Abstract
Bacteria and fungi are thought to degrade cellulose through the activity of either a complexed or a noncomplexed cellulolytic system composed of endoglucanases and cellobiohydrolases. The marine bacterium Saccharophagus degradans 2-40 produces a multicomponent cellulolytic system that is unusual in its abundance of GH5-containing endoglucanases. Secreted enzymes of this bacterium release high levels of cellobiose from cellulosic materials. Through cloning and purification, the predicted biochemical activities of the one annotated cellobiohydrolase Cel6A and the GH5-containing endoglucanases were evaluated. Cel6A was shown to be a classic endoglucanase, but Cel5H showed significantly higher activity on several types of cellulose, was the highest expressed, and processively released cellobiose from cellulosic substrates. Cel5G, Cel5H, and Cel5J were found to be members of a separate phylogenetic clade and were all shown to be processive. The processive endoglucanases are functionally equivalent to the endoglucanases and cellobiohydrolases required for other cellulolytic systems, thus providing a cellobiohydrolase-independent mechanism for this bacterium to convert cellulose to glucose.
The microbial degradation of cellulose is of interest due to applications in the sugar-dependent production of alternative biofuels (25). There are well-characterized cellulolytic systems of bacteria and fungi that employ multiple endo-acting glucanases and exo-acting cellobiohydrolases in the degradation of cellulose (12). For example, the noncomplexed cellulase system of the wood soft rot fungus Hypocrea jecorina (anamorph Trichoderma reesei), the source for most commercially available cellulase preparations, produces up to eight secreted β-1,4-endoglucanases (Cel5A, Cel5B, Cel7B, Cel12A, Cel45A, Cel61A, Cel61B, and Cel61C), two cellobiohydrolases (Cel6A and Cel7A), and several β-glucosidases (e.g., Bgl3A) (21). Cellobiohydrolases are critical to the function of these systems, as, for example, Cel7A comprises in excess of 50% of the cellulases secreted by this organism (11). Another well-characterized noncomplexed cellulase system is found in Thermobifida fusca, a filamentous soil bacterium that is a major degrader of organic material found in compost piles (32). This bacterium also secretes several endoglucanases and end-specific cellobiohydrolases to degrade cellulose (32). An alternative mechanism for degradation of cellulose is found in microorganisms producing complexed cellulolytic systems, such as those found in cellulolytic clostridia. In these microorganisms, several β-1,4-endoglucanases and cellobiohydrolases assemble on surface-associated scaffoldin polypeptides to form cellulose-degrading multiprotein complexes known as cellulosomes (2, 6). The unifying theme in both complexed and noncomplexed systems is the importance of cellobiohydrolases in converting cellulose and cellodextrins to soluble cellobiose.
Recently, a complete cellulolytic system was reported to occur in the marine bacterium Saccharophagus degradans 2-40 (28, 31). This bacterium is capable of growth on both crystalline and noncrystalline celluloses as sole carbon sources and produces multiple glucanases that can be detected in zymograms of cell lysates (28). The genome sequence of this bacterium predicts that the cellulolytic system of this bacterium consists of 10 GH5-containing β-1,4-endoglucanases (Cel5A, Cel5B, Cel5C, Cel5D, Cel5E, Cel5F, Cel5G, Cel5H, Cel5I, and Cel5J), two GH9 β-1,4-endoglucanases (Cel9A and Cel9B), one cellobiohydrolase (Cel6A), five β-glucosidases (Bgl1A, Bgl1B, Bgl3C, Ced3A, and Ced3B), and a cellobiose phosphorylase (Cep94A) (28, 31). The apparent absence of a homolog to a scaffoldin in the genome sequence and to dockerin-like domains in the proposed glucanases suggests that this bacterium produces a noncomplexed cellulolytic system. Two unusual features of this cellulolytic system are the large number of GH5 endoglucanases and the presence of only one annotated cellobiohydrolase, Cel6A (28, 31). The apparent deficiency of cellobiohydrolases in this system raised the question as to the mechanism by which this bacterium degrades cellulose.
To understand the mechanism for degradation of cellulose, the biochemical activities for the predicted cellobiohydrolase Cel6A and each of the GH5 glucanases predicted for the S. degradans cellulolytic system were evaluated. Cel6A exhibited properties of a classic endoglucanase, but three of the originally annotated endoglucanases, Cel5G, Cel5H, and Cel5J, were shown to be processive, forming cellobiose as the end product. Processive endoglucanases substitute for cellobiohydrolases in this system to play a major role in the degradation of cellulose.
MATERIALS AND METHODS
Bacterial growth media and conditions.
Saccharophagus degradans 2-40T (ATCC 43961) was grown at 30°C in minimal medium containing (per liter) 2.3% Instant Ocean (Aquarium Systems, Mentor, OH), 0.05% yeast extract, 0.5% (wt/vol) ammonium chloride, and 16.7 mM Tris-HCl, pH 8.6, supplemented with 0.2% carbon source by use of standard protocols. Escherichia coli DH5α (Invitrogen, Frederick, MD) and Rosetta2 (DE3) (Novagen, Madison, WI) strains were grown at 37°C in Luria-Bertani broth or agar supplemented with the appropriate antibiotics. Antibiotics were added to media at the following concentrations (in μg/ml): chloramphenicol, 30; and kanamycin, 50.
Bioinformatic analyses.
Similarities to the S. degradans 2-40 sequences were based on local alignments obtained through the BLAST program (1). Domain architectures were ascertained using SMART (http://smart.embl-heidelberg.de/) (27). Multiple sequence alignments were created using ClustalX 1.81 (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/) (29) and manually adjusted where appropriate. The neighbor-joining algorithm was executed via ClustalX 1.81, using the “Exclude Positions with Gaps” and “Correct for Multiple Substitutions” options. Statistical support for tree topology was provided by 1,000 bootstrap trials.
Molecular cloning of S. degradans GH5 glucanases.
S. degradans genomic DNA was isolated by using a commercial genomic DNA purification kit (Promega, Madison, WI). Sequences of annotated GH5 glucanase-encoding genes were extracted from the S. degradans genome sequence (http://maple.lsd.ornl.gov/), and tailed primers were designed to amplify the coding sequences for each gene by PCR (see Table S1 in the supplemental material). Amplified fragments were ligated as BamHI-EcoRI (Cel5A, Cel5B, Cel5C, Cel5D, Cel5F, Cel5G, Cel5I, and Cel5J), BamHI-HindIII (Cel5E), or BamHI-XhoI (Cel5H) fragments into pET28b (Novagen) to create in-frame amino- and carboxy-terminal six-His fusions and transformed into E. coli DH5α. After confirmation of the correct construct in Kanr transformants by nested PCR and/or sequencing, the resulting plasmid constructs were isolated and transformed into E. coli Rosetta2 (DE3) (Novogen) for expression.
