Abstract
Data from the genome sequence of the aerobic, marine bacterium Roseovarius nubinhibens ISM were interpreted such that 3-sulfolactate would be degraded as a sole source of carbon and energy for growth via a novel bifurcated pathway including two known desulfonative enzymes, sulfoacetaldehyde acetyltransferase (EC 2.3.3.15) (Xsc) and cysteate sulfo-lyase (EC 4.4.1.25) (CuyA). Strain ISM utilized sulfolactate quantitatively with stoichiometric excretion of the sulfonate sulfur as sulfate. A combination of enzyme assays, analytical chemistry, enzyme purification, peptide mass fingerprinting, and reverse transcription-PCR data supported the presence of an inducible, tripartite sulfolactate uptake system (SlcHFG), and a membrane-bound sulfolactate dehydrogenase (SlcD) which generated 3-sulfopyruvate, the point of bifurcation. 3-Sulfopyruvate was in part decarboxylated by 3-sulfopyruvate decarboxylase (EC 4.1.1.79) (ComDE), which was purified. The sulfoacetaldehyde that was formed was desulfonated by Xsc, which was identified, and the acetyl phosphate was converted to acetyl-coenzyme A by phosphate acetyltransferase (Pta). The other portion of the 3-sulfopyruvate was transaminated to (S)-cysteate, which was desulfonated by CuyA, which was identified. The sulfite that was formed was presumably exported by CuyZ (TC 9.B.7.1.1 in the transport classification system), and a periplasmic sulfite dehydrogenase is presumed. Bioinformatic analyses indicated that transporter SlcHFG is rare but that SlcD is involved in three different combinations of pathways, the bifurcated pathway shown here, via CuyA alone, and via Xsc alone. This novel pathway involves ComDE in biodegradation, whereas it was discovered in the biosynthesis of coenzyme M. The different pathways of desulfonation of sulfolactate presumably represent final steps in the biodegradation of sulfoquinovose (and exudates derived from it) in marine and aquatic environments.
Sulfolactate (Fig. 1A) is a widespread natural product, which contains the stable C-SO3− bond. The compound is known to be (i) a component (5% of dry weight) of bacterial endospores (5), (ii) an intermediate in the biosynthesis of coenzyme M in archaea (55), (iii) in equilibrium with (S)-cysteate in mammals (54), (iv) involved in the metabolism of sulfoquinovose (6-deoxy-6-sulfo-d-glucopyranose, the polar moiety of the plant sulfolipid) in plants and algae (e.g., see reference 48), and (v) an intermediate in the bacterial degradation of sulfoquinovose (44).
FIG. 1.
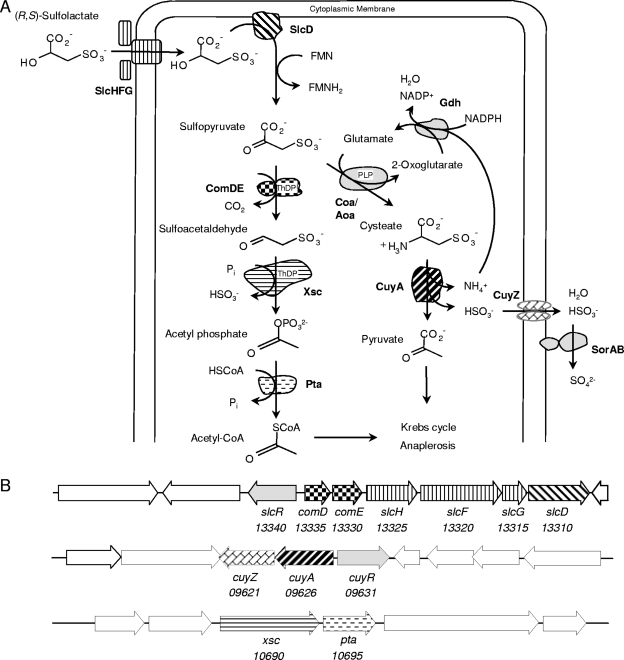
Hypothesized bifurcated degradative pathway of sulfolactate in R. nubinhibens ISM (A) and the corresponding gene clusters (B), with our annotation of the locus tags. Abbreviations of enzyme names are in the main text. ThDP, thiamine diphosphate; PLP, pyridoxal-5-phosphate; CoA, coenzyme A. Taurine has several degradative pathways, but that in Ruegeria pomeroyi DSS-3 (21) corresponds to that found in strain ISM.
Research on the biodegradation of organosulfonates has concentrated on compounds containing one to four carbon atoms (C1, C2, C3, or C4 sulfonates), because where appropriate, larger molecules all seemed to be processed via one of the five desulfonative reactions that have been elucidated. Pathways from (i) sulfoquinovose yield, e.g., sulfoacetate or sulfolactate and 2,3-dihydroxy-1-sulfopropane (36, 44), (ii) taurocholate and N-acetyltaurine yield taurine (37, 43), and (iii) N-methyltaurine yield sulfoacetaldehyde (52). The five desulfonation reactions are two oxygenolyses for the C1 and C4 sulfonates (30, 39), phosphatolysis of sulfoacetaldehyde by sulfoacetaldehyde acetyltransferase (Xsc) (EC 2.3.3.15) (8, 45), dehydratase-like elimination of sulfite from sulfolactate (sulfolactate sulfo-lyase SuyAB) (EC 4.4.1.24) (42), and the pyridoxal phosphate chemistry involved in (S)-cysteate sulfo-lyase (CuyA) (EC 4.4.1.25) (9, 14). Although not relevant here, the range of known desulfonation reactions is wider when enzymes involved in the assimilation of sulfonate-sulfur are considered (e.g., see references 17, 27, and 32).
Whereas sulfoacetaldehyde and sulfolactate are described in the preceding paragraph as degradative intermediates in different, independent pathways, they are also known as biosynthetic intermediates in one pathway, the formation of coenzyme M (55), during which (R)-3-sulfolactate is oxidized to sulfopyruvate by (R)-2-hydroxyacid dehydrogenase (ComC) (EC 1.1.1.272). The latter compound is decarboxylated to sulfoacetaldehyde by sulfopyruvate decarboxylase (EC 4.1.1.79) (ComDE) (55). Bioinformatic analyses of the genome of the bacterium Roseovarius nubinhibens ISM revealed the presence of gene candidates to encode ComDE, Xsc, and CuyA, which, with other gene products, allowed a novel, bifurcated degradative pathway for sulfolactate to be proposed (Fig. 1). A possible alternative pathway via SuyAB was considered to be unlikely, because no candidate suyAB genes were detected on the genome.
We now report that R. nubinhibens ISM utilizes sulfolactate inducibly as a sole source of carbon and energy for growth. There is extensive experimental support for the bifurcated pathway shown in Fig. 1A.
MATERIALS AND METHODS
Chemicals.
Racemic sulfolactate was synthesized as described elsewhere (44), as were sulfopyruvate and the bisulfite addition complex of sulfoacetaldehyde (13). Commercial chemicals were of the highest purity available and purchased from Sigma-Aldrich, Roth, Merck, Biomol, or Fluka. Taq DNA polymerase, Moloney murine leukemia virus reverse transcriptase, and RNase-free DNase were from MBI-Fermentas.
Organisms, growth, harvesting of cells, and preparation of cell extracts.
