation but that the majority of vegetative cell-separating activity is performed by LytF.
FIG. 2.
The lytF mutant has a severe defect in cell separation. Phase-contrast microscopy was performed with cells having the genotypes indicated grown to mid-log phase (OD600, 0.5). Scale bar = 5 μm. The bar graph shows a quantitative measure of chaining. Cells were stained with FM4-64, visualized by fluorescence microscopy, and manually counted, and the proportions of the populations that grew in chains (>4 connected cells) (black bars) and not in chains (≤4 connected cells) (gray bars) were determined. More than 800 cells were counted for each strain. The following strains were used in the experiment: PY79 (swrA), 3610 (wild type), DS3819 (lytC), DS108 (lytD), DS3821 (lytC lytD), DS1447 (lytF), DS1499 (lytD lytF), DS3820 (lytC lytF), DS3822 (lytC lytD lytF), and DS323 (sigD).
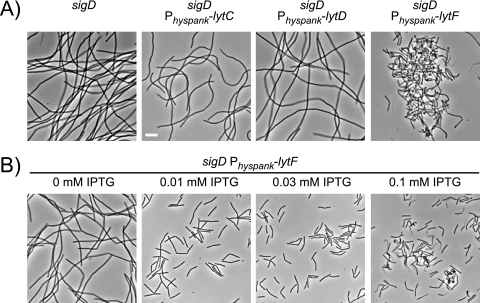
To determine which, if any, of the σD-dependent autolysins was sufficient for cell separation, the gene encoding each autolysin was individually cloned downstream of the IPTG-inducible Physpank promoter and integrated at the ectopic amyE locus of a sigD null mutant whose mutation eliminated native expression of the autolysins and which grew exclusively in chains (Fig. 3A). Induction of either lytC or lytD with 1 mM IPTG was not sufficient to dissolve the chains of the sigD mutant (Fig. 3A). In contrast, induction of lytF with 1 mM IPTG resulted in cell separation but also caused severe cell morphology defects and cell lysis (Fig. 3A). When the expression of lytF was titrated by induction with various amounts of IPTG, however, 0.03 mM IPTG promoted cell separation without the accompanying effects on morphology and viability (Fig. 3B). We conclude that LytF is the only autolysin under σD control that is both necessary and sufficient for cell separation.
FIG. 3.
Expression of lytF is sufficient to dissolve chains. (A) Phase-contrast microscopy of cells grown to mid-log phase (OD600, 0.5) in the presence of 1 mM IPTG. sigD was mutated in all cells. The strains indicated also contained IPTG-inducible copies of either lytC (DS3519), lytD (DS3521), or lytF (DS3520) fused to the Physpank promoter. Scale bar = 5 μm. (B) Phase-contrast microscopy of the sigD Physpank-lytF strain (DS3520) grown to mid-log phase in the presence of the amounts of IPTG indicated.
Autolysin mutants are defective for swarming motility but synthesize flagella.
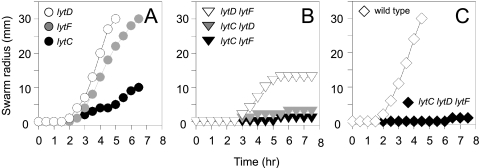
Mutation of the σD-dependent autolysins has been shown to impair the swimming motility of laboratory strains (3, 36). Undomesticated strains of B. subtilis preferentially exhibit swarming motility, which is a rapid, robust, and easily quantifiable manifestation of flagellum-mediated motility that occurs only on solid surfaces (17). In the undomesticated strain, mutation of lytC, but not mutation of lytD or lytF, resulted in a reduction in the rate of swarming motility (Fig. 4A). Swarming motility was completely abolished in cells with double mutations in lytC and either lytD or lytF (Fig. 4B) and in the lytC lytD lytF triple mutant (Fig. 4C). In general, the autolysin mutants showed a comparable reduction in a swimming motility assay performed with 0.3% agar, except that the lytF mutant was slightly more impaired, likely due to a failure of the long chains to navigate pores in the agar (see Fig S1 in the supplemental material). We conclude that LytC is required for efficient swarming motility. We note that the requirement for LytC for motility was not related to cell separation as the lytC lytD double mutant did not swarm despite growing predominantly as single cells.
FIG. 4.