Expression of S. degradans glucanases.
S. degradans was cultured on glucose to give an optical density at 600 nm (OD600) of 0.3 to 0.4, harvested, and transferred to the same volume of medium containing Avicel. After 10 h, the RNA was isolated using RNA Protect Bacteria Reagent (Qiagen) and an RNeasy minikit (Qiagen). The cDNA was synthesized using a QiantiTect reverse transcription kit. The 120- to 200-bp fragments of each indicated gene or the genes for guanylate kinase and dihydrofolate reductase were amplified using a SYBR green master mix kit (Roche) and a LightCycler 480 system (Roche). The primers used are given in Table S2 in the supplemental material. Transcript levels were estimated using instrument software and normalized using the expression of guanylate kinase.
Purification of glucanases.
Twenty milliliters of overnight E. coli Rosetta2 (DE3) (pHZ-Cel) culture was inoculated into 500 ml Luria-Bertani broth and grown at 37°C for 2 h. Expression of cloned genes was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to give a final concentration of 0.1 mM when the OD600 approached 0.6. The culture was then incubated overnight at 15°C with mild shaking. Cells from induced cultures were harvested and resuspended in 50 mM sodium phosphate, 300 mM sodium chloride, 10 mM imidazole, pH 8.0, and 0.5 mM phenylmethylsulfonyl fluoride. Lysozyme was added to give a concentration of 1 mg/ml and the cell suspension incubated on ice for 30 min. Lysis of cells was completed using five 30-s cycles in a bead beater (Biospec Products). The lysate was clarified by centrifugation at 10,000 RPM for 20 min. The expressed proteins in cleared lysates were purified using chelated Ni-nitrilotriacetic acid chromatography according to the manufacturer's recommendations (Qiagen). The protein concentration was determined by a Pierce microBCA protein assay reagent kit (23235), using bovine serum albumin as the standard.
Zymograms.
A modification of the procedures of Taylor et al. was used to prepare zymograms (28). Samples and gels were prepared as in standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with the indicated substrate incorporated directly into the resolving gel at a final concentration of 0.1% (wt/vol) for barley β-glucan (medium viscosity; Megazyme) or hydroxyethyl (HE)-cellulose. After electrophoretic fractionation of the proteins, gels were washed twice in distilled water and incubated in 30 ml of refolding buffer {20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer [pH 6.8], 2.5% Triton X-100, 2 mM dithiothreitol, 2.5 mM CaCl2} for 1 h at 20°C and then held overnight in fresh refolding buffer at 4°C. The gels were washed twice in 20 mM PIPES buffer, pH 6.8, and incubated for 12 h at 37°C in fresh PIPES buffer. Residual substrate was visualized by staining with 0.25% Congo red.
Enzyme assays.
Most assays were performed at pH 6.5 and 50°C with a reaction mixture containing 1% Instant Ocean, 20 mM PIPES buffer, and 0.01 to 1.0 nmol purified enzyme for soluble substrates and 0.1 to 2.0 nmol purified enzyme for insoluble substrates. For alternative pH conditions, 20 mM MES [2-(N-morpholino)ethanesulfonic acid] for pH 3 to 6, 20 mM PIPES (1,4-piperazinediethanesulfonic acid) for pH 6 to 7, or 20 mM Tris for pH 7 to 8.5 was employed. The ionic strength of the reaction mixture was adjusted by the addition of 0 to 10% (wt/vol) Instant Ocean. All assays were performed in triplicate, and results were reported as the mean ± standard deviation.
The carboxymethyl cellulose (CMC) assay was performed with 1% substrate in a total volume of 0.4 ml for a 15-min reaction time. Phosphoric acid-swollen cellulose (PASC) was prepared as described previously (37). The PASC (1 mg) assay was performed with a total volume of 0.15 ml for 30 min. The dinitrosalicylic acid assay (9) was used to detect product formation in these reactions. Digestion of Avicel (1 mg) or Whatman no. 1 filter paper (3 mg) was assayed with a total volume of 0.15 ml for 2 h. The reaction was stopped by incubation for 3 min at 95°C and the substrate separated by centrifugation at 10,000 rpm. The products of filter paper or Avicel digestions were then digested with 0.45 nanomoles of β-glucosidase S. degradans Bgl1A in a total volume of 0.2 ml at 50°C for 1 h. After 1 h at 50°C, the β-glucosidase was inactivated by incubation for 3 min at 95°C. Glucose oxidase (Sigma GAGO-20) was added to give a volume of 0.6 ml and incubated at 37°C for 30 min. Sulfuric acid (12 N; 0.4 ml) was added, and the glucose concentration was measured at OD540 in comparison with a glucose standard curve (22). Release of cellobiose was calculated at 50% the rate of glucose accumulation.
pNP-cellobioside activity was measured using 0.1 ml of 125 mM pNP-cellobioside in a total volume of 0.4 ml at 50°C for 15 min. After incubation, the OD400 was used to calculate the rate of nitrophenol release. Thin-layer chromatography was used to determine products.
Assessment of synergy.
Evaluation of synergy between enzymes employed the filter paper assay as described above. T. fusca Cel6A, T. fusca Cel9B, and T. fusca Cel6B were used as reference enzymes of known activity (gifts from David Wilson, Cornell University). Each assay utilized 1.0 nmol of S. degradans enzyme and 0.1 nmol of the indicated T. fusca enzyme. The reaction conditions were as described above for the filter paper assay.
Viscosity measurements.
Viscosity was monitored using a cross-arm viscometer (ASTM D455 and D2170). T. fusca Cel6B (0.01 nmol), S. degradans Cel5H (0.01 nmol), or T. fusca Cel9B (0.001 nmol) was added to 2.0 ml 1% CMC in assay buffer. The viscosity of the reaction mixture was measured periodically between 0.5 and 20 min.
Thin-layer chromatography.
One-microliter samples were spotted onto Fisher (5729-6) silica gel 60 plates and air dried. Chromatograms were developed using nitromethane, 1-propanol, and water (2:5:1.5) (vol/vol/vol) (17). Two ascents of the solvent were used to ensure high resolution. The plate was dipped in 5% (vol/vol) sulfuric acid in methanol and heated to 140°C for 5 min to visualize resolved products.
Processivity.
The processivity was evaluated by measuring the ratio of soluble to insoluble reducing sugar by using a modification of the procedure of Zhang et al. (36). The substrate was filter paper. Reducing sugar in the soluble fractions was measured using the glucose oxidase method described above. After conversion to halve the rate, results were reported in μmol amounts of cellobiose released. The insoluble reducing sugar was determined using a modified 2,2′-bicinchoninate assay as described by Doner and Irwin (7). At the end of the assay period, the filter paper was washed with 6 M guanidine hydrochloride to remove bound protein. The filter paper disc was then washed four times with assay buffer and water (36). The retained reducing sugar was measured using the Pierce microBCA reagent kit, using glucose standards. The processivity was determined using 0.1 to 1 nmol of the indicated S. degradans glucanase and 0.1 nmol of T. fusca Cel6A and T. fusca Cel9A.
