Abstract
Chromosome replication in Caulobacter crescentus is tightly regulated to ensure that initiation occurs at the right time and only once during the cell cycle. The timing of replication initiation is controlled by both CtrA and DnaA. CtrA binds to and silences the origin. Upon the clearance of CtrA from the cell, the DnaA protein accumulates and allows loading of the replisome at the origin. Here, we identify an additional layer of replication initiation control that is mediated by the HdaA protein. In Escherichia coli, the Hda protein inactivates DnaA after replication initiation. We show that the Caulobacter HdaA homologue is necessary to restrict the initiation of DNA replication to only once per cell cycle and that it dynamically colocalizes with the replisome throughout the cell cycle. Moreover, the transcription of hdaA is directly activated by DnaA, providing a robust feedback regulatory mechanism that adjusts the levels of HdaA to inactivate DnaA.
The events involved in the initiation of chromosomal replication are similar in eubacteria, eukaryotes, and archaea: replication starts with the binding of specific initiator protein(s) to chromosomal origins, resulting in the unwinding of the DNA duplex. Bacteria replicate their chromosome(s) from a single replication origin, and replication is initiated by DnaA, which binds to DnaA boxes within the chromosomal origin (34). In general, replication initiation takes place only once per cell cycle, although in rapidly dividing cells in some bacterial families, the next round of replication can start before the ongoing round has terminated (6, 9).
The gammaproteobacterium Escherichia coli has been a long-standing model system to study the regulation of DNA replication initiation. In this bacterium, at least three mechanisms prevent the reinitiation of DNA replication from the newly replicated origins (21, 34): (i) the inhibition of DnaA activity (22, 35), (ii) the titration of free DnaA molecules by the datA locus (23, 24), and (iii) the sequestration of the chromosomal origin by the SeqA protein (3, 42). When bound to ATP, DnaA is in its active form. ATP-DnaA initiates DNA replication, leading to the loading of the DNA polymerase III holoenzyme. The regulated inactivation of DnaA (RIDA) occurs by the conversion of ATP-DnaA into ADP-DnaA (22, 35) that is stimulated by a protein complex composed of the β-clamp subunit of the DNA polymerase (DnaN) and the Hda protein. The requirement of the DNA-loaded clamp can ensure the timely inactivation of DnaA by Hda, only once DNA replication has initiated (45). The Hda protein is homologous to the ATPase domain of DnaA, and it directly interacts with DnaN through a β-clamp binding motif located at its N terminus (25, 44). E. coli strains for which the hda gene is deleted accumulate suppressor mutations, suggesting that the hda gene is essential for viability or that Hda deficiency leads to severe growth impairment in E. coli (12, 22, 39). The overproduction of DnaA in the absence of Hda leads to growth retardation or cell death, due to the overinitiation of DNA replication by ATP-DnaA (39).
In the aquatic alphaproteobacterium Caulobacter crescentus, the initiation of DNA replication takes place only once per cell cycle, and only at a specific time of the cell cycle (29). Caulobacter divides asymmetrically, yielding a swarmer cell and a stalked cell. The stalked cell immediately initiates DNA replication, while the swarmer cell is unable to initiate the replication of its chromosome until it has differentiated into a stalked cell. The DnaA protein is essential for the initiation of DNA replication in Caulobacter (14). Although E. coli DnaA is stable for more than 24 h (47), Caulobacter DnaA is actively degraded (15) so that levels of DnaA can rapidly change as a function of the cell cycle (5). The transcription of dnaA is also cell cycle regulated, peaking in swarmer cells prior to the initiation of DNA replication (27, 49). The transcription of dnaA is regulated by the methylation state of the dnaA promoter: the dnaA promoter is preferentially transcribed when it is in the fully methylated state prior to the initiation of replication (4). Chromosomal loci are fully methylated at the beginning of the cell cycle. The loci then become hemimethylated by the passage of the replication fork and are not fully methylated again until just before cell division, because the cell cycle-regulated DNA methylase CcrM accumulates only in late predivisional cells. Once DNA replication is initiated and the replication fork passes through dnaA, the two copies of dnaA are maintained in the hemimethylated state until the completion of DNA replication, resulting in decreased levels of transcription from the dnaA promoter. This DNA methylation-dependent regulation of dnaA transcription contributes to the transient high levels of DnaA observed at the swarmer-to-stalked cell transition.
In swarmer cells, replication initiation is inhibited by the CtrA∼P response regulator that binds directly to five sites within the Caulobacter origin of replication (Cori) (37). At the swarmer-to-stalked cell transition, CtrA is degraded by the ClpXP protease (7, 18, 31), which releases the Cori for replication initiation by DnaA. Following the initiation of DNA replication, ctrA transcription is activated (7, 8). The accumulation of CtrA then contributes to the prevention of replication reinitiation. We asked if CtrA silencing of the origin of replication is sufficient to restrict the initiation of DNA replication to the swarmer-to-stalked cell stage of the cell cycle and to only once per cell cycle. To address this question, we identified the Caulobacter homologue of the E. coli Hda protein, named HdaA, and showed that it is required to confine the initiation of DNA replication to specific times in the cell cycle. Cells deficient in HdaA overinitiate DNA replication, and they exhibit severe growth defects. HdaA dynamically colocalizes with the β-clamp of DNA polymerase throughout the S phase, suggesting that HdaA is a component of the Caulobacter replisome. The stimulation of hdaA transcription by DnaA provides a feedback loop that fine-tunes the timing of DNA replication initiation. Thus, controlled dnaA transcription mediated by the differential methylation state of its promoter (4), the inhibition of DNA replication initiation by HdaA (this report), the temporally controlled proteolysis of DnaA (15), and the restriction of the CtrA silencer of DNA replication initiation to swarmer and predivisional cells (7) together provide robust restriction of DnaA function as an initiator of DNA replication to once per cell cycle.
MATERIALS AND METHODS
Bacterial strains, synchronization, and growth conditions.
Caulobacter crescentus strains were grown in peptone yeast extract (PYE) complex medium or M2 minimal salts plus 0.2% glucose (M2G) minimal medium (10) at 28°C. The plasmids and strains used are listed in Table 1. The antibiotics used for the Caulobacter liquid cultures include rifampin (rifampicin) (15 μg/ml), chloramphenicol (1 μg/ml), kanamycin (5 μg/ml), and oxytetracycline (1 μg/ml). The antibiotics used for the E. coli liquid cultures include chloramphenicol (20 μg/ml), kanamycin (30 μg/ml), oxytetracycline (12 μg/ml), and ampicillin (100 μg/ml). Plasmids were mobilized from E. coli S17-1 (41) into Caulobacter strains by bacterial conjugation or introduced by transformation. Bacteriophage φCR30 was used for general transduction into Caulobacter. Synchronized cell cultures were obtained by centrifugation in a Ludox density gradient, followed by the isolation of swarmer cells (11). Swarmer cells were resuspended into M2G medium and allowed to proceed synchronously through their cell cycle.