Cells in which lytC is mutated have a swarming motility defect. Quantitative swarm expansion assays were performed with single (A), double (B), and triple (C) mutants with mutations in the lytC, lytD, and lytF autolysin genes. Each symbol indicates the average of measurements from three experiments. The following strains were used in the experiment: 3610 (wild type), DS3819 (lytC), DS108 (lytD), DS3821 (lytC lytD), DS1447 (lytF), DS1499 (lytD lytF), DS3820 (lytC lytF), and DS3822 (lytC lytD lytF).
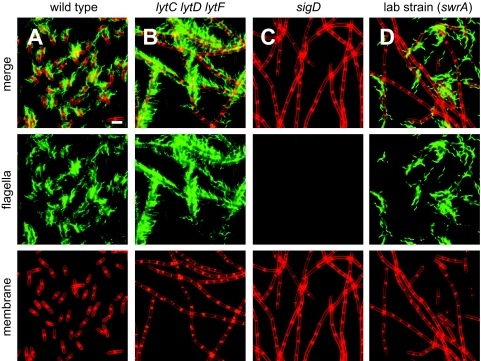
Autolysins may be required for motility because they remodel the cell wall and this may permit flagellum assembly (8, 12). To monitor flagellum assembly, a version of flagellin that may be fluorescently labeled was inserted at the ectopic amyE locus in various genetic backgrounds [amyE::Phag-hag(T209C)] (4). Flagellar staining and fluorescence microscopy revealed assembly of copious flagella in the wild type (Fig. 5A). All single autolysin mutants and all combinations of double mutants produced flagella at a density comparable to that produced by the wild type (data not shown), and the lytC lytD lytF triple mutant grew in chains but was nonetheless proficient for flagellum biosynthesis (Fig. 5B). In contrast, the chains of either sigD or swrA null mutants failed to produce flagella because, in these cases, the flagellin gene was not expressed (Fig. 1A, 1C, 5C, and 5D). Consistent with previous reports, the single cells in a swrA mutant population synthesized flagella, and there appeared to be fewer flagella per cell in this population than in a wild-type population (5) (Fig. 5D). We conclude that the σD-dependent autolysins are required for swarming motility but are not required for flagellum biosynthesis.
FIG. 5.
Cells with simultaneous lytC, lytD, and lytF mutations are proficient for flagellar assembly. The images are fluorescence microscopy images of cells grown to mid-log phase (OD600, 0.5) and stained with FM4-64 (membrane; false color, red) and Alexa Fluor C5 488 maleimide (flagella; false color, green). Each strain expresses the modified flagellar filament protein Hag(T209C). The following strains were used in the experiment: (A) wild-type strain DS1916, (B) lytC lytD lytF mutant DS3824, (C) sigD mutant DS3159, and (D) swrA mutant DS1895. Scale bar = 2 μm.
The motility defect of a lytC lytD double mutant could not be explained by either a failure to synthesize flagella or a failure to separate cells from a chain. Therefore, we took an unbiased genetic approach to identify suppressor mutations that could restore motility to the lytC lytD double mutant. The lytC lytD double mutant was mutagenized with transposons, and seven strains in which swarming motility was rescued were independently isolated. Each transposon insertion was backcrossed into the lytC lytD parental background by using SPP1-mediated generalized transduction, and in each case the rescue of the motility phenotype was found to be inseparable from the insertion.
Four isolates had disruptions in the lonA gene encoding the LonA protease (37, 41) (Fig. 6A). The lonA gene is potentially cotranscribed and translationally coupled with the ysxC gene immediately downstream as the start codon of ysxC overlaps the stop codon of lonA (35). The ysxC gene is reportedly essential in B. subtilis and encodes a putative GTPase that is thought to be required for ribosome assembly (35, 44). To determine whether the transposon insertions in lonA were directly responsible for rescuing motility in the lytC lytD double mutant or whether the transposons were polar for ysxC expression, an in-frame markerless deletion of lonA was generated to minimize polar effects. Like the transposon insertion in lonA, the lonA deletion restored motility to the lytC lytD double mutant (Fig. 6B). Next, a complementation construct that ectopically restored lonA expression was generated. The lonA gene was cloned with 500 bp immediately upstream (PlonA-lonA) and inserted at the ectopic thrC locus (thrC::PlonA-lonA) (Fig. 6A). When the lonA complementation construct was introduced into the lytC lytD lonA triple mutant, motility inhibition was restored (Fig. 6B). We conclude that the absence of lonA rescues motility in the lytC lytD double mutant and that this rescue is not due to polar effects on ysxC.
FIG. 6.