RESULTS
Cellulose degradation by S. degradans.
The genome of S. degradans is annotated to produce a multicomponent cellulolytic system (28, 31), but the mechanism of cellulose degradation was not clear. To estimate the products formed as a result of the activity of the secreted components of the S. degradans cellulolytic system, S. degradans was grown on Avicel to induce expression of glucanases and the degradation products were released from cellulose by the activity of the secreted glucanases determined. Irrespective of the cellulosic substrate or reaction time, cellobiose was the primary product released by the glucanase activity of S. degradans culture filtrates, with a smaller amount of cellotriose also observed (see Fig. S1 in the supplemental material). Larger cellodextrin products were not apparent. This suggests that the cellulolytic system of S. degradans has an efficient mechanism for release of cellobiose during degradation of cellulose.
Activity of Cel6A.
Cellobiose can be formed by the processive exoglucanase activity of a cellobiohydrolase. The only annotated cellobiohydrolase encoded by the S. degradans genome is Cel6A (28). The cel6A gene was amplified by PCR and cloned into pET28b. Only the construct expressing an independently translated polypeptide lacking His tags exhibited activity in E. coli Rosetta2 (DE3) transformants, thus limiting purification. After electrophoretic fractionation, the expressed enzyme migrated at the expected mass and degraded glucan, as indicated by a zone of clearing in Congo red-stained zymograms (data not shown). Cell lysates containing Cel6A released reducing sugar from CMC, consistent with the annotation as a glucanase. The processivity ratio (ratio of soluble to insoluble products) of Cel6A activity (1.02 ± 0.11) and the synergisms had values expected for an endoglucanase. The degrees of synergy for S. degradans Cel6A plus T. fusca Cel6A (endoglucanase) and S. degradans Cel6A plus T. fusca Cel6B (exoglucanase) were 0.64 and 1.58, respectively. The independent yields of glucose for S. degradans Cel6A, T. fusca Cel6A, and T. fusca Cel6B were 0.87 ± 0.15 μg, 7.57 ± 0.44 μg, and 0.34 ± 0.02 μg, respectively. The activity of S. degradans Cel6A was corrected for the basal activity of E. coli Rosetta 2 (DE3) lysates. The combined yields of glucose for S. degradans Cel6A plus T. fusca Cel6A cellulases and S. degradans Cel6A plus T. fusca Cel6B cellulases were 5.43 ± 0.27 μg and 1.91 ± 0.26 μg, respectively. Furthermore, the activity reduced the viscosity of CMC. S. degradans Cel6A caused a 53% decrease in viscosity. In parallel experiments, the exoglucanase T. fusca Cel6B caused a 30% increase in viscosity, whereas the endoglucanase T. fusca Cel9B caused an 84% decrease in viscosity. The activity of Cel6A is typical of a classic endoglucanase, indicating that other enzymes must be contributing to the formation of cellobiose.
Expression of glucanases.
To estimate their potential contribution to cellobiose formation, the transcription of each gene annotated to encode a glucanase was monitored by quantitative reverse transcription-PCR. The most highly expressed gene during exponential growth on cellulose appeared to be cel5H (Table 1). With the exceptions of cel5F and cel5I, transcript levels for cel5H were at least 10-fold higher than the genes for the other glucanases. For example, the transcript level for cel6A was only 2.42% that for cel5H. Thus, the high apparent expression level of Cel5H indicates that this enzyme could be a major contributor to the S. degradans cellulolytic system.
TABLE 1.
Expression of S. degradans cellulolytic system genes
| Genea | Gene no. | Transcript levelb | Relative expression level (%)c |
|---|---|---|---|
| cel5A | 3003 | 4 | 1 |
| cel5B | 2490 | 94 | 10 |
| cel5C | 0325 | 1 | 0 |
| cel5D | 2636 | 1 | 0 |
| cel5E | 2929 | 55 | 6 |
| cel5F | 1572 | 440 | 46 |
| cel5G | 3239 | 1 | 0 |
| cel5H | 3237 | 955 | 100 |
| cel5I | 3420 | 182 | 19 |
| cel5J | 2494 | 113 | 12 |
| cel6A | 2272 | 23 | 2 |
| cel9A | 0636 | 82 | 9 |
| cel9B | 0649 | 39 | 4 |
The gene corresponding to the respective S. degradans endoglucanase described by Taylor et al. (28), with its gene number from the current annotation.
The transcript level after 10 h of culture on Avicel was estimated using quantitative reverse transcription-PCR as described in Materials and Methods. The expression level of the housekeeping gene for guanylate kinase (gene 3695) was used for normalization.
Percent transcript level relative to that of cel5H.
Activity of Cel5H.
For determination of the biochemical activity of Cel5H, the gene was amplified from the S. degradans 2-40 genome by PCR and cloned into the T7 expression system carried by pET28b to create N- and C-terminal His tag fusions. After transformation into E. coli Rosetta2 (DE3), expression was induced by IPTG. The expressed Cel5H protein migrated at the expected mass during SDS-PAGE and hydrolyzed β-glucan and HE-cellulose in zymograms consistent with its annotation (Fig. 1). This enzyme appeared to be very active as lysates of E. coli Rosetta2 (DE3) transformants expressing Cel5H had to be diluted to picomolar concentrations before a well-defined zone of activity typical of an individual polypeptide could be resolved. After purification from Rosetta2 (DE3) transformants to near homogeneity by use of nickel-nitrilotriacetic acid affinity chromatography, the glucanase activity of the purified Cel5H protein exhibited a pH optimum of 6.5 and retained more than 84% of its activity in the range between pH 6.0 and 7.0. Activity increased linearly up to 50°C but decreased at higher temperatures. Addition of 1% Instant Ocean enhanced the activity of the enzyme 2.5-fold. The specific activity of purified Cel5H was highest on PASC and CMC, but statistically significant activity was retained on filter paper and Avicel relative to control levels (Table 2). The enzyme released cellobiose from pNP-cellobioside (data not shown).
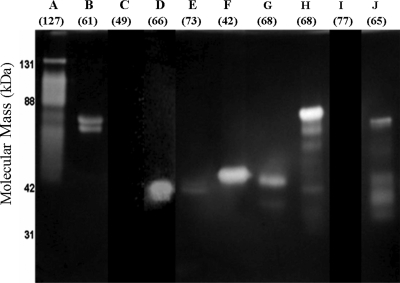
FIG. 1.