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Relevant characteristics, construction, or genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pXGFP4C1 | Integrating plasmid | D. Alley, unpublished data |
| pX-DnaA | dnaA under the control of the xylX promoter in pXGFP4C1 | This study |
| pNPTS138 | Integrating plasmid containing the sacB gene | D. Alley, unpublished |
| pNPTS138-ΔhdaA | The regions upstream and downstream of the hdaA coding sequence cloned into pNPTS138 | This study |
| pMR20 | Low-copy-number replicating plasmid | R. Roberts, unpublished data |
| pX-HdaA | hdaA under the control of the xylX promoter in pMR20 | This study |
| pXGFP-HdaA | gfp-hdaA under the control of the xylX promoter in pXGFP4C1 | This study |
| pCHYC-1 | Integrating plasmid containing the mcherry gene | 46 |
| pDnaN-RFP | dnaN-mcherry under the control of the dnaN promoter once integrated into the chromosome | This study |
| pJS14 | High-copy-number replicating plasmid | J. Skerker, unpublished data |
| pJSX-DnaA | dnaA under the control of the xylX promoter in pJS14 | This study |
| placZ290 | Low-copy-number plasmid to create transcriptional fusions with lacZ | 13 |
| placZ290-hdaAP(WT) | lacZ gene under the control of the wild-type hdaA promoter in placZ290 | This study |
| placZ290-hdaAP(box3) | lacZ gene under the control of the mutant hdaA promoter (box3) in placZ290 | This study |
| pET21a | Overexpression plasmid to clone histidine-tagged proteins | Novagen |
| pET21a-HdaA | hdaA cloned into pET21a to overexpress His6-HdaA | This study |
| Strains | ||
| E. coli | ||
| S17-1 | 294::RP4-2(Tc::Mu)(Km::Tn7) | 41 |
| Rosetta (DE3)/pLysS | Designed to enhance the expression of proteins that contain rare codons | Novagen |
| C. crescentus | ||
| NA1000 | Synchronizable derivative of wild-type strain CB15 (CB15N) | 11 |
| JC249 | pX-DnaA integrated at xylX into NA1000 | This study |
| JC353 | NA1000 ΔhdaA pX-HdaA | This study |
| JC208 | pXGFP-HdaA integrated at xylX into NA1000 | This study |
| JC388 | pDnaN-RFP integrated at dnaN into JC208 | This study |
| LS1 | PlacZ::ccrM bla6 rsaA2 | 48 |
| JC362 | NA1000 PlacZ::ccrM | This study |
| GM2471 | ΝΑ1000 ΔdnaA::Ω PxylX::dnaA | 14 |
Plasmid constructions.
For the construction of the pX-DnaA plasmid, oligonucleotides 5′-CCCATATGACCATGAAGGGCGGGGTTGCC-3′ and 5′-CCGGATCCTTAGCCCCGCAGCTTGCGCGT-3′ were used to amplify the dnaA coding sequence by PCR. The corresponding PCR product was digested with NdeI and BamHI and cloned into NdeI-BamHI-digested pXGFP4C1 (with the gfp gene eliminated), giving the pX-DnaA plasmid.
For the construction of the pNPTS138-ΔhdaA plasmid, the hdaA downstream region was PCR amplified using primers 5′-CCGGATCCGAGGGGGATGAGGGGTAGGC-3′ and 5′-CCGGGCTAGCAGGCGTTGATGCGGGTCAGCT-3′. The 500-bp product was digested with NheI and BamHI and cloned into an NheI-BamHI-digested pNPTS138 plasmid, giving pNPTS138-hdaAdown. The hdaA upstream region was PCR amplified using primers 5′-GGTAAGCTTACCGGAAGGCGAAATGCCACT-3′ and 5′-CCGGATCCTTTGAACTGGGTGGACAATCCT-3′. The 500-bp product was then digested with HindIII and BamHI and cloned into BamHI-HindIII-digested pNPTS138-hdaAdown, giving the pNPTS138-ΔhdaA plasmid.
For the construction of the pX-HdaA plasmid, the hdaA coding sequence was PCR amplified using primers 5′-CCCCATATGTTGTCCACCCAGTTCAAACTGCCGC-3′ and 5′-CCGGATTCCTACCCCTCATCCCCCTCGAAC-3′. The product was digested by NdeI and BamHI and cloned into an NdeI-BamHI-digested pXGFP4C1 plasmid (with the gfp gene eliminated). The resulting plasmid was used as a template to amplify a DNA region containing the hdaA gene under the control of the xylX promoter using primers 5′-CCTCTAGACTACCCCTCATCCCCCTCGAAC-3′ and 5′-AAGGTACCCAGCCGATCAGGCGGAACTGG-3′. This second product was digested by KpnI and XbaI and cloned into a KpnI-XbaI-digested pMR20 low-copy-number plasmid, giving the pX-HdaA plasmid.
For the construction of the pXGFP-HdaA plasmid, the hdaA coding sequence was PCR amplified using primers 5′-CCAAGCTTTGTCCACCCAGTTCAAACTGCC-3′ and 5′-CCGGATCCCTACCCCTCATCCCCCTCGAAC-3′. The product was digested by BamHI and HindIII and cloned into a BamHI-HindIII-digested pXGFP4C1 plasmid, giving pXGFP-HdaA.
For the construction of the pDnaN-RFP plasmid, the 3′ half of the dnaN coding sequence was PCR amplified using primers 5′-CCCATATGCCGAGGGCGCGGTCGGCATC-3′ and 5′-AAGGTACCGACCCGCAGCGGCATCAGCAC-3′. The product was digested by NdeI and KpnI and cloned into NdeI-KpnI-digested pCHYC-1.
For the construction of the pJSX-DnaA plasmid, the pX-DnaA plasmid was used to amplify a DNA region containing the dnaA gene under the control of the xylX promoter by using primers 5′-AAGGTACCCAGCCGATCAGGCGGAACTGG-3′ and 5′-ACGCGCAAGCTGCGGGGCTAAGGATCCGG-3′. The product was digested by KpnI and BamHI and cloned into a KpnI-BamHI-digested pJS14 plasmid, giving pJSX-DnaA.
For the construction of the placZ290-hdaAP(WT) and placZ290-hdaAP(box3) plasmids, the wild-type hdaA promoter region [hdaAP(WT)] was PCR amplified using primers 5′-CCGGATTCACCGGAAGGCGAAATGCCACTT-3′ and 5′-GGTAAGCTTGTTTGAACTGGGTGGACAATCCT-3′. The product was digested by HindIII and BamHI and cloned into a HindIII-BamHI-digested placZ290 plasmid, giving placZ290-hdaAP(WT). The targeted mutagenesis of the DnaA box 3 in the hdaA promoter [hdaAP(box3)] was generated by PCR using the mutagenic primers 5′-CGCGGCCTAACCCCCATCCGTGCCTCCTCCGCCCC-3′ and 5′-GGGGCGGAGGAGGCACGGATGGGGGTTAGGCCGCG-3′. The resulting product was also cloned into a HindIII-BamHI-digested placZ290 plasmid, giving placZ290-hdaAP(box3).
For the construction of the pET21a-HdaA plasmid, the hdaA coding sequence was amplified using primers 5′-CCCCATATGTTGTCCACCCAGTTCAAACTGCCGC-3′ and 5′-CCCTCGAGCCCCTCATCCCCCTCGAACCC-3′. The product was digested by NdeI and XhoI and cloned into an NdeI-XhoI-digested pET21a plasmid, giving pET21a-HdaA.
Strain constructions.
For the construction of the JC249 strain, plasmid pX-DnaA was integrated into the xylX promoter (32) of strain NA1000 by a single integration event.