Mutations in lonA and smiA restore motility to a lytC lytD double mutant. (A) lonA genetic region and lonA complementation construct. The large open arrows indicate open reading frames. The bent arrows indicate promoters. The solid triangles indicate sites of transposon insertions that restored motility to the lytC lytD double mutant. (B) Quantitative swarm expansion assays for lytC lytD mutant DS3961, lytC lytD lonA mutant DS5300, and lytC lytD lonA (PlonA-lonA) mutant DS5318. Each symbol indicates the average of measurements from three experiments. (C) smiA genetic region and smiA complementation constructs. The large open arrows indicate open reading frames. The bent arrows indicate promoters. The solid triangles indicate sites of transposon insertions that restored motility to the lytC lytD double mutant. (D) Quantitative swarm expansion assays for lytC lytD mutant DS3961 and lytC lytD smiA (amyE::Physpank-smiA) mutant DS5036 in the presence (+IPTG) and absence (−IPTG) of 1 mM IPTG. Each symbol indicates the average of measurements from three experiments. (E) The bypass suppression model proposes that the motility defect of the lytC lytD double mutant is due to the uncontrolled activity of the motility inhibitors LonA and SmiA. The compensatory suppression model proposes that swarming motility is reduced in the absence of LytC and LytD, but swarming motility may be improved by mutating either of the swarming motility inhibitors LonA and SmiA. (F) Quantitative swarm expansion assays for wild-type strain 3610, ΔsmiA mutant DS4987, and ΔlonA mutant DS5286. Each symbol indicates the average of measurements from three experiments.
The remaining three transposon insertions were found to be immediately downstream of an operon containing the flagellar biosynthesis genes fliD, fliS, and fliT, but the region into which the transposon inserted was a noncoding region in the published B. subtilis genome sequence (6, 19). Closer inspection revealed that the transposons disrupted an unannotated open reading frame oriented in the same direction as fliT (Fig. 6C). Comparison of the predicted amino acid sequence using BLAST analysis indicated that the putative protein was conserved only in the closely related genomes of Bacillus licheniformis, Bacillus amyloliquefaciens, and Bacillus pumilus (see Fig. S2 in the supplemental material). In-frame markerless deletion of the open reading frame restored motility to the lytC lytD double mutant (Fig. 6D). The unannotated open reading frame was designated smiA because mutation of this gene improved swarming motility in the lytC lytD mutant and thus the product, SmiA, behaved as a swarming motility inhibitor.
In an attempt to complement smiA, the smiA gene was separately cloned downstream of approximately 500-bp regions immediately upstream of smiA (PsmiA) or upstream of fliD (PfliD) and introduced at the ectopic amyE locus (Fig. 6C) (6). Neither complementation construct restored motility inhibition when it was introduced into the lytC lytD mutant with an in-frame markerless deletion of the smiA gene (see Fig S3 in the supplemental material). We hypothesized that complementation was not successful either because we had failed to clone the smiA cognate promoter or because insertions in smiA were polar for downstream gene expression. In a second attempt to complement smiA, the smiA gene was cloned downstream of the nonnative, IPTG-inducible promoter Physpank and inserted at the ectopic thrC locus (Physpank-smiA) (Fig. 6C). The lytC lytD smiA triple mutant containing the Physpank-smiA construct swarmed readily in the absence of IPTG, but motility was inhibited to the level of the lytC lytD double mutant in the presence of IPTG (Fig. 6D). We conclude that the smiA mutation may be complemented in trans but that the native promoter of smiA remains unknown.
Mutation of either lonA or smiA restored swarming motility to a lytC lytD double mutant. To further determine the consequences of mutations in lonA or smiA, streaming video microscopy was conducted. Whereas wild-type cells were vigorously motile in liquid broth (see Movie S1 in the supplemental material), the majority of cells in the lytC lytD mutant population were nonmotile (see Movie S2 in the supplemental material). Mutation of either lonA or smiA restored vigorous swimming motility to the lytC lytD double mutant (see Movies S3 and S4 in the supplemental material). We conclude that the flagellar function of the lytC lytD mutant is impaired in a manner that can be bypassed by mutation of lonA or smiA.