Zymogram of S. degradans GH5 glucanase activities. After fractionation by SDS-PAGE and renaturing, retained barley glucan substrate was stained using Congo red after a 12-h digestion. Zones of clearing represent glucanase activity. Similar results were obtained for zymograms containing HE-cellulose. The expected molecular mass of each polypeptide as cloned is shown in parentheses under each lane label. Letters A through J refer to the Cel5 enzyme. With the exceptions of Cel5E and Cel5J, the expression of each protein in the Rosetta 2 (DE3) host appeared to be equivalent in Coomassie blue-stained gels, as the peak areas of each expressed protein were similar. To resolve the activity of polypeptides in the zymograms, the samples were diluted. The amount of protein in each well was equivalent to that in the original cell culture, as follows: Cel5A, 100 nl; Cel5B, 1,000 nl; Cel5C, 7,000 nl; Cel5D, 7,000 nl; Cel5E, 1,000 nl; Cel5F, 100 nl; Cel5G, 1 nl; Cel5H, 10 nl; Cel5I, 7,000 nl; and Cel5J, 10 nl. Precision Plus molecular mass markers (Bio-Rad, Hercules, CA) were used as molecular mass markers.
TABLE 2.
Specific activity of S. degradans Cel5H on cellulosic substratesa
| Enzyme | Sp act on indicated substrate (μmol reducing sugar/minute/μmol enzyme)
|
|||
|---|---|---|---|---|
| CMC | PASC | Filter paper | Avicel | |
| Cel5H | 643 ± 148 | 792 ± 119 | 2.21 ± 0.1 | 1.45 ± 0.1 |
| Cel5H′ | 500 ± 10 | 545 ± 82 | 0.92 ± 0.1 | 0.66 ± 0.1 |
The amounts of reducing sugar released from CMC and swollen cellulose relative to a cellobiose standard curve were measured using a DNS assay, whereas the amounts of cellobiose released from filter paper and Avicel were measured using the glucose oxidase method after treatment with an excess of β-glucosidase. Means ± standard deviations are reported. For measurements of protein, BSA was used as the reference. The approximate percents crystallinity for the substrates (38) were as follows: CMC, 0%; PASC, 10 to 20%; filter paper, 50%; and Avicel, 70%.
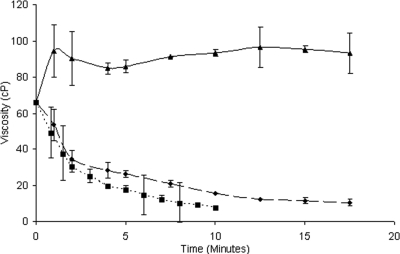
To evaluate whether Cel5H is an endoglucanase like other GH5 enzymes (20), its effect on the viscosity of CMC solutions was monitored. S. degradans Cel5H rapidly decreased the viscosity of CMC similarly to the endoglucanase T. fusca Cel9B (a gift from D. Wilson). In contrast, the exo-acting T. fusca Cel6B (a gift from D. Wilson) increased the viscosity of CMC as expected (Fig. 2).
FIG. 2.
Effect of endo- and exoglucanases on CMC viscosity. S. degradans Cel5H (♦), T. fusca Cel9B (▪), and T. fusca Cel6B (▴) were added to a 1% CMC solution. The viscosity was measured as a function of time as described in Materials and Methods. The viscosity is given in centipoise (cP) units.
Synergistic interactions between glucanases also provide an indication of the activity of the enzymes. S. degradans Cel5H acted synergistically with the exoglucanase T. fusca Cel6B, producing at least 30% more product when acting together than each individually (Table 3). In contrast, antisynergism was observed with the endoglucanase T. fusca Cel9B, with combined activities 15 to 25% lower than theoretical levels, as seen previously in the interaction of classical endoglucanases (16).
TABLE 3.
Synergy of S. degradans and T. fusca glucanases
| Enzyme | Activity | Amt (nmol) | Yield of glucose (μg)
|
|||
|---|---|---|---|---|---|---|
| Independenta | Theoreticalb | Observedc | DoSd | |||
| S. degradans Cel5H plus: | 1 | 12.6 ± 0.9 | ||||
| T. fusca Cel9B | Endoglucanase | 0.1 | 5.9 ± 0.5 | 18.5 | 14.3 ± 0.4 | 0.77 |
| T. fusca Cel6B | Exoglucanase | 0.1 | 2.6 ± 0.0 | 15.3 | 20.1 ± 0.9 | 1.31 |
| S. degradans Cel5G plus: | 1 | 10.5 ± 1.3 | ||||
| T. fusca Cel9B | Endoglucanase | 0.1 | 5.9 ± 0.5 | 16.4 | 14.1e | 0.86 |
| T. fusca Cel6B | Exoglucanase | 0.1 | 2.6 ± 0.0 | 13.2 | 20.4 ± 0.9 | 1.55 |
The yields were measured after 2 hours as described for the filter paper assay. Means ± standard deviations of results from three trials are rounded to tenths.
The sum of glucose produced by the indicated S. degradans and T. fusca glucanases during independent digestions.
The yield obtained under the same conditions as the independent reactions in a digestion involving Cel5H or Cel5G together with T. fusca Cel9B or Cel6B.
DoS, degree of synergy (the product formed when both enzymes are present in the reaction mixture, divided by the sum of the yields from equivalent amounts of the same enzymes under identical conditions).
Representative of a single trial, due to a lack of T. fusca endoglucanase.
Cellobiose was the primary product released from PASC, filter paper, and Avicel by the activity of purified Cel5H in heat-stopped reactions, irrespective of the length of digestion (Fig. 3). Products larger than cellotriose were not apparent during 45-s digestions or thereafter. The amount of Cel5H used did not affect this product distribution.
FIG. 3.
Products formed by S. degradans Cel5H activity. Controls: E, enzyme alone; A, Avicel alone; FP, filter paper alone; and SC, PASC alone. Products formed by the activity of purified Cel5H on the indicated substrate (A, Avicel; FP, filter paper; and SC, PASC) are shown. For both the control and the digestion trials, reaction mixtures were incubated at 50°C for 16 h. Two-microliter aliquots were spotted on Silica Gel G plates and resolved using a nitromethane-propanol-water solvent system as described in Materials and Methods. The markers G1 to G4 represent the migration of glucose, cellobiose, cellotriose, and cellotetraose, respectively. The time courses of products released by the activity of S. degradans Cel5H on PASC are shown. The reaction conditions were as described in Materials and Methods and the products resolved as described above. Time is indicated in minutes.
Cel5H is processive.