For the construction of the JC353 strain, plasmid pNPTS138-ΔhdaA was introduced into strain NA1000 by conjugation, selecting for kanamycin-resistant colonies with the plasmid integrated at the hdaA locus by PCR. Plasmid pNPTS138-ΔhdaA was integrated into the Caulobacter chromosome by single homologous recombination. Plasmid pX-HdaA was then introduced into the resulting strain by transformation. The resulting strain was grown to stationary phase in PYE medium lacking kanamycin. The cells were plated on PYE medium plus 3% sucrose and incubated at 28°C. Single colonies were picked and transferred in parallel onto plain PYE plates and PYE plates containing kanamycin. Kanamycin-sensitive clones, which had lost the integrated plasmid due to a second recombination event, were then tested for the presence of the mutated allele by colony PCR.
For the construction of the JC208 strain, plasmid pXGFP-HdaA was integrated at the xylX promoter of strain NA1000 by a single integration event.
For the construction of the JC388 strain, plasmid pDnaN-RFP was integrated at the dnaN locus of strain JC208 by a single integration event.
For the construction of the JC362 strain, transduction was used for introducing the Plac::ccrM construct from phage lysates grown on strain LS1 into NA1000.
HdaA antibody preparation.
The HdaA protein tagged with hexahistidine was overproduced using pET21a-HdaA in the Rosetta E. coli strain. The tagged protein was purified by nickel affinity chromatography under standard nondenaturing conditions (Qiagen) and used to immunize rabbits for the production of polyclonal antibodies (Josman).
Immunoblot analysis.
DnaA and CcrM proteins were resolved on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (40). CtrA and HdaA proteins were resolved on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. The gels were electrotransferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA). Immunodetection was performed with polyclonal antibodies. Anti-DnaA sera, anti-CtrA sera, and donkey anti-rabbit sera conjugated to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA) were diluted 1:10,000. Anti-CcrM serum was diluted 1:5,000. Anti-HdaA serum was diluted 1:2,000. A chemiluminescent reagent (PerkinElmer, Wellesley, MA) and Kodak (Rochester, NY) Bio-Max MR films were used. The images were processed with Photoshop (Adobe, Mountain View, CA), and the relative band intensities were determined by using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Flow cytometry analysis.
Rifampin-treated cells were fixed and stained with DNA-binding Vybrant DyeCycle orange (Invitrogen), as previously described (28). Fixed cells were analyzed on a Becton Dickinson FACScan fluorescence-activated cell sorter. Flow cytometry data were analyzed using FlowJo software (Tree Star).
Analysis of the copy-number ratio of Cori to ter by Southern hybridization.
Chromosomal DNA was prepared using the Puregene Yeast/Bact. kit B (Qiagen, Valencia, CA). Chromosomal DNA (1 to 3 μg) was digested by BamHI and analyzed by Southern hybridization using digoxigenin (DIG)-labeled probes obtained by PCR (Roche, Mannheim, Germany) with primers 5′-CTGAGGACACGACAGCGACCTC-3′ and 5′-CGCGGCGTAGCAGGGCATTTC-3′ for the Cori probe and primers 5′-CTCAACATGCTTGACCGCCAGAT-3′ and 5′-ACCCAGGTCCTCGCCAAAGCTG-3′ for the ter probe. DIG-labeled probes were detected using anti-DIG-AP antibodies and CSPD (disodium 3-[4-methoxyspiro{l,2-dioxetane-3,2′- [5′-chloro]tricyclo[3.3.1.13,7]decan}-4-yl]phenylphosphate) following the protocol recommended by the manufacturer (Roche, Mannheim, Germany), and Bio-Max MR films (Kodak, Rochester, NY). The images were processed with Photoshop (Adobe, Mountain View, CA), and the relative band intensities were determined using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Microscopy.
Cells were immobilized using a thin layer of medium plus 1% agarose. Normarski differential interference contrast (DIC) and fluorescence microscopy images were taken with a 100× DIC objective on an E800 microscope (Nikon, East Rutherford, NJ) with a 5-MHz Micromax 5600 cooled charge-coupled-device camera controlled through Metamorph (Universal Imaging, Downingtown, PA). The images were processed using Adobe Photoshop and Metamorph version 4.5.
Promoter activity assays.
The β-galactosidase activities of strains containing placZ290 derivatives were assayed in log-phase cultures in PYE medium, as previously described (33). β-Galactosidase activities represent the averages of the results from at least three independent assays.
RESULTS
Control of the timing of replication initiation is maintained in cells that overexpress dnaA throughout the cell cycle.
Caulobacter permits detailed analysis of the cell cycle timing of chromosome replication because one can easily obtain synchronized cell populations by differential density centrifugation, without perturbing normal physiology. Immunoblot analysis of the relative levels of DnaA and CtrA as a function of the cell cycle revealed that there is a significant period of the cell cycle when the DnaA replication initiator is detectable, while the CtrA inhibitor of replication initiation drops to undetectable levels (5). This period lasts for about 40 min during the swarmer-to-stalked cell transition and in stalked cells, and it is far more than the time needed to initiate the replication of a bacterial chromosome. This observation suggested that CtrA action may not be sufficient to restrict the initiation of DNA replication to only once per cell cycle. It was also recently shown that DNA replication does not overinitiate when CtrA binding sites are eliminated from the Cori by targeted mutagenesis of the Caulobacter chromosome (2). The function of CtrA in replication control is more likely to prevent chromosome replication in swarmer cells.
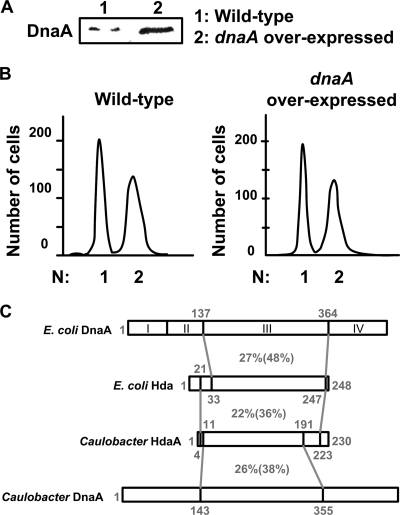
We observed that the levels of DnaA decrease after the initiation of DNA replication (5), even before the accumulation of CtrA in predivisional cells, suggesting that the levels of DnaA may become too limiting to allow the reinitiation of DNA replication. To test if the changes in the expression of dnaA over the cell cycle are necessary for the correct timing of the initiation of DNA replication, we constructed a mutant strain (JC249) that transcribes dnaA constitutively throughout the cell cycle and measured the DNA content in these cells by flow cytometry. The dnaA gene was expressed from the xylose-inducible xylX promoter so that the level of DnaA in JC249 cells grown in rich medium supplemented with xylose (PYEGX medium) for 4 h was significantly higher than that observed in wild-type cells (Fig. 1A). To examine the effect of high levels of DnaA on the initiation of chromosome replication, cultures of mutant and wild-type cells were examined by flow cytometry (Fig. 1B). JC249 cells and wild-type cells were treated with the RNA polymerase inhibitor rifampin for 3 h, which allows the completion of DNA replication but not cell division or the reinitiation of DNA replication (28). Flow cytometry analysis revealed that both wild-type cells and JC249 cells accumulate only one or two complete chromosomes, demonstrating that the control of replication initiation remains efficient when dnaA is overexpressed throughout the cell cycle. Thus, the temporal regulation of dnaA transcription is not solely responsible for restricting the initiation of DNA replication to once and only once during the cell cycle.
FIG. 1.