DISCUSSION
Autolysins in B. subtilis, particularly LytC, LytD, and LytF, have been studied previously, but to the best of our knowledge, this is the first report that compares the functions of LytC, LytD, and LytF simultaneously in a common genetic background and in a background in which the majority of cells express the three proteins. For the first time, we used fluorescence microscopy and swarming motility to quantitatively determine the effects of the autolysins on cell separation and flagellar function. Here we rule out two previous models for the role of autolysins in motility and provide genetic evidence that autolysin mutants are defective in motility for functional reasons that can be genetically suppressed. Finally, we demonstrate that the σD-dependent autolysins are expressed in a subpopulation of cells, and our findings strengthen the argument that a heterogeneous population of B. subtilis motile and chaining cell types is a form of bacterial development.
Historically, it has been difficult to identify specific physiological roles for individual autolysins for three reasons. First, B. subtilis encodes as many as 35 different autolysins, and all of them act at different places on the same substrate, peptidoglycan. Second, functional redundancy may phenotypically compensate for the lack of an individual autolysin. Third, many of the autolysins are differentially regulated, and we now report that the σD-dependent autolysins are expressed in only a subpopulation of cells. To complicate matters further, the physiological roles of LytC, LytD, and LytF have traditionally been studied using domesticated lab strains in which the genes that encode these autolysins are not expressed in a majority of the cells (Fig. 1A). To mitigate the problems of heterogeneity, we reinvestigated autolysin function using an undomesticated genetic background in which the autolysins are expressed in a majority of the population. With this approach we were able to identify LytF as the primary autolysin involved in vegetative daughter cell separation and LytC as the primary autolysin required for efficient swarming motility. Consistent with these roles, LytF has been reported to localize to the division septa and LytC localizes uniformly to the cell wall, into which the peritrichous flagella are randomly inserted (45). The specific role of LytD remains unclear.
Autolysins have been implicated in B. subtilis motility, and two models have been invoked to explain the autolysins requirement (38). The first model invokes the role of autolysins in peptidoglycan remodeling to allow flagellum biosynthesis (8). In Salmonella enterica serovar Typhimurium, an autolysin, FlgJ, is mounted on the end of the extending flagellar rod to remodel the peptidoglycan and permit rod penetration through the cell wall during flagellum assembly (30). The B. subtilis genome does not encode an FlgJ homolog, but the flagellum must nonetheless transit the thick gram-positive layer of peptidoglycan. It has been suggested that instead of FlgJ, one or more of the B. subtilis σD-dependent autolysins might create holes in the cell wall to enable flagellum biosynthesis (12). If flagellum assembly requires remodeling of the peptidoglycan, this function is not carried out by the σD-dependent autolysins because a lytC lytD lytF triple mutant was fully proficient for flagellum synthesis.
A second model to explain the motility defect of autolysin mutants invokes the idea that uncoordinated flagellar rotation in cells in a chain would impair force generation (3). Our genetic analysis demonstrated that swarming motility was poorly correlated with cell separation. For example, the lytC lytD double mutant had a minor defect in cell separation, but its swarming was severely impaired (Fig. 2 and 4B). Conversely, the lytF mutant had a severe defect in cell separation but exhibited swarming that was like that of the wild type (Fig. 2 and 4B). Our data suggest that the requirement for the σD-dependent autolysins in motility is related neither to flagellum biosynthesis nor to cell separation. Instead, we propose a third model to explain the motility defect of autolysin mutants in which the autolysins, particularly LytC, are somehow required for proper flagellum function.
Mutations either in the lonA gene, encoding the Lon protease, or in the smiA gene, encoding the previously unannotated 128-amino-acid SmiA protein with an unknown function, restored swarming motility to the lytC lytD double mutant. The lonA and smiA mutations may be either bypass or compensatory suppressors (Fig. 6E). For example, the absence of lytC and lytD may result in uncontrolled inhibition of motility by LonA and SmiA, and thus mutations in lonA and smiA bypass this regulatory block. Alternatively, LonA and SmiA may be motility inhibitors that are independent of the LytC and LytD function, and mutation of LonA and SmiA may enhance motility in general. Thus, mutation of either lonA or smiA would compensate for a reduction in motility in the absence of the autolysins. Consistent with the compensatory model, mutations in either lonA or smiA enhanced the motility of B. subtilis independent of specific autolysin defects. Mutation of either gene abolished the lag period of otherwise wild-type cells (Fig. 6F) and improved the swarming of double mutants with any combination of LytC, LytD, and LytF autolysin mutations (Fig. 6; see Fig. S4 in the supplemental material). The proteolytic target of LonA involved in motility inhibition is not known, but mutation of the Lon protease has also been shown to enhance the swarming motility of Proteus mirabilis and Vibrio parahaemolyticus (7, 40). SmiA has no predicted transmembrane domains and no helix-turn-helix motif, and it is conserved only in close relatives of B. subtilis (see Fig. S1 in the supplemental material). The mechanism of SmiA activity is unknown.