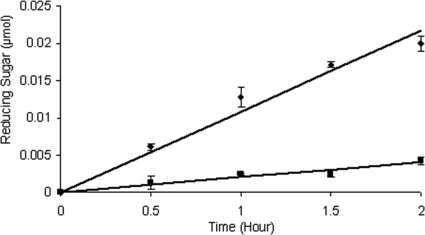
The production of cellobiose by Cel5H activity could be an indication of processivity. To evaluate the processivity of Cel5H, the ratio of soluble to insoluble products was measured and compared to those for T. fusca Cel9A (processive endoglucanase; a gift from D. Wilson) and T. fusca Cel6A (classical endoglucanase) (32). As with the processive T. fusca Cel9A, and in contrast to the classical endoglucanase T. fusca Cel6A (35), 82% of the products formed by the activity of the S. degradans Cel5H protein were soluble (Table 4). For both T. fusca Cel9A and S. degradans Cel5H, the rates of soluble product formation exceeded those of insoluble products throughout a 2-hour time course (Fig. 4).
TABLE 4.
Processivity of S. degradans Cel5H
| Enzymea | Sp actb | Processive ratioc | % Soluble reducing sugard | % Insoluble reducing sugard |
|---|---|---|---|---|
| T. fusca Cel9A (processive endoglucanase) | 0.70 ± 0.01 | 4.72 ± 0.43 | 82.5 | 17.5 |
| T. fusca Cel6A (endoglucanase) | 1.33 ± 0.14 | 2.55 ± 0.28 | 71.9 | 28.1 |
| S. degradans Cel5H | 1.66 ± 0.07 | 4.26 ± 0.71 | 81.4 | 18.6 |
| S. degradans Cel5H′ | 0.92 ± 0.01 | 4.42 ± 1.07 | 81.1 | 18.9 |
A total of 0.1 nmol of each endoglucanase was used in each 2-h digestion of filter paper.
The specific activity is reported as μmol reducing sugar/minute/μmol enzyme.
The processive ratio is defined as μmol soluble reducing sugar (cellobiose standard) divided by μmol insoluble sugar (glucose standard), as described in Materials and Methods.
The mass fractions of soluble and insoluble products, given as percentages.
FIG. 4.
Processivity of Cel5H activity. Purified Cel5H was incubated with filter paper for the indicated times, and products formed as reducing sugar were determined as described in Materials and Methods. Diamonds (top line) indicate soluble reducing sugar detected (μmol cellobiose). Squares (bottom line) indicate insoluble reducing sugar (μmol glucose). Linear trend lines were calculated using y = mx. For release of soluble reducing sugar, m is 0.0108, with a goodness of fit R2 value of 0.97. For formation of insoluble reducing sugar, m is 0.0020 and R2 is 0.94.
The contribution of the resident CBM6 module of Cel5H to its activity and processivity was evaluated by comparing the activity of the full-length polypeptide to that of a truncated derivative consisting of the GH5 catalytic domain constructed by specific amplification (Cel5H′). For these experiments, the GH5 domain of Cel5H was amplified by PCR and ligated into pET28b as before. Cel5H′ was purified from Rossetta2 (DE3) transformants. The specific activity of Cel5H′ was 69% that of Cel5H on amorphous PASC and 78% on soluble CMC. The ratio of soluble to insoluble products, however, remained indistinguishable from that of Cel5H (Table ). Thus, the data suggest that the processivity of the enzyme resides with the catalytic domain.
Phylogenetic analysis of the S. degradans GH5 domains.
The processivity of a GH5 endoglucanase was unexpected. To understand the relationships between Cel5H and the other GH5 endoglucanases, phylogenetic relationships were determined by nearest neighbor joining. Significant differences were apparent in the structural organization of S. degradans GH5 glucanases relative to those of other GH5 endoglucanases that precluded the use of full-length polypeptides in the phylogenetic analyses. Instead, the GH5 domain of each endoglucanase, together with that of its closest homolog in the nonredundent database, was used in the analyses. Two distinct clades of GH5 endoglucanases were apparent (Fig. 5). The majority of the endoglucanases carry a GH5 domain near the carboxy terminus of the host polypeptide and segregate into a single clade with strong bootstrap support. The first clade included Cel5AN, Cel5B, Cel5C, Cel5D, Cel5E, CelF, and Cel5I as well as their homologs. The second clade comprised Cel5AC, Cel5G, Cel5H, and Cel5J. Most of the enzymes associated with the second clade carry a GH5 domain near the amino terminus of the polypeptide and a CBM6 module near the carboxy terminus. Of particular interest were Cel5G and Cel5H. Both enzymes exhibited strong similarities in sequence and domain organization, including the seven conserved catalytic residues of the GH5 catalytic domain (8, 30), differing only in the length and sequence of the linker region. The distinct domain organization and the phylogenetic segregation into a separate clade opened the possibility that the enzymes of the second clade represent a distinct subclass of GH5 endoglucanases.
FIG. 5.
Phylogenetic analysis of S. degradans GH5 domains and their processivity on filter paper. The sequences of the GH5 domains found in the S. degradans glucanases identified by Taylor et al. (28), together with the closest homologs identified in another organism by BLAST, were extracted and subjected to nearest-neighbor analysis as described in Materials and Methods. The resulting phylogenetic tree and the domain organization of each enzyme as predicted by the SMART algorithm are shown. The larger gray-filled boxes represent the location of the GH5 domain in each polypeptide. The red triangles denote the location of carbohydrate binding modules. The processivities of the indicated enzymes were determined as described for Table 4 and are shown in the embedded tables. The enzymes included in the upper-right embedded table have processivity values typical of classic endoglucanases, whereas the enzymes included in the lower-left embedded table have processivity values in excess of 4. The GenBank accession numbers for the genes, with the closest homologs shown in parentheses, are as follows: for the Cel5A-N gene, YP_435061 (endoglucanase [Hahella chejuensis KCTC 2396]); for the Cel5A-C gene, YP_528706 (Sde_3237 and Cel5H); for the Cel5B gene, ZP_00510594 (glycoside hydrolase family 5, Clostridium cellulosome enzyme dockerin type I, and carbohydrate binding domain family 11 [Clostridium thermocellum ATCC 27405]); for the Cel5C gene, ZP_01246425 (glycoside hydrolase family 5 [Flavobacterium johnsoniae UW101]); for the Cel5D gene, ZP_01115721 (endoglucanase family 5 [Reinekea sp. strain MED297]); for the Cel5E gene, ABD81750 (Sde_2490 and Cel5B); for the Cel5F gene, ABA02176 (Cellvibrio japonicus Ueda107); for the Cel5G gene, YP_528706 (Sde_3237 and Cel5H); for the Cel5H gene, YP_528708 (Sde_3239 and Cel5G); for the Cel5I gene, ZP_01113981 (endo-1,4-beta-glucanase [Reinekea sp. strain MED297]); and for the Cel5J gene, YP_435061 (endoglucanase [Hahella chejuensis KCTC 2396]).