R. nubinhibens ISM, a marine alphaproteobacterium (19), was kindly made available by M. A. Moran (Georgia, United States). Cells were grown aerobically at 30°C in a modified Silicibacter basal medium (14) containing 0.05% yeast extract; the sole added source of carbon and energy was 10 mM (R,S)-sulfolactate or (S)-cysteate, 20 mM taurine, or acetate. Precultures (3 ml) were grown in 30-ml screw-cap tubes in a roller. Growth experiments were done on the 30-ml scale in 300-ml Erlenmeyer flasks shaken at 30°C; 6 mM sulfolactate was used. Samples were taken at intervals to measure optical density (580 nm), to assay protein, and to determine the concentration of sulfolactate, sulfate, or sulfite. Similar cultures were used to generate small amounts of cells for enzyme assays. Cultures (1 liter) for protein purification were grown in 5-liter Erlenmeyer flasks on a shaker. Cells were harvested at an optical density at 580 nm of 0.5 (about 170 mg protein liter−1) by centrifugation (15,000 × g, 20 min, 4°C); washed with 50 mM potassium phosphate buffer, pH 7.5 (containing 5 mM MgCl2); and stored frozen. The same buffer served as extraction buffer. Cell extracts free of nucleic acids (0.05 mg DNase I ml−1) were generated by disruption via four passages through a French pressure cell set at 140 MPa (26) or by ultrasonication. The membrane/particulate fraction was sedimented by ultracentrifugation (220,000 × g, 30 min, 4°C), and the supernatant fluid was called the soluble fraction. Solubilization of membranes was done by stirring with Triton X-100 (0.5 mg [mg protein]−1) on ice. After ultracentrifugation (220,000 × g, 30 min, 4°C), the clear supernatant fluid was called the membrane fraction.
Chromohalobacter salexigens DSM 3043 was obtained from the German Culture Collection (Braunschweig, Germany). Roseobacter sp. strain MED193 (http://www.roseobase.org/roseo/med193.html) was kindly supplied by J. Pinhassi (University of Kalmar, Kalmar, Sweden). Roseovarius sp. strain 217 (46) was kindly made available by J. C. Murrell (University of Warwick, Coventry, United Kingdom). These three organisms were grown in Tris-buffered artificial seawater (33). Ruegeria (Silicibacter) pomeroyi DSS-3 (19, 58) was kindly provided by M. A. Moran and grown in modified Silicibacter basal medium.
Enzyme assays.
Sulfoacetaldehyde acetyltransferase (Xsc, EC 2.3.3.15) was assayed as the formation of sulfite or acetylphosphate from sulfoacetaldehyde (45). (S)-Cysteate sulfo-lyase (CuyA, EC 4.4.1.25) was assayed as the release of sulfite from (S)-cysteate (14). 3-Sulfolactate sulfo-lyase (SuyAB, EC 4.4.1.24) was assayed as the release of sulfite from sulfolactate; an extract of Paracoccus pantotrophus NKNCYSA (42) was used as a positive control. The photometric assay for sulfolactate dehydrogenase (SlcD) contained 50 mM Tris-HCl (pH 8 or pH 9), 20 mM sulfolactate, 1 mM ferricyanide (ɛ420 = 0.9 mM−1 cm−1), or 0.1 mM dichlorophenol indophenol (ɛ600 = 16.1 mM−1 cm−1) and 0.05 to 0.5 mg protein ml−1. To demonstrate substrate disappearance and product formation by ion chromatography, the assay consisted of 0.5 mM sulfolactate, 5 mM ferricyanide, and membrane fraction (0.6 mg protein ml−1) in 50 mM Tris-HCl, pH 8.0. The assay for cysteate:2-oxoglutarate aminotransferase (Coa, EC 2.6.1.-) was adapted from that used by Mikosch et al. (38), and the formation of glutamate was measured discontinuously by high-pressure liquid chromatography (HPLC) after derivatization. Glutamate dehydrogenase (Gdh, EC 1.4.1.4) was assayed photometrically (47). The assay for sulfopyruvate decarboxylase (ComDE; EC 4.1.1.79) was contained in 50 mM Tris-HCl (pH 7.5), 1 mM thiamine diphosphate, 1 to 5 mM sulfopyruvate, and protein (0.1 to 1 mg ml−1). Routinely the disappearance of sulfopyruvate was followed by ion chromatography. Occasionally the formation of sulfoacetaldehyde was measured by HPLC after derivatization. Phosphate acetyltransferase (Pta, EC 2.3.1.8) was assayed photometrically in fresh extracts according to standard methods (4). Sulfite dehydrogenase (Sor, EC 1.8.2.1) was assayed photometrically with K3Fe(CN)6 as an electron acceptor (41), with modifications that are described elsewhere (16). Taurine:pyruvate aminotransferase (Tpa, EC 2.6.1.77) was assayed discontinuously by HPLC for alanine formation after derivatization (53). Simultaneous operation of both branches of the bifurcated pathway was explored with 3-sulfopyruvate as a substrate in crude extract of sulfolactate-grown cells at 37°C; the reaction mixture contained 50 mM Tris-HCl buffer (pH 8.0), 10 mM sulfopyruvate, and 5 mM glutamate, and samples were taken at zero time and 15 min to determine the concentrations of cysteate and sulfoacetaldehyde.
Separation and purification of enzymes.
The soluble fraction of R. nubinhibens ISM in 50 mM potassium phosphate buffer, pH 7.2, was loaded onto an anion-exchange chromatography column (Mono Q, HR 10/10; Pharmacia) at a flow rate of 1.0 ml min−1. A step gradient of sodium sulfate up to 0.5 M was applied, and active separated fractions of CuyA, Xsc, and ComDE were eluted.
Active fractions of CuyA were pooled, rebuffered in 50 mM Tris-sulfate (pH 9.0) on PD10 columns (Sephadex G-25; GE Healthcare, München, Germany), and subjected to a second anion-exchange chromatography step at pH 9.0 (see above). The same increasing sodium sulfate gradient was applied, and CuyA eluted at about 100 mM sodium sulfate.
For purification of ComDE, the soluble fraction was brought to 1.7 M ammonium sulfate, the precipitate was spun off, and the supernatant was subject to hydrophobic interaction chromatography on Phenyl Superose HR 10/10 (Pharmacia). A linear decreasing gradient of ammonium sulfate in potassium phosphate buffer, pH 7.2, was applied, and ComDE eluted at 0 mM ammonium sulfate. The rebuffered active fraction was loaded onto the anion-exchange chromatography column as a second purification step, and ComDE eluted at about 250 mM sodium sulfate. Active fractions were concentrated using Vivaspin units (10-kDa cutoff, PES membrane; Sartorius, Göttingen, Germany). Gel filtration was used as a third purification step (Superose 12 HR 10/30; Pharmacia) in 50 mM potassium phosphate buffer, pH 7.2, including 150 mM sodium sulfate at a flow rate of 0.4 ml min−1. Standard high-molecular-weight proteins (conalbumin, aldolase, catalase, and ferritin) were used to generate a calibration curve, and the molecular weight of native ComDE was estimated by interpolation.