The temporal control of dnaA transcription is not necessary for the correct timing of the initiation of DNA replication. (A) DnaA accumulates to high levels in cells bearing a chromosomal copy of dnaA that is transcribed from an inducible xylX promoter (dnaA overexpressed, strain JC249). Cells were grown for 4 h in PYE medium plus 0.2% glucose and 0.3% xylose (PYEGX medium), and then immunoblotting of the cell extracts was performed using antibodies to DnaA. (B) Cells from strain JC249 do not overinitiate DNA replication. Cells were grown for 4 h in PYEGX medium and then treated with rifampin for 3 h. Cells were fixed and stained with Vybrant DyeCycle orange, before analyzing their DNA content by flow cytometry. The horizontal axis indicates the number of complete chromosomes. (C) Regions of homology between the E. coli DnaA and Hda proteins and the Caulobacter DnaA and HdaA proteins are shown. The amino acid positions are shown in gray. Identities and similarities (in parentheses) between protein regions are indicated by percent values. DnaA domains I, II, III, and IV are indicated (44). DnaA domain III includes the AAA+ motifs for the ATPase activity of DnaA.
HdaA is required for normal cell growth, for cell division, and for restricting the initiation of DNA replication to once per cell cycle.
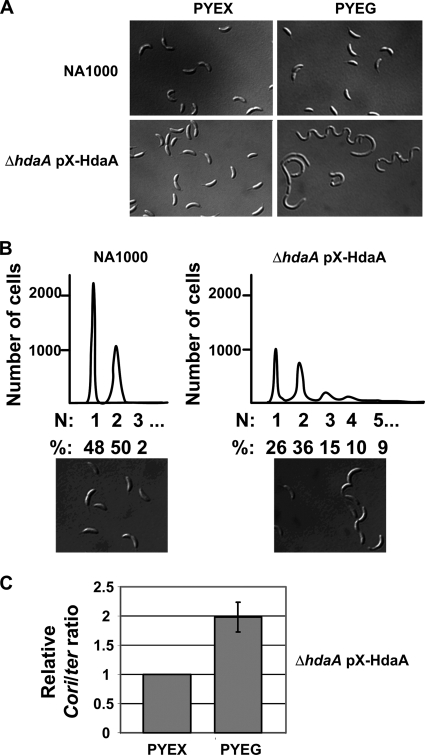
The observation that the initiation of DNA replication still occurs only once per cell cycle in cells that express dnaA throughout the cell cycle (Fig. 1B), despite the absence of CtrA for about 40 min in stalked cells, suggested that there exists a second negative regulator of the initiation of DNA replication in Caulobacter cells. One possibility is that the activity of DnaA is temporally regulated by a mechanism similar to the RIDA mechanism found in E. coli. The inactivation of DnaA after the initiation of DNA replication is mediated by the Hda protein in E. coli (22). We identified the Caulobacter homologue of the hda gene, named the hdaA gene (CC_1711), encoding a putative protein that is similar to both the E. coli Hda protein and to the Caulobacter and the E. coli DnaA proteins (Fig. 1C). We were not able to obtain an hdaA deletion strain, unless we added a complementing low-copy-number plasmid that expressed hdaA under the control of the inducible xylX promoter. When the resulting strain (JC353) was grown in rich medium in the presence of glucose (PYEG medium) at all times, conditions that prevented the expression of hdaA from the xylX promoter, we observed that cells grew very slowly with a generation time of ∼235 min, instead of ∼90 min for a wild-type strain (data not shown). DIC microscopy showed that HdaA-depleted cells are filamentous (Fig. 2A), indicating that HdaA is required for normal cell growth. This phenotype is similar to that of mutant cells which overinitiate DNA replication when expressing a thermosensitive allele of ctrA (37), consistent with HdaA functioning as an inhibitor of replication initiation. When JC353 was grown in the presence of xylose (PYEX medium), the wild-type phenotype was restored (Fig. 2A).
FIG. 2.
The Caulobacter hdaA gene is required for cell division and for the temporal control of the initiation of DNA replication. (A) Nomarski DIC microscopy images of NA1000 (wild-type) and JC353 (ΔhdaA pX-HdaA) cells grown in PYE medium plus xylose (PYEX; hdaA expressed) or PYE medium plus glucose (PYEG; hdaA not expressed) at all times. (B) Flow cytometry analyses of the NA1000 and JC353 strains grown for four generations in PYEG medium. Cells were treated with rifampin for 3 h prior to fixing and staining with Vybrant DyeCycle orange. The horizontal axis indicates the number of complete chromosomes. The percentages correspond to the proportion of cells containing the indicated number of chromosomes per cell. Nomarski DIC microscopy images of NA1000 and JC353 cells at the time of rifampin addition are shown in the bottom panels. (C) The JC353 strain was grown in PYEX or PYEG medium at all times, and samples were harvested to quantify the copy numbers of the chromosomal Cori and ter sites in a Southern hybridization experiment using specific probes. The ratio of the Cori sites to ter sites in cells grown in PYEX medium is defined as 1.0, and the relative value for cells grown in PYEG medium is shown. Results are the averages of data from independently duplicated experiments. The error bar indicates the standard deviation.
To test if the initiation of DNA replication was deregulated in cells depleted for HdaA, we performed flow-cytometry experiments to observe the number of chromosomes in wild-type and JC353 cells grown for four generations in PYEG medium and treated with rifampin (Fig. 2B). DIC microscopy showed that cells were only slightly filamentous at the time when rifampin was added to the culture medium (Fig. 2B, bottom panels). We found that ∼35% of the HdaA-depleted cells contained more than two chromosomes, suggesting that these cells had initiated a new round of DNA replication before the end of the previous round of replication, which happened only very rarely in wild-type Caulobacter cells (∼2% of the cells). We also performed Southern hybridizations to examine the copy-number ratio of Cori to ter (the replication termination region) in JC353 cells grown in PYEX or PYEG medium. As shown in Fig. 2C, the Cori to ter ratio was 1.98 (±0.25)-fold higher in cells depleted of HdaA (PYEG medium) than in cells that contained HdaA (PYEX medium), indicating that HdaA-depleted cells often overinitiate DNA replication. Cumulatively, these results suggest that HdaA is a second inhibitor of the initiation of DNA replication in Caulobacter.
HdaA dynamically colocalizes with the replisome throughout the cell cycle.