Purification and properties of other GH5 endoglucanases.
As before, each gene for the other GH5 glucanases of S. degradans was amplified by PCR and ligated into pET28 for expression in E. coli Rosetta2 (DE3). Most of the expressed GH5 enzymes exhibited endoglucanase activity in β-glucan and HE-cellulose zymograms, consistent with their annotation as glucanases. Cel5B, Cel5D, Cel5F, and Cel5H were comparatively stable, with a single full-length polypeptide of the expected size having glucanase activity (Fig. 1). In contrast, Cel5A, Cel5E, Cel5G, and Cel5J were unstable; either they produced multiple degradation products with activity or the active fragment migrated at a mass lower than expected. Full-length polypeptides as well as the fragments of Cel5A, Cel5D, Cel5G, and Cel5J sufficient to carry the catalytic domain retained glucanase activity (see Fig. 5). None of the fragments derived from Cel5C or Cel5I exhibited evidence of glucanase activity on any substrate.
Each polypeptide was purified to near homogeneity from Rosetta2 (DE3) transformants as previously either as the full-length polypeptide (Cel5B, Cel5E, and Cel5F) or as an apparent catalytic domain (Cel5D, Cel5G, and Cel5J) upon the basis of the retention of activity and size correspondence with the structural predictions of each polypeptide (28, 31) (Fig. 5). The glucanase activity of the expressed enzymes had pH optima near 6.5, did not require salts, and functioned up to 50°C. In contrast to fungal endoglucanases (18), Cel5B, Cel5D, Cel5E, Cel5F, and Cel5G retained at least 60% of the optimal activity at pH 8.0. The glucanase activity was inversely proportional to the crystallinity of the cellulose substrate, with the highest activity observed on CMC (Table 5). Although Cel5F is expressed at 46% that of Cel5H, its specific activity on the tested substrates was significantly lower than that of Cel5H on the tested substrates, suggesting that this enzyme makes a minor contribution to the degradation of cellulose.
TABLE 5.
Specific activities of the S. degradans GH5 endoglucanasesa
| Enzyme | Sp act on indicated substrate (μmol reducing sugar/min/mg protein)
|
|||
|---|---|---|---|---|
| CMC | PASC | Filter paper | Avicel | |
| Cel5B | 2.23 ± 0.36 | 0.07 ± 0.02 | (1.84 ± 0.01) × 10−3 | (1.44 ± 0.1) × 10−3 |
| Cel5D | (8.59 ± 2.23) × 10−4 | (1.13 ± 0.06) × 10−4 | (2.68 ± 0.75) × 10−5 | ND |
| Cel5E | 0.05 ± 0.02 | (5.96 ± 0.05) × 10−3 | (4.01 ± 0.76) × 10−6 | ND |
| Cel5F | 0.37 ± 0.08 | 0.18 ± 0.04 | (6.60 ± 0.46) × 10−4 | (6.26 ± 0.75) × 10−4 |
| Cel5G | 4.62 ± 0.60 | 11.70 ± 1.75 | (5.12 ± 0.20) × 10−3 | (4.88 ± 0.44) × 10−3 |
| Cel5H | 9.61 ± 2.21 | 11.80 ± 1.77 | (3.29 ± 0.16) × 10−2 | (2.18 ± 0.15) × 10−2 |
| Cel5J | 6.97 ± 0.56 | 4.06 ± 0.61 | (6.67 ± 0.40) × 10−3 | (2.18 ± 0.04) × 10−3 |
The amounts of reducing sugar released from CMC and swollen cellulose were measured using a DNS assay and a cellobiose standard curve. For activities on filter paper and Avicel, the values for released cellobiose were converted to those for glucose by using an excess of β-glucosidase, and the activities are calculated at 50% the rate of glucose release as measured using the glucose oxidase method. For measurements of protein, BSA was used as the reference. The approximate percents crystallinity for the substrates (38) were as follows: for CMC, 0%; for PASC, 10 to 20%; for filter paper, 50%; and for Avicel, 70%. ND, activity not detected in a 15-h digestion period.
The activity of Cel5B, Cel5D, Cel5E, and Cel5F formed products typical of classical endoglucanases, with ratios of soluble to insoluble products similar to those for other endoglucanases. The processivity ratios for Cel5G and Cel5J, however, were all found to be significantly greater than 4 (Fig. 5), and each degraded filter paper, Avicel, and pNP-cellobioside like Cel5H. Interestingly, the activity of these processive endoglucanases was not dependent upon the activity of classic endoglucanases. For example, Cel5H was not synergistic with Cel5D, Cel5F, and Cel5G at three different molar ratios (1:4, 1:1, and 4:1) (see Table S3 in the supplemental material), indicating that the enzyme does not recognize the free nonreducing ends of cellulose polymers like cellobiohydrolases.
DISCUSSION
The noncomplexed and complexed cellulolytic systems of microorganisms generally rely upon the activity of endoglucanases and cellobiohydrolases to solubilize cellulose. Some exceptions to this model had been noted to occur in bacterial systems that appear to lack or have deficiencies in cellobiohydrolases (33). An example is the S. degradans cellulolytic system that is predicted to produce an unusual abundance of GH5 endoglucanases but has a comparative deficiency in annotated cellobiohydrolases (28, 31). Experimental analysis showed that the single annotated cellobiohydrolase was in fact a classic endoglucanase, yet cellobiose was an early product of cellulose degradation by the S. degradans cellulolytic system. Thus, it was not apparent how the enzymes of the S. degradans cellulolytic system interact to produce cellobiose. The results presented here indicate that the S. degradans cellulolytic system utilizes a novel set of processive GH5 endoglucanases (Cel5G, Cel5H, and Cel5J) to substitute for the apparent deficiency in cellobiohydrolase activity. These processive enzymes were not dependent on endoglucanases for activity, as unlike with cellobiohydrolases, there was an absence of synergism with endoglucanases. Therefore, in contrast to other cellulolytic systems dependent upon endoglucanases and cellobiohydrolases, a processive endoglucanase coupled with the activity of β-glucosidases or a phosphorylase is sufficient for this bacterium to metabolize cellulose.