To separate SlcD, the solubilized membrane fraction was loaded onto an anion-exchange chromatography column with the buffers described above, including 0.1% (vol/vol) Triton X-100, or onto a gel filtration column with the buffer system described above, including 0.1% (vol/vol) Triton X-100 at a flow rate of 0.2 ml min−1.
Analytical methods.
Protein in whole cells was quantified by a Lowry-type method (31) without the initial acid treatment to avoid precipitation of salts originating from the medium. Soluble protein was assayed by protein dye binding (6). Denatured proteins were separated by 16% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and stained with Coomassie-brillant blue R250 (34). Stained protein bands were cut out of the gel and subjected to peptide mass fingerprinting to identify the corresponding genes, which was done under contract at TopLab (Martinsried, Germany). Sulfolactate, sulfopyruvate, and isethionate were quantified by ion chromatography with suppression (15, 50). Sulfate release during growth was measured as turbidity of a suspension of insoluble BaSO4 (49); a 1:10 dilution of the sample was necessary to avoid a precipitate with other components of the medium. Sulfite was quantified as the fuchsin adduct (12). Acetylphosphate was determined chemically as iron(III) acetyl hydroxamate (40). Reversed-phase HPLC was used to quantify derivatized alanine, glutamate, or cysteate (35) or derivatized sulfoacetaldehyde (11). Reverse transcription-PCR (RT-PCR) was done as detailed elsewhere (33) using the primer pairs shown in Table S1 in the supplemental material. The RNA was tested prior to use for residual DNA with primer pair ISM-pta-F and ISM-pta-R. Chromosomal DNA from R. nubinhibens ISM was used as a positive control for PCRs.
Bioinformatic analyses.
Analysis of the draft genome sequence (accession no. NZ_AALY00000000) of R. nubinhibens ISM was done using the BLAST algorithm on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). The BLAST server on the Transport Classification Database (http://www.tcdb.org), which is coupled to a predictor of transmembrane helices, was also used. Sequence data up to 2 April 2009 were used. Sequence data were manipulated with different subroutines from the LASERGENE program package (DNAStar, Madison, WI). Alignments were made using ClustalX and plotted in NJPlot (51). Primers for RT-PCR and PCR were designed using the program Amplify (version 1.2).
RESULTS
Growth of R. nubinhibens ISM in minimal medium.
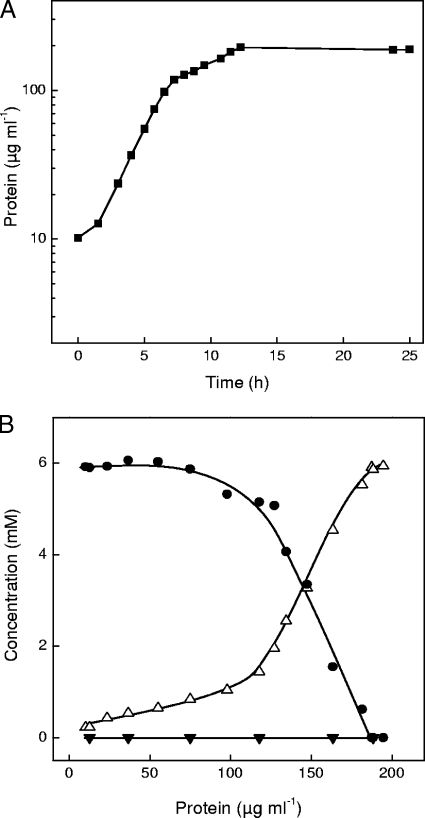
Given the genome sequence of R. nubinhibens ISM, we postulated the utilization of at least three organosulfonates as sole sources of carbon and energy for growth. Preliminary experiments in minimal medium gave nonreproducible growth, but the supplement of yeast extract solved the problem. Strain ISM then grew reproducibly with (R,S)-sulfolactate, (S)-cysteate, taurine, or acetate as the sole source of carbon and energy, with similar molar growth yields (about 6 g protein [mol C]−1), typical for the quantitative utilization of carbon (7). In all appropriate cases, quantitative recovery of the sulfonate moiety as sulfate was observed. The growth rate (μ) with 0.05% yeast extract alone was about 0.23 h−1, and about 0.3 mM sulfate was excreted (not shown). When sulfolactate was present, the growth rate was initially about 0.23 h−1, at which a low release of sulfate was observed, although no significant disappearance of sulfolactate was detected (Fig. 2). The growth rate then dropped to 0.10 h−1, and both substrate disappearance and sulfate release were concomitant with growth and quantitative (Fig. 2). Under these last conditions, the growth yield was 6 g protein (mol C)−1, so with the growth yield, a specific turnover rate of 1.4 mkat (kg protein)−1 for sulfolactate could be calculated. We attributed the initial rapid growth largely to the utilization of organic components in the yeast extract, while the slower growth (after 6 h) obviously involved the utilization of both enantiomers of sulfolactate.
FIG. 2.
Growth of R. nubinhibens ISM with sulfolactate as the carbon source (A) and changes in concentrations of substrate and product as a function of growth (B). •, sulfolactate; ▵, sulfate; ▾, sulfite.
C. salexigens DSM 3043 utilized sulfolactate quantitatively with stoichiometric release of sulfate. We confirmed that Roseovarius sp. strain 217 (2) and R. pomeroyi DSS-3 (14) utilized (S)-cysteate quantitatively, but neither organism utilized sulfolactate extensively (i.e., ∼10%). Strains 217 and DSS-3 involve Xsc in taurine metabolism, as observed previously (2, 21). Roseobacter sp. strain MED193 did not grow with sulfolactate but did grow quantitatively with taurine as the sole source of carbon and energy.
Induction of desulfonative and other enzymes.
R. nubinhibens ISM was grown with (R,S)-sulfolactate, (S)-cysteate, taurine, or acetate as the sole source of carbon and energy; the cells were harvested and disrupted; and cell extracts were prepared. A set of 10 enzymes in the soluble and particulate fractions of these extracts was assayed (Table 1). No activity of Xsc or CuyA, each a desulfonative enzyme, was detected with extracts of acetate-grown cells, whereas Xsc was found in extracts of taurine-grown cells and of sulfolactate-grown cells, and CuyA was found in extracts of cysteate-grown cells and of sulfolactate-grown cells. Xsc and CuyA were thus inducible. Further, no activity of SuyAB was detected (Table 1).
TABLE 1.
Specific activities of enzymes in crude extracts of R. nubinhibens ISM grown with different sole sources of carbon and energy
| Enzyme | Specific enzyme activity (mkat [kg protein]−1) in extracts from cells grown with:
|
|||
|---|---|---|---|---|
| Sulfolactate | Taurine | Cysteate | Acetate | |
| Xsc | 0.9 | 1.5 | 0.1 | BLDa |
| CuyA | 1.0 | BLD | 5.2 | BLD |
| SuyAB | BLD | BLD | BLD | BLD |
| SlcDb | 0.8 | 0.1 | 0.1 | 0.1 |
| Coa | 1.2 | 1.0 | 0.8 | 1.3 |
| Gdh | 0.4 | 0.5 | 0.5 | 0.4 |
| ComDE | 0.9 | BLD | BLD | BLD |
| Pta | 6.9 | 0.5 | BLD | BLD |
| Sor | 0.4 | 0.4 | 0.3 | 0.2 |
| Tpa | 0.5 | 4.9 | 0.2 | 0.1 |
BLD, below the limit of detection.