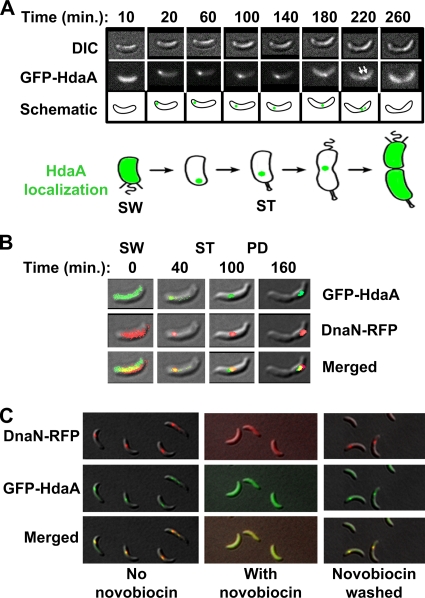
It has been shown in E. coli that Hda must interact with DnaN bound to DNA to inactivate DnaA. Thus, DnaA is inactivated by HdaA only once DNA replication has initiated (20, 22, 45). The E. coli Hda protein binds directly to DnaN in vitro (25, 44). In Caulobacter, DNA polymerase III is dynamically localized during the cell cycle (19). The replisome is assembled on the Cori, is positioned at the future stalked-cell pole during the swarmer-to-stalked cell transition, and then gradually moves to mid-cell as DNA replication progresses. Once DNA replication is complete in late predivisional cells, the replisome disassembles. To determine if the HdaA protein is in a complex with the Caulobacter replisome in vivo, we constructed a strain that would allow us to observe the subcellular localization of HdaA in live cells. We visualized HdaA in live cells by integrating an in-frame fusion between the N terminus of HdaA and the green fluorescent protein (GFP), under the control of the native xylX promoter. We isolated a pure population of swarmer cells from this strain (JC208) grown in minimal medium containing xylose (M2GX medium) for 3 h before synchronization. The intracellular location of GFP-HdaA was examined in individual cells by time-lapse fluorescence microscopy, as the cells progressed through their cell cycle. Figure 3A shows representative time-lapse images of cells bearing both wild-type HdaA and HdaA-GFP. At the 10-min time point, prior to the initiation of replication, we observed cells with fluorescence diffuse in the cytoplasm. Once a tight fluorescent focus was formed upon replication initiation, it proceeded to migrate toward mid-cell (between the 20-min and the 180-min time points), before disappearing in predivisional cells. During the migration process, we sometimes observed cells with two closely spaced GFP-HdaA foci. The pattern of HdaA cellular localization during the cell cycle is the same as that described for the localization of multiple replisome components in Caulobacter (19), suggesting that HdaA interacts with the replisome throughout the cell cycle.
FIG. 3.
HdaA dynamically colocalizes with the replisome. (A) Time-lapse fluorescence microscopy analysis of GFP-HdaA-expressing cells. Strain JC208 was cultivated in M2G medium plus 0.3% xylose (M2GX) for 3 h prior to synchronization of the culture. Swarmer cells were isolated and placed on a thin layer of agarose containing nutrients, and images of the same cells were acquired at the indicated time points as the cells progressed through their cell cycle. Cell division occurs at ∼280 min under these growth conditions. DIC images, fluorescence images, and schematics of the same cells are shown. White arrows indicate cells with two closely spaced GFP-HdaA foci. The green color in the schematics indicates the intracellular position of GFP-HdaA. SW, swarmer cell; ST, stalked cell. (B) Time-lapse fluorescence microscopy analysis of GFP-HdaA and DnaN-RFP colocalization. Strain JC388 was cultivated in M2GX medium for 3 h prior to synchronization of the culture. Swarmer cells were isolated and placed on a thin layer of agarose containing nutrients, and images of the same cells were acquired at the indicated time points. Top panels, GFP-HdaA (green) over DIC image; middle panels, DnaN-RFP (red) over DIC image; bottom panels, GFP-HdaA and DnaN-RFP over DIC image to show colocalization of the fluorescence markers (yellow). SW, swarmer cell; ST, stalked cell; PD, predivisional cell. (C) Strain JC388 was synchronized, and novobiocin was added at the late stalked-cell stage (65 min into the cell cycle). Some cells were washed and resuspended in fresh M2GX medium 42 min later. Top row, DnaN-RFP (red) over DIC; middle row, GFP-HdaA (green) over DIC; bottom row, DnaN-RFP and GFP-HdaA (colocalization in yellow) over DIC; left column, untreated stalked cells 65 min into the cell cycle; middle column, cells 12 min after the addition of novobiocin; right column, cells treated with novobiocin for 42 min, washed, and incubated in fresh medium for 10 min.
To determine if HdaA colocalizes with DnaN in vivo, we observed a double-labeled strain by time-lapse fluorescence microscopy. In the strain containing the GFP-HdaA construct, we replaced the wild-type allele of DnaN by an in-frame fusion between the red fluorescent protein (RFP) and the C terminus of DnaN under the control of the endogenous dnaN promoter (strain JC388). The intracellular location of GFP-HdaA and DnaN-RFP was examined in individual JC388 cells as they progressed through the cell cycle (Fig. 3B). We observed that GFP-HdaA and DnaN-RFP colocalize throughout the whole replication cycle, suggesting that HdaA is a component of the replisome.
It was previously shown that ongoing replication is required for the formation of replisome foci in Caulobacter cells (19). The DNA gyrase inhibitor novobiocin blocks the elongation of DNA replication in Caulobacter cells (19). To determine if the dynamic localization of HdaA also requires active replication, we treated a population of replication-competent stalked JC388 cells with novobiocin and examined the cells for DnaN and HdaA foci (Fig. 3C, middle panels). Upon the inhibition of DNA replication, DnaN-RFP foci were not detected, but rather a diffuse fluorescent signal was observed, confirming that the presence of replisome foci requires ongoing replication. Under these conditions, GFP-HdaA also appeared diffuse. When the cells were washed and resuspended in fresh medium without novobiocin, both DnaN-RFP and GFP-HdaA foci reappeared and colocalized, providing additional evidence that HdaA is a component of the Caulobacter replisome.
The levels of HdaA are adjusted to the needs of the cell to inhibit the initiation of DNA replication.
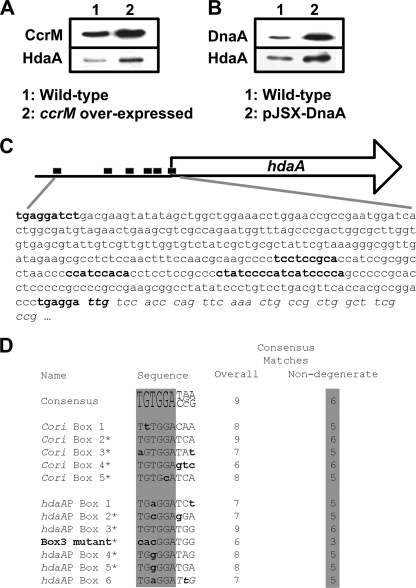
We compared the levels of HdaA in the wild-type strain and in two mutant strains that accumulate excess DnaA. In the first mutant strain tested, the gene encoding the CcrM DNA methyltransferase is expressed constitutively throughout the cell cycle (data not shown). Because the dnaA gene is preferentially transcribed from a fully methylated promoter, high levels of DnaA are maintained throughout the cell cycle in such mutant strains (4). As for hdaA transcription, it is not directly controlled by CcrM, since the hdaA promoter region does not contain GANTC sequences that become methylated by CcrM in predivisional cells. Immunoblots of wild-type and mutant cell extracts, using antibodies raised against HdaA, showed a greater accumulation of HdaA in the mutant strains than in the isogenic wild type (Fig. 4A). To test whether it is the increase in DnaA levels that promotes the accumulation of HdaA, we constructed a second strain in which a high-copy-number plasmid expressed the dnaA gene under the control of the inducible xylX promoter (pJSX-dnaA). A strain carrying this plasmid and grown in PYEX medium for 4 h accumulated significantly more DnaA and HdaA than the isogenic wild-type strain (Fig. 4B). Thus, in Caulobacter, the levels of HdaA are adjusted to the levels of DnaA.
FIG. 4.