Here we fully explain the basis for the morphological heterogeneity of single motile cells and nonmotile chains observed in exponentially growing populations of B. subtilis. Cells that are ON for σD express late-class flagellar genes, express LytC, and are motile. The same cells that complete flagellar biosynthesis also express LytF that remodels the cell wall and separate from their siblings. Cells that are OFF for σD express neither flagella nor the autolysins and grow in long nonmotile chains. Thus, heterogeneity is the consequence of σD bistable gene expression that coordinately activates motility and chain separation in a subpopulation of cells. We conclude that the single motile cells and nonmotile chains are bona fide developmental states as each morphologically and functionally distinct subpopulation is differentiated by the activity of an alternative sigma factor and the expression of a broad regulon of genes.
Supplementary Material
Acknowledgments
We thank Amy Camp, Natalie Campo, Loralyn Cozy, Rich Losick, Andrew Phillips, and David Rudner for sharing genetic constructs.
This work was supported by National Science Foundation grant MCB-0721187 to D.B.K.
Footnotes
Published ahead of print on 19 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ayusawa, D., Y. Yoneda, K. Yamane, and B. Maruo. 1975. Pleiotropic phenomena in autolytic enzyme content, flagellation, and simultaneous hyperproduction of extracellular alpha-amylase and protease in a Bacillus subtilis mutant. J. Bacteriol. 124459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Yehuda, S., D. Z. Rudner, and R. Losick. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299532-536. [DOI] [PubMed] [Google Scholar]
- 3.Blackman, S. A., T. J. Smith, and S. J. Foster. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 14473-82. [DOI] [PubMed] [Google Scholar]
- 4.Blair, K., L. Turner, J. T. Winkelman, H. C. Berg, and D. B. Kearns. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 3201636-1638. [DOI] [PubMed] [Google Scholar]
- 5.Calvio, C., F. Celandroni, E. Ghelardi, G. Amati, S. Salvetti, F. Ceciliani, A. Galizzi, and S. Senesi. 2005. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J. Bacteriol. 1875356-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L., and J. D. Helmann. 1994. The Bacillus subtilis sigma D-dependent operon encoding the flagellar proteins FliD, FliS, and FliT. J. Bacteriol. 1763093-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemmer, K. M., and P. N. Rather. 2008. The Lon protease regulates swarming motility and virulence gene expression in Proteus mirabilis. J. Med. Microbiol. 57931-937. [DOI] [PubMed] [Google Scholar]
- 8.Dijkstra, A. J., and W. Keck. 1996. Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 1785555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubnau, D., and R. Losick. 2006. Bistability in bacteria. Mol. Microbiol. 61564-572. [DOI] [PubMed] [Google Scholar]
- 10.Fan, D. P., and M. M. Beckman. 1971. Mutant of Bacillus subtilis demonstrating the requirement of lysis for growth. J. Bacteriol. 105629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fein, J. E., and H. J. Rogers. 1976. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J. Bacteriol. 1271427-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fein, J. E. 1979. Possible involvement of bacterial autolysin enzymes in flagellar morphogenesis. J. Bacteriol. 137933-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guérout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic resistance cassettes for Bacillus subtilis. Gene 167335-336. [DOI] [PubMed] [Google Scholar]
- 14.Guérout-Fleury, A.-M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 18057-61. [DOI] [PubMed] [Google Scholar]
- 15.Helmann, J. D., L. M. Márquez, and M. J. Chamberlin. 1988. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J. Bacteriol. 1701568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearns, D. B., F. Chu, R. Rudner, and R. Losick. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 52357-369. [DOI] [PubMed] [Google Scholar]
- 17.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49581-590. [DOI] [PubMed] [Google Scholar]
- 18.Kearns, D. B., and R. Losick. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 193083-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390249-256. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda, A., and J. Sekiguchi. 1991. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J. Bacteriol. 1737304-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 1381949-1961. [DOI] [PubMed] [Google Scholar]