Activities in the particulate fraction.
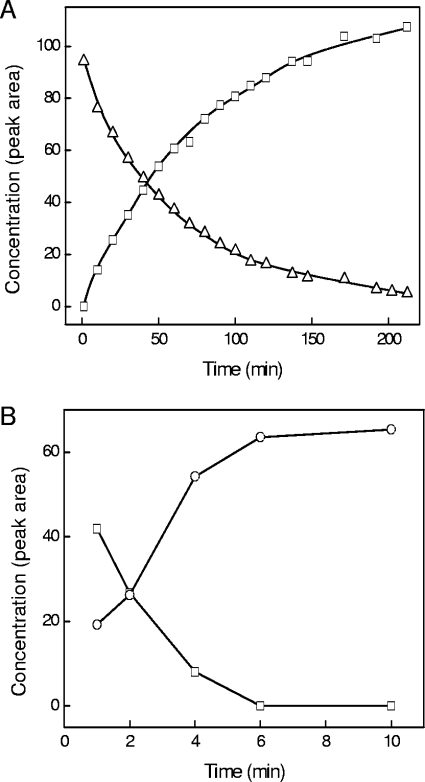
The postulated bifurcated pathway to degrade sulfolactate involves a sulfolactate dehydrogenase to generate sulfopyruvate (Fig. 1A). Sulfolactate dehydrogenase (SlcD) activity, which was dependent on an artificial electron acceptor (ferricyanide), was detected with sulfolactate-grown cells only (Table 1). The enzyme is thus inducible. The reaction observed was a conversion of sulfolactate to sulfopyruvate (Fig. 3A). There was no sulfolactate dehydrogenase activity with NADP+ (i.e., orthologous to the ComC of Methanocaldococcus jannaschii [MJ1425] [24]), NAD+, flavin adenine dinucleotide, flavine mononucleotide (FMN), or cytochrome c.
FIG. 3.
Transformations of sulfolactate (▵) to sulfopyruvate (□) during the reaction of sulfolactate dehydrogenase (SlcD) (A) and of sulfopyruvate (□) to sulfoacetaldehyde (○) during the reaction of sulfopyruvate decarboxylase (ComDE) (B) from R. nubinhibens ISM.
SlcD was found in both the soluble and the particulate fractions, but the specific activity in the particulate fraction (0.8 mkat [kg protein]−1; Table 1) was fivefold higher than that in the soluble fraction, so the enzyme was considered to be membrane associated. SlcD was attributed to the (membrane-bound) 2-hydroxyacid dehydrogenases (see below); there was no activity with (R,S)-mandelate or (R,S)-malate, but (S)-lactate was oxidized with about 10% of the rate with sulfolactate.
The CuyA branch of the pathway to degrade sulfopyruvate requires a cysteate transaminase. A constitutive (S)-cysteate:2-oxoglutarate aminotransferase (Coa) in bacteria is known (38), and this enzyme was found to be constitutive in R. nubinhibens ISM (Table 1). Correspondingly, a constitutive glutamate dehydrogenase (Gdh) was observed, and it recycled the amino group to generate (S)-cysteate from sulfopyruvate (Fig. 1A; Table 1). No pyruvate-coupled (S)-cysteate transaminase was detected.
The Xsc branch of the pathway requires activity of the sulfopyruvate decarboxylase (ComDE) noted in the introduction (Fig. 1A). Activity of ComDE could be detected as substrate disappearance in extracts of sulfolactate-grown cells only (Table 1), so the enzyme was considered to be inducible.
Phosphate acetyltransferase (Pta) is usually essential in the degradation of sulfoacetaldehyde (Fig. 1A) (8; see also reference 2). Inducible enzyme activity was detected (Table 1).
The second product from Xsc and also a product from CuyA is sulfite (Fig. 1A). This is presumed to be oxidized periplasmically by a sulfite dehydrogenase, some of which seems to be difficult to assay (16, 28). This enzyme was detected with extracts of cells from all cultures examined (Table 1).
The pathway predicted for the degradation of taurine involves taurine:pyruvate aminotransferase (Tpa), which is encoded separately from Xsc. Activity of this enzyme in extracts of acetate-grown, (S)-cysteate-grown, and sulfolactate-grown cells was detectable, but the enzyme was strongly induced in taurine-grown cells (Table 1).
An experiment was done with the crude extract of sulfolactate-grown cells to which sulfopyruvate and glutamate were added. A representative intermediate of each branch of the pathway, cysteate and sulfoacetaldehyde (0.1 mM), was formed. Both branches of the bifurcated pathway were thus in operation simultaneously.
Separation, purification, and identification of enzymes in the bifurcated pathway.
Proteins in the soluble extract of sulfolactate-grown cells were loaded onto an anion-exchange column. Separated fractions with activities of Xsc, CuyA, or ComDE were examined.
All fractions containing activity of Xsc contained the characteristic 63-kDa band (SDS-PAGE) known from earlier work (e.g., reference 45); other fractions did not. The protein was subject to peptide mass fingerprinting (see Fig. S1 in the supplemental material), which confirmed that it represented the gene product of the candidate xsc gene (ISM_10690) (Fig. 1B; Table 2).
TABLE 2.
Organisms with orthologues of SlcD, and orthologues of relevant enzymes of the bifurcated sulfolactate degradative pathwayc
| Organism | Organism tag | Locus tag encoding indicated enzyme:
|
||||
|---|---|---|---|---|---|---|
| SlcD | ComE | ComD | Xsc | CuyA | ||
| Roseovarius nubinhibens ISM | ISM_ | 13310 | 13330 | 13335 | 10690 | 09626 |
| Phaeobacter gallaeciensis BS107 | RGBS107_ | 10911 | 10916 | 10921 | 03088 | 05569a |
| Roseobacter denitrificans OCh 114 | RD1_ | 3814 | 3813 | 3812 | 0826 | 0819 |
| Roseobacter litoralis Och 149 | RLO149_ | 14678 | 14683 | 14688 | 15453 | 15243 |
| Roseobacter sp. strain SK 209-2-6 | RSK20926_ | 07142 | 07147 | 07152 | 10419 | 14094 |
| Rhodobacterales bacterium HTCC2083 | RB2083_ | 2743 | 2808 | 708 | 3832 | 3184 |
| Rhodobacterales bacterium Y41 | RBY41_ | 4170 | 4207 | 4138 | 3832 | 1128 |
| Roseovarius sp. strain 217 | ROS217_ | 11241 | None | None | 11936 | 09350 |
| Roseovarius sp. strain TM1035 | RTM1035_ | 17047 | None | None | 16562 | 19346 |
| Ruegeria pomeroyi DSS-3 | SPO | 0598 | None | None | 3561 | A0158 |
| Jannaschia sp. strain CCS1 | Jann_ | 1403 | 1402 | 1401 | 2846 | None |
| Octadecabacter antarcticus 238 | OA238_ | 2138 | —b | 1847 | 2599 | None |
| Octadecabacter antarcticus 307 | OA307_ | 2251 | —b | 3307 | 4001 | None |
| Phaeobacter gallaeciensis 2.10 | RG210_ | 02412 | 02407 | 02402 | 10327 | None |
| Rhodobacterales bacterium HTCC2150 | RB2150_ | 17119 | 17114 | 17109 | 15441 | None |
| Rhodobacterales bacterium KLH11 | RKHL11_ | 3493 | 3454 | 3575 | 2875 | None |
| Roseobacter sp. strain CCS2 | RCCS2_ | 12604 | 12609 | 12614 | 04724 | None |
| Roseobacter sp. strain MED193 | MED193_ | 17034 | 17039 | 17044 | 12208 | None |
A paralogous hypothetical protein (19978) is also present.