S. degradans Cel5G, Cel5H, and Cel5J exhibit endoglucanase activity as indicated by the effect of these enzymes on the viscosity of CMC solutions and the synergisms detected with a known exoglucanase. The identification of Cel5G, Cel5H, and Cel5J as processive endoglucanases acting on the β-1,4 bonds linking cellobiose units is supported by the constant ratio of soluble to insoluble products formed during reaction time courses and the phylogenetic segregation of these enzymes from classic GH5 endoglucanases. Unlike classical endoglucanases that randomly cleave cellulose polymers to form a variety of degradation products, these enzymes appeared to primarily release cellobiose from a variety of cellulose substrates. Although this is not demonstrative of processivity, due to the turnover rates of many endoglucanases (13), the processivity values for Cel5G, Cel5H, and Cel5J exceeded 4, irrespective of the reaction time. The processivity values reported for T. fusca Cel9A, an extensively characterized processive endoglucanase, range from 3.1 to 7.0 under similar conditions (15, 19) as well as in our own experiments. In contrast, the T. fusca classic endoglucanase Cel6A released only twice as much soluble sugar as insoluble sugar (35). Therefore, the processivity values for S. degradans Cel5G, Cel5H, and Cel5J were most similar to that of the processive T. fusca Cel9A protein. While a cloning artifact resulting in the observed processivity cannot be completely excluded, the cloning strategy was designed to take advantage of the modularity of these enzymes such that functional domains were retained in the expressed protein.
The processivity of most endoglucanases is dependent upon their associated CBM module. For example, the processivity of several bacterial GH9 endoglucanases is dependent upon the resident CBM3 module (10, 26). The processive GH5 endoglucanases identified in this study are all linked to CBM6 modules via flexible linkers (14). These CBM6 modules are expected to exhibit properties typical of a type B CBM module that binds to individual polysaccharide chains (3). The CBM6 modules of Cel5H or Cel5G, however, were not necessary for activity or processivity. A specifically designed derivative of Cel5H lacking the CBM6 module as well as the expressed form of Cel5G that, due to its size, appeared to lack its CBM6 module retained significant activity. The processivity ratio of each enzyme was not affected by the absence of the CBM6 module.
The processivity of the GH5 domains of S. degradans Cel5G, Cel5H, and Cel5J is unusual. In bacterial systems, processive endoglucanases have almost exclusively been found in the GH9 family (33). Processive endoglucanase activity, however, has been reported for another member of the GH5 family, Cel5A, produced by the brown rot basidiomycete Gloeophyllum trabeum (5). Because this is an atypical activity for GH5 enzymes, the processive endoglucanases of S. degradans should have distinctive structures. The catalytic sites of processive enzymes are typically associated with tunnel conformations that either enclose the substrate during catalysis or are located in deep clefts that partially enclose the substrate (4). The catalytic sites of GH5 enzymes are clefts, and the seven residues that form the active site and cleft of the GH5 domains of Cel5G, Cel5H, and Cel5J are conserved (8, 30). Phylogenetic analysis, however, revealed that other aspects of the primary sequences of these enzymes are sufficiently divergent to allow their segregation from the other families of GH5 endoglucanases found in S. degradans and other microorganisms. This implies that these enzymes have a distinct structure relative to those of other GH5 enzymes. As the GH5 cleft appears to be conserved in these enzymes, an induced fit with the substrate could explain the processivity of these enzymes.
Other systems lacking obvious cellobiohydrolases where the role of processive endoglucanases is not well understood have also been reported (33). For example, the cellulolytic system of Cytophaga hutchinsonii appears to be composed of nine candidate endoglucanases containing either a GH5 or a GH9 domain and four candidate β-glucosidases (34). Cellobiohydrolases are not obvious in this system. A similar system that contains five endoglucanases, including Cel9D, appears to be found in Fibrobacter succinogenes (23). Cel9D is interesting in that it exhibits a wide range of synergistic interactions with other members of its own family (24). C. hutchinsonii, F. succinogenes, and S. degradans all share the property of having a large number of predicted endoglucanases, with relatively few cellobiohydrolases. Processive enzymes would enable these organisms to more efficiently solubilize and metabolize cellulose.
We conclude from this work that S. degradans degrades cellulose by a mechanism based upon secreted processive endoglucanases. S. degradans produces at least three processive endoglucanases during growth on celluloses but lacks the cellobiohydrolase activity typical of other cellulolytic systems. The majority of the processive endoglucanase activity appears to be associated with Cel5H. Of the tested endoglucanases, this enzyme has the highest specific activity and expression. Although the processive activity of the annotated S. degradans GH9 endoglucanase cannot yet be excluded, their expression levels appear to be less than 9% that of Cel5H. The high expression and processive activity levels of Cel5H thus provide the best explanation for the formation of cellobiose by the S. degradans cellulolytic system. The cellobiose released by the activity of Cel5H can then be converted to glucose by the activity of cytoplasmic β-glucosidases Bgl1A or Bgl1B. A homolog to a cellobiose phosphorylase is also present in the genome of S. degradans, opening the possibility of phosphorylytic cleavage to glucose 1-phosphate and glucose. Thus, the processive endoglucanases in the S. degradans cellulolytic system can substitute for endoglucanases and cellobiohydrolases found in other cellulolytic systems to form a simple two-enzyme mechanism for the degradation of cellulose. This mechanism, coupled with the diversity of enzymes annotated to degrade the hemicellulose and pectic components of the higher plant cell wall, provides an explanation for the robust digestion of whole-plant material by this bacterium.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Science Foundation (DEB0621297) and the University of Maryland Energy Research Center.
We express our sincere gratitude to David Wilson for the gifts of T. fusca enzymes and to Rajeev Kumar for his valuable comments.