DnaA promotes HdaA accumulation and the hdaA promoter region contains multiple putative DnaA boxes. (A) Strain JC362, with an extra chromosomal copy of ccrM under the control of the constitutive lacZ promoter (ccrM overexpressed) and the wild-type strain, with a single cell cycle-regulated copy of ccrM, were grown in PYE medium and used to perform immunoblotting using antibodies raised against CcrM and HdaA. (B) A wild-type strain, with or without pJSX-DnaA, was grown in PYE medium plus 0.2% glucose and 0.3% xylose (PYEGX) for 4 h and used to perform immunoblotting using antibodies raised against DnaA and HdaA. (C) The nucleotide sequence of the hdaA promoter region (345 nucleotides) is shown. The nucleotides corresponding to the six putative DnaA boxes are in bold. Italics indicate the beginning of the hdaA coding sequence. The black boxes in the schematic above indicate the DnaA boxes. (D) Consensus sequence for DnaA boxes in Caulobacter (17) and the putative DnaA boxes within the Cori (30) and the hdaA promoter. Reverse-complement sequences are presented for boxes marked with asterisks. Nucleotide positions differing from the consensus are denoted in lowercase bold lettering. The six nondegenerate nucleotides of the consensus DnaA box are highlighted in gray. The number of nucleotide matches to the overall and nondegenerate consensus DnaA box are shown for each sequence. The sequence of the mutated hdaAP(box3) is displayed, with lowercase bold letters indicating nucleotide changes compared to the wild type.
DnaA directly activates the transcription of hdaA.
DnaA is a dual-function protein that acts both as an initiator of DNA replication and as a transcription factor by binding to DnaA boxes found in the origin of replication and in promoters of specific genes, respectively (14, 17). In Caulobacter, DnaA directly regulates the transcription of at least 40 genes, including the gene encoding the GcrA master regulator (5, 17).
Because we found that the cellular levels of HdaA increased upon an increase in the levels of DnaA (Fig. 4A and B), we tested the possibility that DnaA could directly activate the transcription of hdaA. An examination of the 350-base sequence upstream of the translational start site of hdaA revealed a surprisingly high number of DnaA boxes (Fig. 4C); six motifs matched a minimum of seven of the nine nucleotides of the consensus DnaA box from Caulobacter (17) and a minimum of five of the six nondegenerate nucleotides of the consensus. These motifs look as similar to the consensus DnaA box as the five putative DnaA boxes found in the Cori (Fig. 4D) (30).
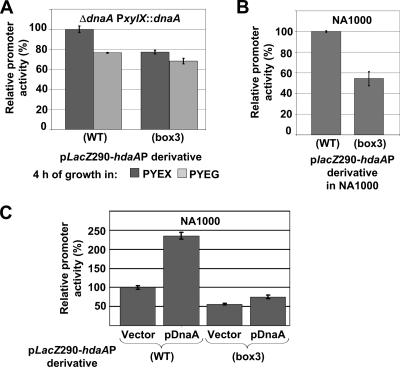
To test if the transcription of hdaA is activated by DnaA, we constructed a transcriptional fusion of the hdaA promoter and the lacZ gene on a low-copy-number plasmid. The resulting plasmid placZ290-hdaAP(WT) was first introduced into strain GM2471, in which the sole copy of dnaA is under the control of the chromosomal xylX promoter. We measured β-galactosidase activity, in the presence or the absence of the xylose inducer, as an indication of hdaA promoter activity (Fig. 5A). Four hours after this strain was shifted from PYEX to PYEG medium to deplete DnaA, β-galactosidase activity decreased by ∼25%. This change of activity is significant, since β-galactosidase is a very stable enzyme that takes a long time to disappear in cells when its synthesis is arrested. To confirm this result, we also introduced plasmid placZ290-hdaAP(WT) into an NA1000 strain containing the pJSXDnaA plasmid or an empty control vector. The activity of the hdaA promoter was 2.35-fold higher in the strain containing the pJSXDnaA plasmid than in the strain containing the control vector, when cells were grown in PYEX medium to overproduce DnaA from pJSXDnaA (Fig. 5C). We conclude that DnaA significantly activates transcription from the hdaA promoter.
FIG. 5.
DnaA directly activates the transcription of hdaA. (A) The graph shows the relative β-galactosidase activities from placZ290-hdaAP(WT) and placZ290-hdaAP(box3) in a GM2471 (ΔdnaA PxylX::dnaA) strain upon the depletion of DnaA 4 h after a shift from PYE medium plus xylose (PYEX) to PYE medium plus glucose (PYEG). (B) The graph shows the relative β-galactosidase activities from placZ290-hdaAP(WT) and placZ290-hdaAP(box3) in an NA1000 (WT) strain grown in PYE medium. Activities in Miller units were normalized so that the activity of hdaAP(WT) equals 100% in PYE (NA1000) or PYEX (GM2471) medium to facilitate comparison. (C) The graph shows the relative β-galactosidase activities from placZ290-hdaAP(WT) and placZ290-hdaAP(box3) in an NA1000 strain containing the DnaA-overexpressing plasmid pJSX-DnaA or the empty control vector pJS14 grown in PYEX medium for 4 h. Activities in Miller units were normalized so that the activity of hdaAP(WT) equals 100% in NA1000 containing pJS14 to facilitate comparison. Error bars indicate the standard deviations.
If DnaA activates the transcription of hdaA by directly binding to the hdaA promoter, then the elimination of one or more DnaA boxes in the hdaA promoter could affect the efficiency of transcription from that promoter. To test this hypothesis, we analyzed the transcription from a mutant hdaA promoter in which the DnaA box with a 9/9 base conservation (box 3 in Fig. 4C and D) was disrupted by targeted mutagenesis. This mutant promoter was fused to lacZ, yielding plasmid placZ290-hdaAP(box3). We introduced this plasmid into wild-type cells and measured β-galactosidase activity. The activity of the mutant promoter was about twofold lower than the activity of the wild-type promoter (Fig. 5B), suggesting that DnaA directly activates the transcription of hdaA. We also introduced the placZ290-hdaAP(box3) plasmid into the GM2471 strain and into the NA1000 strain containing the pJSXDnaA plasmid to test if the hdaAP(box3) mutant promoter is still sensitive to changing levels of DnaA in the cells. The β-galactosidase activity of the mutant promoter decreased by ∼12% when the GM2471 strain was shifted from PYEX to PYEG medium for 4 h to deplete DnaA, compared to the 25% decrease observed for the wild-type hdaA promoter (Fig. 5A). The β-galactosidase activity of the mutant promoter was 1.34-fold higher in the NA1000 strain containing pJSXDnaA than in the NA1000 strain containing the pJS14 empty vector when the cells were grown in PYEX medium, compared to 2.35-fold for the wild-type hdaA promoter. The hdaAP(box3) mutant promoter is therefore less sensitive to changes in DnaA levels in the cell than the wild-type promoter (Fig. 5C).
Cumulatively, these results suggest that DnaA directly activates the transcription of hdaA by binding at minimum to DnaA box 3 within the hdaA promoter.
DISCUSSION
We have determined that the Caulobacter HdaA protein functions as a negative regulator of replication initiation, in concert with CtrA, to ensure that the initiation of DNA replication takes place only once per cell cycle. The HdaA protein is an apparent component of the Caulobacter replisome and it inhibits the initiation of DNA replication. The accumulation of HdaA is regulated by the activation of hdaA transcription by DnaA, providing an interesting feedback loop.
Model for the temporal regulation of DNA replication initiation in Caulobacter.