—, comE is present but not annotated.
Contiguous clusters are shown in bold type. Abbreviations of enzyme names are given in the text.
All fractions containing activity of CuyA contained the characteristic 35-kDa band (SDS-PAGE) known from earlier work (14); other fractions did not. Active fractions were combined and subjected to a second purification step by anion-exchange at a higher pH value. An enrichment factor of about 10-fold was achieved; the prominent 35-kDa band was excised and subjected to peptide-mass fingerprinting (not shown), which confirmed that it represented the gene product of the candidate cuyA gene (ISM_09626) (Fig. 1B; Table 2).
The activity of ComDE, detected as substrate disappearance (described above), was confirmed to involve the concomitant release of sulfoacetaldehyde (Fig. 3B). This product was tentatively identified, after derivatization, by cochromatography (HPLC) with derivatized authentic material. The identification was confirmed by its reaction with the specific sulfoacetaldehyde reductase (EC 1.1.1.-) of Chromohalobacter salexigens DSM 3043 (Z. Krejčík and A. M. Cook, unpublished data) to form isethionate, which was identified by ion chromatography.
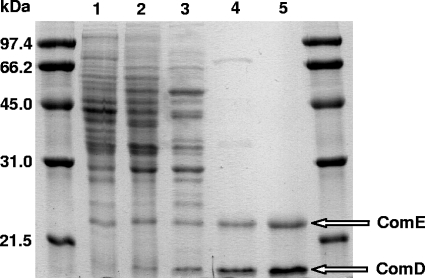
A three-step purification involving hydrophobic interaction chromatography, anion-exchange chromatography, and gel filtration chromatography yielded the two subunits of ComDE purified to homogeneity (Fig. 4). The proteins were subject to peptide mass fingerprinting (not shown), which confirmed that the subunits were indeed the products of the candidate comDE genes (ISM_13335 and ISM_13330, respectively) (Fig. 1B; Table 2).
FIG. 4.
Electropherogram of denatured proteins of R. nubinhibens ISM at different stages of purification of ComDE. The sizes of the molecular mass markers are indicated on the left. Lane 1, soluble fraction of acetate-grown cells; lane 2, soluble fraction of sulfolactate-grown cells; lane 3, active fraction after hydrophobic-interaction chromatography; lane 4, active fraction after anion-exchange chromatography; lane 5, purified enzyme after gel filtration chromatography.
The enrichment factor was about 150-fold. The native enzyme was estimated by gel filtration chromatography to be about 235 kDa (not shown), which suggests the same α6β6 structure (calculated as 230 kDa) found in the enzyme from the archaeon M. jannaschii (25). The purified enzyme was stable under an air atmosphere; it could be stored at 4°C for at least a month without loss of activity.
Membrane-associated SlcD could be solublized from the membrane and applied to an anion-exchange column, and activity was eluted from the column. However, the separated fractions represented no purification, in that the pattern of protein bands was largely identical with that of the starting material. A 10-fold increase in the specific activity of SlcD after gel filtration still did not allow a reasonable candidate protein to be postulated directly (not shown), but the presence of inducible, membrane-bound sulfolactate dehydrogenase activity and annotation of a gene as (membrane-bound) 2-hydroxy-acid dehydrogenase allowed a gene candidate to be nominated (see below).
Inducible transcription of relevant genes shown by RT-PCR.
The biochemical identification of four gene products (ComDE, Xsc, and CuyA) in sulfonate degradation (Fig. 1) was complemented by observing inducible transcription of the genes (comDE, xsc, and cuyA) by RT-PCR (not shown). The three genes downstream of comDE were designated slcHFG, and the gene products were postulated to be a sulfolactate transporter in the tripartite tricarboxylate transporter (TTT) family (TC 2.A.80.-.- in the transport classification system) after using the BLASTP server on the Transport Classification Database. We could observe inducible transcription of slcH and slcG and detect the transcript of slcF (not shown), which we interpreted as support for the presence and presumed function of SlcHFG. The next and last gene in this cluster was designated slcD, because the annotation (membrane-bound 2-hydroxy-acid dehydrogenase) corresponded to properties of SlcD (membrane-bound sulfolactate dehydrogenase). The slcD gene was transcribed inducibly (see Fig. S2 in the supplemental material).
SlcD was considered to be another key to the bifurcated pathway shown in Fig. 1, so all orthologues, which could be identified with the BLASTP algorithm on the NCBI website, were tabulated (Table 2). The 18 sequences formed a tight cluster in a dendrogram (not shown).
The gene downstream of xsc is annotated as pta. Both xsc and pta are transcribed inducibly, corresponding to the enzyme data shown in Table 1 and supporting the role of these enzymes in sulfolactate metabolism in strain ISM (Fig. 1).
The gene downstream of cuyA is cuyZ. Each is transcribed inducibly (not shown). We conclude that CuyZ is a sulfite exporter (Fig. 1), as deduced previously (10, 14). Sulfite dehydrogenase (SorAB), which is under further study, is currently attributed to ISM_01020 and ISM_01015 (see reference 16).
DISCUSSION
The growth curve showing utilization of sulfolactate by a bacterium (Fig. 2A) is apparently the first to be published. The need to synthesize sulfolactate, which is not commercially available, presumably explains this oddity. The phase representing growth with sulfolactate shows that substrate utilization and product (sulfate) formation are concomitant with growth (Fig. 2B), which proves the stoichiometry and the mass balances in this system. Further, the derived, specific turnover rate for sulfolactate, 1.4 mkat (kg protein)−1, is of the same order of magnitude as specific activities of relevant enzymes in the degradative process (Table 1).
Two pathways including sulfolactate degradation have been sketched in the literature. The first involves desulfonation via SuyAB (42), which is absent here (introduction; Table 1). The second involves transformation to (S)-cysteate (9) via oxidation and transamination. (S)-Cysteate is desulfonated to pyruvate by CuyA, and the export of sulfite is attributed to CuyZ (14). This second pathway is obviously functional in R. nubinhibens ISM (Fig. 1A, right; Table 1).
In addition, a novel (third) pathway is described above. It involves the same oxidation (to sulfopyruvate) found in the second pathway, but decarboxylation to sulfoacetaldehyde and desulfonation by Xsc are involved (Fig. 1A, left; Table 1). The oxidation (Fig. 3A), decarboxylation (Fig. 3B), and desulfonation (Table 1) have clearly been demonstrated; the sulfopyruvate decarboxylase has been purified (Fig. 4). This degradative pathway (Fig. 1) involves some previously known components (see below), but for the first time, there is a candidate uptake system for sulfolactate, SlcHFG.