Footnotes
Published ahead of print on 17 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer, E. A., H. Chanzy, R. Lamed, and Y. Shoham. 1998. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 8548-557. [DOI] [PubMed] [Google Scholar]
- 3.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breyer, W. A., and B. W. Matthews. 2001. A structural basis for processivity. Protein Sci. 101699-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, R., M. R. Suzuki, and K. E. Hammel. 2005. Processive endoglucanase active in crystalline cellulose hydrolysis by the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 712412-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: Plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2541-551. [DOI] [PubMed] [Google Scholar]
- 7.Doner, L. W., and P. L. Irwin. 1992. Assay of reducing end-groups in oligosaccharide homologs with 2,2′-bicinchoninate. Anal. Biochem. 20250-53. [DOI] [PubMed] [Google Scholar]
- 8.Ducros, V., M. Czjzek, A. Belaich, C. Gaudin, H. P. Fierobe, L. P. Belaich, G. J. Davies, and R. Haser. 1995. Crystal-structure of the catalytic domain of a bacterial cellulase belonging to family-5. Structure 3939-949. [DOI] [PubMed] [Google Scholar]
- 9.Ghose, T. K. 1987. Measurement of cellulase activities. Pure Appl. Chem. 59257-268. [Google Scholar]
- 10.Gilad, R., L. Rabinovich, S. Yaron, E. A. Bayer, R. Lamed, H. J. Gilbert, and Y. Shoham. 2003. Ce1I, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gusakov, A. V., A. P. Sinitsyn, T. N. Salanovich, F. E. Bukhtojarov, A. V. Markov, B. B. Ustinov, C. van Zeijl, P. Punt, and R. Burlingame. 2005. Purification, cloning and characterisation of two forms of thermostable and highly active cellobiohydrolase I (Cel7A) produced by the industrial strain of Chrysosporium lucknowense. Enzyme Micob. Technol. 3657-69. [Google Scholar]
- 12.Himmel, M. E., S. Y. Ding, D. R. Johnson, W. S. Adney, M. R. Nimlos, J. W. Brady, and T. D. Foust. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315804-807. [DOI] [PubMed] [Google Scholar]
- 13.Horn, S. J., P. Sikorski, J. B. Cederkvist, G. Vaaje-Kolstad, M. Sorlie, B. Synstad, G. Vriend, K. M. Varum, and V. G. H. Eijsink. 2006. Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. USA 10318089-18094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard, M. B., N. A. Ekborg, L. E. Taylor, S. W. Hutcheson, and R. M. Weiner. 2004. Identification and analysis of polyserine linker domains in prokaryotic proteins with emphasis on the marine bacterium Microbulbifer degradans. Protein Sci. 131422-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irwin, D. C., M. Spezio, L. P. Walker, and D. B. Wilson. 1993. Activity studies of 8 purified cellulases—specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 421002-1013. [DOI] [PubMed] [Google Scholar]
- 16.Jeoh, T., D. B. Wilson, and L. P. Walker. 2006. Effect of cellulase mole fraction and cellulose recalcitrance on synergism in cellulose hydrolysis and binding. Biotechnol. Prog. 22270-277. [DOI] [PubMed] [Google Scholar]
- 17.Kang, M. S., I. C. Kang, S. M. Kim, H. C. Lee, and J. S. Oh. 2007. Effect of Leuconastoc spp. on the formation of Streptococcus mutans biorilm. J. Microbiol. 45291-296. [PubMed] [Google Scholar]
- 18.Kumar, R., S. Singh, and O. V. Singh. 2008. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 35377-391. [DOI] [PubMed] [Google Scholar]
- 19.Li, Y. C., D. C. Irwin, and D. B. Wilson. 2007. Processivity, substrate binding, and mechanism of cellulose hydrolysis by Thermobifida fusca Ce19A. Appl. Environ. Microbiol. 733165-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo Leggio, L., and S. Larsen. 2002. The 1.62 angstrom structure of Thermoascus aurantiacus endoglucanase: completing the structural picture of subfamilies in glycoside hydrolase family 5. FEBS Lett. 523103-108. [DOI] [PubMed] [Google Scholar]
- 21.Martinez, D., R. M. Berka, B. Henrissat, M. Saloheimo, M. Arvas, S. E. Baker, J. Chapman, O. Chertkov, P. M. Coutinho, D. Cullen, E. G. J. Danchin, I. V. Grigoriev, P. Harris, M. Jackson, C. P. Kubicek, C. S. Han, I. Ho, L. F. Larrondo, A. L. de Leon, J. K. Magnuson, S. Merino, M. Misra, B. Nelson, N. Putnam, B. Robbertse, A. A. Salamov, M. Schmoll, A. Terry, N. Thayer, A. Westerholm-Parvinen, C. L. Schoch, J. Yao, R. Barabote, M. A. Nelson, C. Detter, D. Bruce, C. R. Kuske, G. Xie, P. Richardson, D. S. Rokhsar, S. M. Lucas, E. M. Rubin, N. Dunn-Coleman, M. Ward, and T. S. Brettin. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26553-560. [DOI] [PubMed] [Google Scholar]
- 22.Park, J. K., L. X. Wang, H. V. Patel, and S. Roseman. 2002. Molecular cloning and characterization of a unique beta-glucosidase from Vibrio cholerae. J. Biol. Chem. 27729555-29560. [DOI] [PubMed] [Google Scholar]
- 23.Qi, M., H. S. Jun, and C. W. Forsberg. 2007. Characterization and synergistic interactions of Fibrobacter succinogenes glycoside hydrolases. Appl. Environ. Microbiol. 736098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi, M., H. S. Jun, and C. W. Forsberg. 2008. Cel9D, an atypical 1,4-β-d-glucan glucohydrolase from Fibrobacter succinogenes: characteristics, catalytic residues, and synergistic interactions with other cellulases. J. Bacteriol. 1901976-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin, E. M. 2008. Genomics of cellulosic biofuels. Nature 454841-845. [DOI] [PubMed] [Google Scholar]
- 26.Sakon, J., D. Irwin, D. B. Wilson, and P. A. Karplus. 1997. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4810-818. [DOI] [PubMed] [Google Scholar]
- 27.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 955857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor, L. E., B. Henrissat, P. M. Coutinho, N. A. Ekborg, S. W. Hutcheson, and R. A. Weiner. 2006. Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40T. J. Bacteriol. 1883849-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Violot, S., N. Aghajari, M. Czjzek, G. Feller, G. K. Sonan, P. Gouet, C. Gerday, R. Haser, and V. Receveur-Brechot. 2005. Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by x-ray diffraction and small angle x-ray scattering. J. Mol. Biol. 3481211-1224. [DOI] [PubMed] [Google Scholar]
- 31.Weiner, R. M., L. E. Taylor, B. Henrissat, L. Hauser, M. Land, P. M. Coutinho, C. Rancurel, E. H. Saunders, A. G. Longmire, H. T. Zhang, E. A. Bayer, H. J. Gilbert, F. Larimer, I. B. Zhulin, N. A. Ekborg, R. Lamed, P. M. Richardson, I. Borovok, and S. Hutcheson. 2008. Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40(T). PLOS Genet. 4e100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, D. B. 2004. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 472-82. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, D. B. 2008. Three microbial strategies for plant cell wall degradation. Ann. N. Y. Acad. Sci. 1125289-297. [DOI] [PubMed] [Google Scholar]
- 34.Xie, G., D. C. Bruce, J. F. Challacombe, O. Chertkov, J. C. Detter, P. Gilna, C. S. Han, S. Lucas, M. Misra, G. L. Myers, P. Richardson, R. Tapia, N. Thayer, L. S. Thompson, T. S. Brettin, B. Henrissat, D. B. Wilson, and M. J. McBride. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 733536-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, S., B. K. Barr, and D. B. Wilson. 2000. Effects of noncatalytic residue mutations on substrate specificity and ligand binding of Thermobifida fusca endocellulase Cel6A. Eur. J. Biochem. 267244-252. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, S., D. C. Irwin, and D. B. Wilson. 2000. Site-directed mutation of noncatalytic residues of Thermobifida fusca exocellulase Cel6B. Eur. J. Biochem. 2673101-3115. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, Y. H. P., J. B. Cui, L. R. Lynd, and L. R. Kuang. 2006. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: Evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7644-648. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Y. H. P., M. E. Himmel, and J. R. Mielenz. 2006. Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 24452-481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.