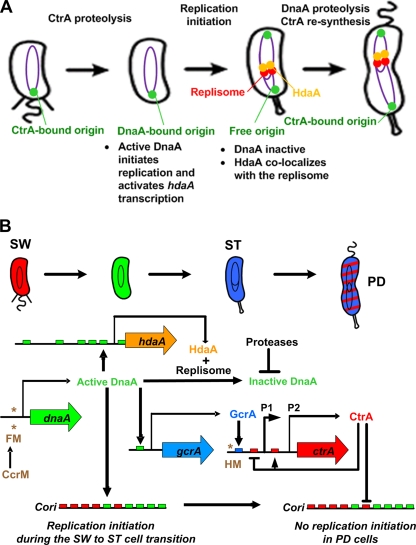
As shown in Fig. 6A, we propose the following multistep control system that limits the initiation of DNA replication to only once per cell cycle: (i) the initiation process is inhibited in swarmer cells by CtrA bound to five sites within the origin region (37); (ii) CtrA is eliminated by targeted proteolysis (7) and DnaA accumulates during the swarmer-to-stalked cell transition (4), allowing the initiation of DNA replication; (iii) a second round of replication initiation is inhibited by an HdaA/DnaN complex bound to the chromosome once DNA replication has initiated in stalked cells (Fig. 2) and by the directed proteolysis of DnaA (15); and (iv) CtrA accumulates again to strengthen the inhibition of replication initiation in predivisional cells.
FIG. 6.
Model for the temporal control of DNA replication initiation in Caulobacter. (A) A schematic of the beginning of the Caulobacter cell cycle is shown. Purple theta structures inside the cells indicate replicating DNA. The single origin of replication (green focus) in swarmer cells is bound to CtrA, which represses the initiation of DNA replication. During the swarmer-to-stalked cell transition, CtrA is rapidly degraded by the ClpXP protease, and active DnaA binds to the origin to initiate DNA replication. The replisome (red foci), associated with HdaA (orange foci), replicates the chromosome and inactivates DnaA once DNA replication is ongoing. CtrA reaccumulates in predivisional cells and binds to the origin to prevent more replication initiation events. (B) A schematic of the beginning of the Caulobacter cell cycle is shown. Red indicates CtrA accumulation, green indicates DnaA accumulation, and blue indicates GcrA accumulation. SW, swarmer cell; ST, stalked cell; PD, predivisional cell. DnaA is synthesized in swarmer cells, when the dnaA promoter is in the fully methylated state (FM, two asterisks). New molecules of DnaA initiate DNA replication and activate the transcription of gcrA and hdaA by directly binding to DnaA boxes (green boxes). Once the replisome is assembled, the replisome-HdaA complex inhibits the initiation of DNA replication, probably by a mechanism similar to the RIDA mechanism in E. coli, and the DnaA protein is degraded by a protease (15) to prevent more initiation events in stalked cells. Soon after the initiation of DNA replication, the dnaA and the ctrA genes are duplicated by the passage of the replication fork and therefore hemimethylated (HM, asterisk). Transcription from the hemimethylated dnaA gene is shut down, while transcription from the hemimethylated ctrA gene is turned on by the binding of GcrA to the ctrA P1 promoter (blue box). Accumulation of CtrA in early predivisional cells then contributes to the inactivation of replication initiation by directly binding to the CtrA sites in the Cori (red boxes), yielding a robust replication control system.
The regulated control of DnaA accumulation and activity is integrated in a wider regulatory network that controls the Caulobacter cell cycle (Fig. 6B) (4). The expression of about 550 genes is temporally regulated during the Caulobacter cell cycle (27). The two master transcriptional regulators CtrA and GcrA together control the expression of ∼145 of these genes. The CtrA response regulator directly controls the transcription of 95 genes (26), while GcrA regulates the transcription of about 50 genes (16). CtrA and GcrA oscillate out of phase temporally and spatially. The DnaA protein is not only the initiator of DNA replication, but it is also a transcription factor that regulates the transcription of at least 40 genes in Caulobacter (17). One of these is the gcrA gene, which is directly activated by DnaA (5). The GcrA master regulator activates one of the two ctrA promoters, ctrA P1, when it is in the hemimethylated state, after the passage of the replication fork through the ctrA locus in late stalked cells (16, 38). CtrA activates its own transcription by binding to the ctrA P2 promoter (8), so that CtrA efficiently accumulates in predivisional cells to inhibit the initiation of DNA replication (37).
Feedback control of the activity of DnaA by HdaA.
Evidence that DnaA activates the transcription of hdaA in vivo includes the observations that (i) the cellular levels of the HdaA protein increased in mutant strains that accumulate excess DnaA (Fig. 4A and B); (ii) the hdaA promoter contains multiple DnaA boxes, similar to those present in the Cori sequence (Fig. 4C and D); and (iii) lacZ transcribed from the hdaA promoter was activated by DnaA in vivo, and a mutation in a conserved 9/9 DnaA box in the hdaA promoter decreased activation by DnaA (Fig. 5). These results suggest that the synthesis of HdaA is proportional to the levels of active DnaA in the cell, providing an interesting feedback mechanism for the control of DnaA activity. We examined the level of HdaA protein as a function of the cell cycle and found that it did not change significantly (data not shown), suggesting that HdaA is a rather stable protein in Caulobacter. However, as is the case in E. coli (45), HdaA in Caulobacter is likely to only inactivate DnaA once it joins the replisome as it is assembled onto the DNA. Thus, we propose that the activity, and not the protein availability of HdaA, is cell cycle regulated so that it functions to inactivate DnaA only after the first round of DNA replication has initiated.
We believe that the main function of the feedback mechanism is not to control the accumulation of HdaA as a function of the cell cycle but to promote HdaA accumulation in cases when DnaA becomes too abundant in the cell and may cause overinitiation defects. This can happen, for example, when the transcription of dnaA or ccrM is mis-regulated (Fig. 4A and B). HdaA contributes to preventing the start of a second round of DNA replication in stalked cells, even when dnaA is transcribed constitutively throughout the cell cycle (Fig. 1A and B). We found previously that DnaA accumulates constitutively throughout the cell cycle when the dnaA locus is maintained in the fully methylated state by moving the position of the dnaA gene on the chromosome from its native location next to the Cori to a location next to the terminus of replication (4). Interestingly, this strain still initiates the replication of its chromosome only once per cell cycle, likely due to the efficient inactivation of DnaA by the HdaA/DnaN complex immediately following the initiation of DNA replication. The feedback regulation of hdaA transcription by DnaA likely makes the cell cycle control system more robust to accidental variations or mutations.
Control of the initiation of DNA replication in different bacteria.
The RIDA mechanism was first identified in E. coli, where it prevents the overinitiation of replication (22). However, there are significant differences between the regulatory networks that control the initiation of chromosome replication in E. coli and in Caulobacter. One striking difference is that the Caulobacter CtrA inhibitor of the initiation of DNA replication is not conserved in E. coli. Instead, E. coli has a unique SeqA protein that binds to the hemimethylated origin of replication to prevent further the overinitiation of DNA replication (42). A second difference is that the DnaA protein is unstable in Caulobacter cells (15), while it is very stable in E. coli cells (47). A third difference is that the promoter of the hda gene in E. coli does not contain any obvious DnaA boxes, whereas in Caulobacter, DnaA activates the transcription of hdaA (Fig. 5). A fourth interesting difference is that the levels of DnaA remain unchanged when hdaA is depleted in Caulobacter cells (data not shown), while the levels of DnaA decrease in Hda-deficient E. coli cells (39), due to the autorepression of dnaA transcription by DnaA-ATP (43). The last known difference is that the overexpression of hdaA is not deleterious in Caulobacter cells (data not shown) unlike the overexpression of hda in E. coli cells, which induces the SOS response and cell division defects (1).