Graham et al. (23) consider that the biogenic sulfonate substituent is present to prevent a molecule from crossing a membrane, so a transport system is essential to allow biodegradation of an extracellular organosulfonate by intracellular enzymes. The candidate sulfolactate transporter, SlcHFG (Fig. 1), whose genes are inducibly transcribed (see above), is apparently a member of TC 2.A.80.-.-, whose sole defined member (TctABC) (56) has components of 504, 144, and 325 amino acid residues, with 12, 4, and 0 transmembrane helices, respectively; the predicted data for SlcHFG are 502 amino acid residues/11 transmembrane helices (SlcF), 152/4 (SlcG), and 305/0 (SlcH, a periplasmic binding protein). SlcHFG is not widespread in the organisms listed in Table 2, being present only in strain ISM and Roseobacter sp. strain CCS2. The slcHFG genes are also found in the genome sequence of C. salexigens DSM 3043 contiguous with the suyAB genes, and C. salexigens is found to utilize sulfolactate; we therefore consider this indirect support for the transport candidate (see the supplemental material). Available organisms lacking the slcHFG genes (Roseovarius sp. strain 217, R. pomeroyi DSS-3, and Roseobacter sp. strain MED193) do not utilize sulfolactate, and as in many organisms listed in Table 2 which contain slcD-comED, the latter genes are located at the end of a long gene cluster. We postulate that the sulfolactate in many organisms is generated intracellularly from a precursor, presumably 2,3-dihydroxysulfopropane (see below), which is utilized by the relevant organisms tested to date, Ruegeria pomeroyi DSS-3 and Roseovarius sp. strain 217 (K. Denger, unpublished data).
The inducible sulfolactate dehydrogenase shown in Fig. 1 is attributed by us to the gene product of ISM_13310 (slcD). Its annotation suggests that FMN is the electron acceptor, but the enzyme was assayed with the artificial electron acceptor, ferricyanide, because the tested flavins elicited no activity. The (R)-sulfolactate dehydrogenase best known in the literature is the NAD(P)-coupled EC 1.1.1.272 (ComC); ComC can also oxidize (S)-malate (22, 24), which is not a substrate for SlcD from strain ISM. The archaeal enzyme is involved in the biosynthesis of coenzyme M, hence the abbreviation “Com.” An unsolved problem is that our chemically synthesized sulfolactate is racemic (42), whereas the compound is degraded completely (Fig. 2). We presume that SlcD is enantiomer specific, and we postulate the presence of an unknown sulfolactate racemase (or an equivalent pathway).
The following enzyme in the novel pathway, ComDE (Fig. 1A, 3B, and 4), was first characterized as an archaeal gene product which is similar in size and structure (α6β6) but oxygen sensitive (25). The enzyme in strain ISM is stable in air, and the orthologues referred to in Table 2 all cluster in a dendrogram apart from the archaeal sequences (see Fig. S3 in the supplemental material). The function of ComDE in this pathway (Fig. 1A) is to convert a C3 compound to a C2 compound, which is the substrate for desulfonative Xsc (Fig. 1A). We presume that the sulfite released by Xsc is exported via CuyZ. The carbon moiety formed by Xsc, acetyl phosphate, is converted to acetyl-coenzyme A by Pta and thus made available for carbon skeletons and energy conservation via the Krebs cycle, anaplerosis via malyl-coenzyme A lyase (EC 4.1.3.24) (1), and fatty acid formation.
The pathway to desulfonation by CuyA, which operates simultaneously with the novel pathway, involves a cysteate aminotransferase (Coa) (Fig. 1A; Table 1). Literature data suggest that Coa is a known enzyme, aspartate:2-oxoglutarate aminotransferase (EC 2.6.1.1) (Aoa) (29, 57), and this hypothesis is currently being tested (J. Mayer, unpublished data).
When CuyA was discovered in several terrestrial organisms, cuyA had no orthologues in genome sequences (14). It is clear from the data shown in Table 2 that cuyA is widespread in marine bacteria. A dendrogram (see Fig. S4 in the supplemental material) indicates that some 14 CuyA orthologues are clearly separated from at least three other enzymes. We presume that CuyZ then exports the sulfite released by CuyA (Fig. 1); cuyZ is subject to inducible transcription under these growth conditions.
Six organisms share with R. nubinhibens ISM the combined genotype of slcD-comDE, with xsc and cuyA (Table 2). These seven organisms are presumed to degrade sulfolactate via the bifurcated pathway. Whereas R. nubinhibens ISM encodes SlcD and ComDE in one cluster separated by the three transporter genes (Fig. 1B), the other six organisms share contiguous slcD-comED genes. Xsc is encoded (with Pta) separately, as in R. nubinhibens ISM. CuyA is also encoded separately.
Three organisms (two Roseovarius spp. and R. pomeroyi) contain the slcD gene, but not the comDE genes, although orthologues of xsc and cuyA are present. We presume that the degradation of sulfolactate proceeds via cysteate only. Roseovarius sp. strain 217 and R. pomeroyi DSS-3 utilize (S)-cysteate quantitatively, but neither organism utilizes sulfolactate extensively (i.e., ∼10%). None of these organisms encodes SlcHFG, so we presume that SlcD catalyzes transformation of internally generated sulfolactate. Roseovarius sp. strain 217 and R. pomeroyi DSS-3 involve Xsc in taurine metabolism (2, 21).
At least six organisms contain the contiguous slcD-comDE genes and the xsc gene, but not the cuyA gene (Table 2); no orthologues of suyAB were detected either. This is interpreted as the degradation of sulfolactate solely via Xsc. Roseobacter sp. strain MED193 grows with taurine, so it presumably expresses Xsc, but the organism does not utilize sulfolactate. No orthologue of SlcHFG is available, so sulfolactate is presumably generated intracellularly from a precursor (e.g., 2,3-dihydroxysulfopropane).
We feel unable to speculate on the “reason” for or the advantages of the presence of the bifurcated pathway, because many organisms grow well with a single pathway (Table 2). Each branch of the pathway is inducible individually (Table 1), so the bifurcated pathway is possibly serendipitous.
Fig. 1B shows a gene (ISM_13340) encoding a putative transcriptional regulator (SlcR; LysR type) of the six upstream genes. There are very few orthologues of this gene (see the supplemental material), possibly because most clusters involving comDE-slcD are larger and presumably under the control of a different regulator. This regulator is also present in R. nubinhibens ISM, presumably encoding enzymes to generate sulfolactate from its precursor.
We suspect that Fig. 1A represents aspects of bacterial sulfoglycolysis and expands the work of Roy et al. (44) by providing information on some fates (e.g., desulfonation) of several C3 sulfonates. In that work, two sulfonates, sulfolactate and 2,3-dihydroxysulfopropane (44), were observed to be excreted, and we postulate that the latter is a precursor of sulfolactate in bacterial sulfoglycolysis. The term sulfoglycolysis was coined by Benson's group (3) for transformations of sulfoquinovose in plants and algae. Benson and Lee (3) describe (S)-cysteate as an intermediate and 2,3-dihydroxysulfopropane as an excretion product of all algae; both compounds derive from sulfoquinovose. We therefore propose that the reactions represented in Fig. 1A are bacterial models for aspects of plant and algal sulfoglycolysis. We also speculate that sulfoquinovose and derived products from phototrophs represent a significant source of carbon in the oceans, where Pelagibacter ubique and the Roseobacter clade, which contain orthologues of, e.g., xsc, cuyA, and suyAB (Table 2), represent >40% of the bacterial population (18). Indeed, the Roseobacter clade was overrepresented in association with an algal bloom (20), which was presumably (3) excreting 2,3-dihydroxysulfopropane.