- 22.Le Breton, Y., N. P. Mohapatra, and W. G. Haldenwang. 2006. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 72327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemon, K. P., and A. D. Grossman. 2000. Movement of replicating DNA through a stationary replisome. Mol. Cell 61321-1330. [DOI] [PubMed] [Google Scholar]
- 24.Levano-Garcia, J., S. Verjovski-Almeida, and A. C. R. da Silva. 2005. Mapping transposon insertion sites by touchdown PCR and hybrid degenerate primers. BioTechniques 38225-229. [DOI] [PubMed] [Google Scholar]
- 25.Lominski, I., J. Cameron, and G. Wyllie. 1958. Chaining and unchaining Streptococcus faecalis—a hypothesis of the mechanism of bacterial cell separation. Nature 1811477. [DOI] [PubMed] [Google Scholar]
- 26.Margot, P., C. Mauel, and D. Karamata. 1994. The gene of the N-acetylglucosaminidase, a Bacillus subtilis 168 cell wall hydrolase not involved in vegetative cell autolysis. Mol. Microbiol. 12535-545. [DOI] [PubMed] [Google Scholar]
- 27.Margot, P., M. Pagni, and D. Karamata. 1999. Bacillus subtilis 168 gene lytF encodes a γ-d-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, σD. Microbiol. 14557-65. [DOI] [PubMed] [Google Scholar]
- 28.Márquez, L. M., J. D. Helmann, E. Ferrari, H. M. Parker, G. W. Ordal, and M. J. Chamberlin. 1990. Studies of σD-dependent functions of Bacillus subtilis. J. Bacteriol. 1723435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirel, D. B., and M. J. Chamberlin. 1989. The Bacillus subtilis flagellin gene (hag) is transcribed by the σ28 form of RNA polymerase. J. Bacteriol. 1713095-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nambu, T., T. Minamino, R. M. Macnab, and K. Kutsukake. 1999. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J. Bacteriol. 1811555-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnishi, R., S. Ishikawa, and J. Sekiguchi. 1999. Peptidoglycan hydrolase LytF plays a role in cell separation with CwlF during vegetative growth of Bacillus subtilis. J. Bacteriol. 1813178-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patrick, J. E., and D. B. Kearns. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 701166-1179. [DOI] [PubMed] [Google Scholar]
- 33.Perez, A. R., A. Abanes-De Mello, and K. Pogliano. 2006. Suppression of engulfment defects in Bacillus subtilis by elevated expression of the motility regulon. J. Bacteriol. 1881159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pooley, H. M., and D. Karamata. 1984. Genetic analysis of autolysin-deficient and flagellaless mutants of Bacillus subtilis. J. Bacteriol. 1601123-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prágai, Z., and C. R. Harbwood. 2000. YsxC, a putative GTP-binding protein essential for growth of Bacillus subtilis 168. J. Bacteriol. 1826819-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rashid, M. H., A. Kuroda, and J. Sekiguchi. 1993. Bacillus subtilis mutant deficient in the major autolytic amidase and glucosaminidase is impaired in motility. FEMS Microbiol. Lett. 112135-140. [DOI] [PubMed] [Google Scholar]
- 37.Riethdorf, S., U. Völker, U. Gerth, A. Winkler, S. Engelmann, and M. Hecker. 1994. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J. Bacteriol. 1766518-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146249-262. [DOI] [PubMed] [Google Scholar]
- 39.Smith, T. J., and S. J. Foster. 1995. Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J. Bacteriol. 1773855-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart, B. J., J. L. Enos-Berlarge, and L. L. McCarter. 1997. The lonS gene regulates swarmer cell differentiation of Vibrio parahaemolyticus. J. Bacteriol. 179107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsilibaris, V., G. Maenhaut-Michel, and L. Van Melderen. 2006. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 157701-713. [DOI] [PubMed] [Google Scholar]
- 42.Vollmer, W., B. Joris, P. Charlier, and S. Foster. 2008. Bacterial peptidoglycan (murien) hydrolases. FEMS Microbiol. Rev. 32259-286. [DOI] [PubMed] [Google Scholar]
- 43.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12259-265. [DOI] [PubMed] [Google Scholar]
- 44.Wicker-Planquart, C., A.-E. Foucher, M. Louwagie, R. A. Britton, and J.-M. Jault. 2008. Interactions of an essential Bacillus subtilis GTPase, YsxC, with ribosomes. J. Bacteriol. 190681-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto, H., S. Kurosawa, and J. Sekiguchi. 2003. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J. Bacteriol. 1856666-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasbin, R. E., and F. E. Young. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J. Virol. 141343-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeigler, D. R., Z. Prágai, S. Rodriguez, B. Chevreux, A. Muffler, T. Albert, R. Bai, M. Wyss, and J. B. Perkins. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 1906983-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.