The use of an inhibitor of the initiation of DNA replication that interacts with the β-clamp of DNA polymerase is not restricted to gram-negative bacteria. Indeed, the conserved YabA protein from Bacillus subtilis also acts as a negative inhibitor of replication initiation and forms a ternary complex with DnaA and DnaN in vivo, although YabA shares no homology with Hda (36). This finding suggests that there is an evolutionary conserved need for a temporally precise mechanism that controls the initiation of bacterial DNA replication.
Acknowledgments
We are grateful to Adam Saunders for constructing plasmids pX-HdaA and pNPTS138-ΔhdaA, to David Parks from the Stanford University FACS facility for helping to develop a method to look at DNA content in Caulobacter cells by using DyeCycle orange, and to Carmen Fernández Fernández for analyzing the intracellular levels of HdaA as a function of the Caulobacter cell cycle. We thank Nathan Hillson and Antonio Iniesta for the critical reading of the manuscript and all the members of the Shapiro and McAdams laboratories for helpful comments during the course of this work.
This work was supported by NIH grants RO1 GM51426 and R24GM073011-04 to L.S. and by Swiss National Science Foundation Fellowship 3100A0_122541 to J.C.
Footnotes
Published ahead of print on 24 July 2009.
REFERENCES
- 1.Banack, T., N. Clauson, N. Ogbaa, J. Villar, D. Oliver, and W. Firshein. 2005. Overexpression of the Hda DnaA-related protein in Escherichia coli inhibits multiplication, affects membrane permeability, and induces the SOS response. J. Bacteriol. 1878507-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastedo, D. P., and G. T. Marczynski. 2009. CtrA response regulator binding to the Caulobacter chromosome replication origin is required during nutrient and antibiotic stress as well as during cell cycle progression. Mol. Microbiol. 72139-154. [DOI] [PubMed] [Google Scholar]
- 3.Brendler, T., A. Abeles, and S. Austin. 1995. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 144083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier, J., H. H. McAdams, and L. Shapiro. 2007. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc. Natl. Acad. Sci. USA 10417111-17116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier, J., S. R. Murray, and L. Shapiro. 2006. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 25346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper, S., and C. E. Helmstetter. 1968. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 31519-540. [DOI] [PubMed] [Google Scholar]
- 7.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90415-424. [DOI] [PubMed] [Google Scholar]
- 8.Domian, I. J., A. Reisenauer, and L. Shapiro. 1999. Feedback control of a master bacterial cell-cycle regulator. Proc. Natl. Acad. Sci. USA 966648-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donachie, W. D. 1968. Relationship between cell size and time of initiation of DNA replication. Nature 2191077-1079. [DOI] [PubMed] [Google Scholar]
- 10.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204372-384. [DOI] [PubMed] [Google Scholar]
- 11.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimitsu, K., M. Su'etsugu, Y. Yamaguchi, K. Mazda, N. Fu, H. Kawakami, and T. Katayama. 2008. Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J. Bacteriol. 1905368-5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbatyuk, B., and G. T. Marczynski. 2001. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 40485-497. [DOI] [PubMed] [Google Scholar]
- 15.Gorbatyuk, B., and G. T. Marczynski. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 551233-1245. [DOI] [PubMed] [Google Scholar]
- 16.Holtzendorff, J., D. Hung, P. Brende, A. Reisenauer, P. H. Viollier, H. H. McAdams, and L. Shapiro. 2004. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304983-987. [DOI] [PubMed] [Google Scholar]
- 17.Hottes, A. K., L. Shapiro, and H. H. McAdams. 2005. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 581340-1353. [DOI] [PubMed] [Google Scholar]
- 18.Iniesta, A. A., P. T. McGrath, A. Reisenauer, H. H. McAdams, and L. Shapiro. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. USA 10310935-10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen, R. B., S. C. Wang, and L. Shapiro. 2001. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 204952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 9461-71. [DOI] [PubMed] [Google Scholar]
- 21.Kato, J. 2005. Regulatory network of the initiation of chromosomal replication in Escherichia coli. Crit. Rev. Biochem. Mol. Biol. 40331-342. [DOI] [PubMed] [Google Scholar]
- 22.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 204253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitagawa, R., H. Mitsuki, T. Okazaki, and T. Ogawa. 1996. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 191137-1147. [DOI] [PubMed] [Google Scholar]
- 24.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 123032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurz, M., B. Dalrymple, G. Wijffels, and K. Kongsuwan. 2004. Interaction of the sliding clamp β-subunit and Hda, a DnaA-related protein. J. Bacteriol. 1863508-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 994632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 2902144-2148. [DOI] [PubMed] [Google Scholar]
- 28.Lesley, J. A., and L. Shapiro. 2008. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J. Bacteriol. 1906867-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marczynski, G. T. 1999. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J. Bacteriol. 1811984-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marczynski, G. T., and L. Shapiro. 1992. Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J. Mol. Biol. 226959-977. [DOI] [PubMed] [Google Scholar]
- 31.McGrath, P. T., A. A. Iniesta, K. R. Ryan, L. Shapiro, and H. H. McAdams. 2006. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell 124535-547. [DOI] [PubMed] [Google Scholar]
- 32.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Mott, M. L., and J. M. Berger. 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5343-354. [DOI] [PubMed] [Google Scholar]
- 35.Nishida, S., K. Fujimitsu, K. Sekimizu, T. Ohmura, T. Ueda, and T. Katayama. 2002. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 27714986-14995. [DOI] [PubMed] [Google Scholar]
- 36.Noirot-Gros, M. F., M. Velten, M. Yoshimura, S. McGovern, T. Morimoto, S. D. Ehrlich, N. Ogasawara, P. Polard, and P. Noirot. 2006. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1032368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 95120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reisenauer, A., and L. Shapiro. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 214969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riber, L., J. A. Olsson, R. B. Jensen, O. Skovgaard, S. Dasgupta, M. G. Marinus, and A. Lobner-Olesen. 2006. Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 202121-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1784-790. [Google Scholar]
- 42.Slater, S., S. Wold, M. Lu, E. Boye, K. Skarstad, and N. Kleckner. 1995. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82927-936. [DOI] [PubMed] [Google Scholar]
- 43.Speck, C., C. Weigel, and W. Messer. 1999. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 186169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su'etsugu, M., T. R. Shimuta, T. Ishida, H. Kawakami, and T. Katayama. 2005. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J. Biol. Chem. 2806528-6536. [DOI] [PubMed] [Google Scholar]
- 45.Su'etsugu, M., M. Takata, T. Kubota, Y. Matsuda, and T. Katayama. 2004. Molecular mechanism of DNA replication-coupled inactivation of the initiator protein in Escherichia coli: interaction of DnaA with the sliding clamp-loaded DNA and the sliding clamp-Hda complex. Genes Cells 9509-522. [DOI] [PubMed] [Google Scholar]
- 46.Thanbichler, M., A. A. Iniesta, and L. Shapiro. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torheim, N. K., E. Boye, A. Lobner-Olesen, T. Stokke, and K. Skarstad. 2000. The Escherichia coli SeqA protein destabilizes mutant DnaA204 protein. Mol. Microbiol. 37629-638. [DOI] [PubMed] [Google Scholar]
- 48.Zweiger, G., G. Marczynski, and L. Shapiro. 1994. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 235472-485. [DOI] [PubMed] [Google Scholar]
- 49.Zweiger, G., and L. Shapiro. 1994. Expression of Caulobacter dnaA as a function of the cell cycle. J. Bacteriol. 176401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]