Supplementary Material
Acknowledgments
We are grateful to Christine Gielisch for data generated in an advanced practical class.
Funding was provided by the University of Konstanz and the German Research Foundation (J.M., M.B., and S.W.) (CO 206/6-1 and CO 206/7-1 to A.M.C. and T.H.M.S.), in part under the auspices of the Konstanz Research School, Chemical Biology (J.M.). We are also grateful to many sequencing organizations, but especially to the Moore Foundation for funding the sequencing of the genome of R. nubinhibens ISM and making the data generally available.
Footnotes
Published ahead of print on 6 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alber, B. E., R. Spanheimer, C. Ebenau-Jehle, and G. Fuchs. 2006. Study of an alternative glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol. Microbiol. 61297-309. [DOI] [PubMed] [Google Scholar]
- 2.Baldock, M. I., K. Denger, T. H. M. Smits, and A. M. Cook. 2007. Roseovarius sp. strain 217: aerobic taurine dissimilation via acetate kinase and acetate-CoA ligase. FEMS Microbiol. Lett. 271202-206. [DOI] [PubMed] [Google Scholar]
- 3.Benson, A. A., and R. F. Lee. 1972. The sulphoglycolytic pathway in plants. Biochem. J. 12829P-30P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmeyer, H. U. 1983. Determination of metabolite concentrations with end-point methods, p. 163-181. In H. U. Bergmeyer (ed.), Methods of enzymic analysis, 3rd ed., vol. 1. Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 5.Bonsen, P. P. M., J. A. Spudich, D. L. Nelson, and A. Kornberg. 1969. Biochemical studies of bacterial sporulation and germination XII. A sulfonic acid as a major sulfur compound of Bacillus subtilis spores. J. Bacteriol. 9862-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cook, A. M. 1987. Biodegradation of s-triazine xenobiotics. FEMS Microbiol. Rev. 4693-116. [Google Scholar]
- 8.Cook, A. M., and K. Denger. 2002. Dissimilation of the C2 sulfonates. Arch. Microbiol. 1791-6. [DOI] [PubMed] [Google Scholar]
- 9.Cook, A. M., K. Denger, and T. H. M. Smits. 2006. Dissimilation of C3-sulfonates. Arch. Microbiol. 18583-90. [DOI] [PubMed] [Google Scholar]
- 10.Cook, A. M., T. H. M. Smits, and K. Denger. 2007. Sulfonates and organotrophic sulfite metabolism, p. 170-181. In C. Dahl and C. G. Friedrich (ed.), Microbial sulfur metabolism. Springer Verlag, Berlin, Germany.
- 11.Cunningham, C., K. F. Tipton, and H. B. F. Dixon. 1998. Conversion of taurine into N-chlorotaurine (taurine chloramine) and sulphoacetaldehyde in response to oxidative stress. Biochem. J. 330939-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denger, K., and A. M. Cook. 2001. Ethanedisulfonate is degraded via sulfoacetaldehyde in Ralstonia sp. strain EDS1. Arch. Microbiol. 17689-95. [DOI] [PubMed] [Google Scholar]
- 13.Denger, K., J. Ruff, U. Rein, and A. M. Cook. 2001. Sulfoacetaldehyde sulfo-lyase (EC 4.4.1.12) from Desulfonispora thiosulfatigenes: purification, properties and primary sequence. Biochem. J. 357581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denger, K., T. H. M. Smits, and A. M. Cook. 2006. l-Cysteate sulfo-lyase, a widespread, pyridoxal 5′-phosphate-coupled desulfonative enzyme purified from Silicibacter pomeroyi DSS-3T. Biochem. J. 394657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denger, K., S. Weinitschke, K. Hollemeyer, and A. M. Cook. 2004. Sulfoacetate generated by Rhodopseudomonas palustris from taurine. Arch. Microbiol. 182254-258. [DOI] [PubMed] [Google Scholar]
- 16.Denger, K., S. Weinitschke, T. H. M. Smits, D. Schleheck, and A. M. Cook. 2008. Bacterial sulfite dehydrogenases in organotrophic metabolism: separation and identification in Cupriavidus necator H16 and in Delftia acidovorans SPH-1. Microbiology (Reading, England) 154256-263. [DOI] [PubMed] [Google Scholar]
- 17.Eichhorn, E., J. R. van der Ploeg, M. A. Kertesz, and T. Leisinger. 1997. Characterization of α-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J. Biol. Chem. 27223031-23036. [DOI] [PubMed] [Google Scholar]
- 18.Giovannoni, S., and M. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, NY.
- 19.González, J. M., J. S. Covert, W. B. Whitman, J. R. Henriksen, F. Mayer, B. Scharf, R. Schmitt, A. Buchan, J. A. Fuhrman, R. P. Kiene, and M. A. Moran. 2003. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 531261-1269. [DOI] [PubMed] [Google Scholar]
- 20.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 664237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorzynska, A. K., K. Denger, A. M. Cook, and T. H. M. Smits. 2006. Inducible transcription of genes involved in taurine uptake and dissimilation by Silicibacter pomeroyi DSS-3T. Arch. Microbiol. 185402-406. [DOI] [PubMed] [Google Scholar]
- 22.Graham, D. E., and R. H. White. 2002. Elucidation of methanogenic coenzyme biosyntheses: from spectroscopy to genomics. Nat. Prod. Rep. 19133-147. [DOI] [PubMed] [Google Scholar]
- 23.Graham, D. E., H. Xu, and R. H. White. 2002. Identification of coenzyme M biosynthetic phosphosulfolactate synthase: a new family of sulfonate biosynthesizing enzymes. J. Biol. Chem. 27713421-13429. [DOI] [PubMed] [Google Scholar]
- 24.Graupner, M., H. Xu, and R. H. White. 2000. Identification of an archaeal 2-hydroxy acid dehydrogenase catalyzing reactions involved in coenzyme biosynthesis in methanoarchaea. J. Bacteriol. 1823688-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graupner, M., H. Xu, and R. H. White. 2000. Identification of the gene encoding sulfopyruvate decarboxylase, an enzyme involved in biosynthesis of coenzyme M. J. Bacteriol. 1824862-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Junker, F., T. Leisinger, and A. M. Cook. 1994. 3-Sulphocatechol 2,3-dioxygenase and other dioxygenases (EC 1.13.11.2 and EC 1.14.12.-) in the degradative pathways of 2-aminobenzenesulphonic, benzenesulphonic and 4-toluenesulphonic acids in Alcaligenes sp. strain O-1. Microbiology (Reading, England) 1401713-1722. [DOI] [PubMed] [Google Scholar]
- 27.Kahnert, A., and M. A. Kertesz. 2000. Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J. Biol. Chem. 27531661-31667. [DOI] [PubMed] [Google Scholar]
- 28.Kappler, U., B. Bennett, J. Rethmeier, G. Schwarz, R. Deutzmann, A. G. McEwan, and C. Dahl. 2000. Sulfite:cytochrome c oxidoreductase from Thiobacillus novellus. Purification, characterization, and molecular biology of a heterodimeric member of the sulfite oxidase family. J. Biol. Chem. 27513202-13212. [DOI] [PubMed] [Google Scholar]
- 29.Kearney, E. B., and P. P. Singer. 1953. Enzymic transformations of l-cysteinesulfinic acid. Biochim. Biophys. Acta 11276-289. [DOI] [PubMed] [Google Scholar]
- 30.Kelly, D. P., and J. C. Murrell. 1999. Microbial metabolism of methanesulfonic acid. Arch. Microbiol. 172341-348. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy, S. I. T., and C. A. Fewson. 1968. Enzymes of the mandelate pathway in bacterium N.C.I.B. 8250. Biochem. J. 107497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kertesz, M. A. 2004. Metabolism of sulphur-containing organic compounds, p. 323-357. In J. Ramos (ed.), Pseudomonas, vol. 3. Wiley, New York, NY. [Google Scholar]
- 33.Krejčík, Z., K. Denger, S. Weinitschke, K. Hollemeyer, V. Pačes, A. M. Cook, and T. H. M. Smits. 2008. Sulfoacetate released during the assimilation of taurine-nitrogen by Neptuniibacter caesariensis: purification of sulfoacetaldehyde dehydrogenase. Arch. Microbiol. 190159-168. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 35.Laue, H., K. Denger, and A. M. Cook. 1997. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl. Environ. Microbiol. 632016-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martelli, H. L., and A. A. Benson. 1964. Sulfocarbohydrate metabolism 1. Bacterial production and utilization of sulfoacetate. Biochim. Biophys. Acta 93169-171. [DOI] [PubMed] [Google Scholar]
- 37.Mayer, J., K. Denger, T. H. M. Smits, K. Hollemeyer, U. Groth, and A. M. Cook. 2006. N-Acetyltaurine dissimilated via taurine by Delftia acidovorans NAT. Arch. Microbiol. 18661-67. [DOI] [PubMed] [Google Scholar]
- 38.Mikosch, C., K. Denger, E.-M. Schäfer, and A. M. Cook. 1999. Anaerobic oxidations of cysteate: degradation via a cysteate:2-oxoglutarate aminotransferase in Paracoccus pantotrophus. Microbiology (Reading, England) 1451153-1160. [DOI] [PubMed] [Google Scholar]
- 39.Quick, A., N. J. Russell, S. G. Hales, and G. F. White. 1994. Biodegradation of sulphosuccinate: direct desulphonation of a secondary sulphonate. Microbiology (Reading, England) 1402991-2998. [DOI] [PubMed] [Google Scholar]
- 40.Racker, E. 1962. Fructose-6-phosphate phosphoketolase from Acetobacter xylinum. Methods Enzymol. 5276-280. [PubMed] [Google Scholar]
- 41.Reichenbecher, W., D. P. Kelly, and J. C. Murrell. 1999. Desulfonation of propanesulfonic acid by Comamonas acidovorans strain P53: evidence for an alkanesulfonate sulfonatase and an atypical sulfite dehydrogenase. Arch. Microbiol. 172387-392. [DOI] [PubMed] [Google Scholar]
- 42.Rein, U., R. Gueta, K. Denger, J. Ruff, K. Hollemeyer, and A. M. Cook. 2005. Dissimilation of cysteate via 3-sulfolactate sulfo-lyase and a sulfate exporter in Paracoccus pantotrophus NKNCYSA. Microbiology (Reading, England) 151737-747. [DOI] [PubMed] [Google Scholar]
- 43.Rösch, V., K. Denger, D. Schleheck, T. H. M. Smits, and A. M. Cook. 2008. Different bacterial strategies to degrade taurocholate. Arch. Microbiol. 19011-18. [DOI] [PubMed] [Google Scholar]
- 44.Roy, A. B., M. J. E. Hewlins, A. J. Ellis, J. L. Harwood, and G. F. White. 2003. Glycolytic breakdown of sulfoquinovose in bacteria: a missing link in the sulfur cycle. Appl. Environ. Microbiol. 696434-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruff, J., K. Denger, and A. M. Cook. 2003. Sulphoacetaldehyde acetyltransferase yields acetyl phosphate: purification from Alcaligenes defragrans and gene clusters in taurine degradation. Biochem. J. 369275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schäfer, H., I. R. McDonald, P. D. Nightingale, and J. C. Murrell. 2005. Evidence for the presence of a CmuA methyltransferase pathway in novel marine methyl halide-oxidizing bacteria. Environ. Microbiol. 7839-852. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, E. 1974. Glutamat-dehydrogenase UV-test, p. 689-696. In H. U. Bergmeyer (ed.), Methoden der enzymatischen analyse. Verlag Chemie, Weinheim, Germany.
- 48.Shibuya, I., T. Yagi, and A. A. Benson. 1963. Sulfonic acids in algae, p. 627-636. In Japanese Society of Plant Physiologists (ed.), Studies on microalgae and photosynthetic bacteria. University of Tokyo Press, Tokyo, Japan.
- 49.Sörbo, B. 1987. Sulfate: turbidimetric and nephelometric methods. Methods Enzymol. 1433-6. [DOI] [PubMed] [Google Scholar]
- 50.Styp von Rekowski, K., K. Denger, and A. M. Cook. 2005. Isethionate as a product from taurine during nitrogen-limited growth of Klebsiella oxytoca Tau-N1. Arch. Microbiol. 183325-330. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinitschke, S., K. Denger, T. H. M. Smits, K. Hollemeyer, and A. M. Cook. 2006. The sulfonated osmolyte N-methyltaurine is dissimilated by Alcaligenes faecalis and by Paracoccus versutus with release of methylamine. Microbiology (Reading, England) 1521179-1186. [DOI] [PubMed] [Google Scholar]
- 53.Weinitschke, S., K. Styp von Rekowski, K. Denger, and A. M. Cook. 2005. Sulfoacetaldehyde is excreted quantitatively by Acinetobacter calcoaceticus SW1 during growth with taurine as sole source of nitrogen. Microbiology (Reading, England) 1511285-1290. [DOI] [PubMed] [Google Scholar]
- 54.Weinstein, C. L., and O. W. Griffith. 1988. Cysteinesulfonate and β-sulfopyruvate metabolism. Partitioning between decarboxylation, transamination, and reduction pathways. J. Biol. Chem. 2633735-3743. [PubMed] [Google Scholar]
- 55.White, R. H. 1986. Intermediates in the biosynthesis of coenzyme M (2-mercaptoethanesulfonic acid). Biochemistry 255304-5308. [Google Scholar]
- 56.Winnen, B., R. N. Hvorup, and M. H. Saier, Jr. 2003. The tripartite tricarboxylate transporter (TTT) family. Res. Microbiol. 154457-465. [DOI] [PubMed] [Google Scholar]
- 57.Yagi, T., H. Kagamiyama, and M. Nozaki. 1979. Cysteine sulfinate transamination activity of aspartate aminotransferases. Biochem. Biophys. Res. Commun. 90447-452. [DOI] [PubMed] [Google Scholar]
- 58.Yi, H., Y. W. Lim, and J. Chun. 2007. Taxonomic evaluation of the genera Ruegeria and Silicibacter: a proposal to transfer the genus Silicibacter Petursdottir and Kristjansson 1999 to the genus Ruegeria Uchino et al. 1999. Int. J. Syst. Evol. Microbiol. 57815-819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.