Abstract
The attaching and effacing (A/E) pathogen enteropathogenic Escherichia coli (EPEC) forms characteristic actin-filled membranous protrusions upon infection of host cells termed pedestals. Here we examine the role of the RNA binding protein CsrA in the expression of virulence genes and proteins that are necessary for pedestal formation. The csrA mutant was defective in forming actin pedestals on epithelial cells and in disrupting transepithelial resistance across polarized epithelial cells. Consistent with reduced pedestal formation, secretion of the translocators EspA, EspB, and EspD and the effector Tir was substantially reduced in the csrA mutant. Purified CsrA specifically bound to the sepL espADB mRNA leader, and the corresponding transcript levels were reduced in the csrA mutant. In contrast, Tir synthesis was unaffected in the csrA mutant. Reduced secretion of Tir appeared to be in part due to decreased synthesis of EscD, an inner membrane architectural protein of the type III secretion system (TTSS) and EscF, a protein that forms the protruding needle complex of the TTSS. These effects were not mediated through the locus of enterocyte effacement (LEE) transcriptional regulator GrlA or Ler. In contrast to the csrA mutant, multicopy expression of csrA repressed transcription from LEE1, grlRA, LEE2, LEE5, escD, and LEE4, an effect mediated by GrlA and Ler. Consistent with its role in other organisms, CsrA also regulated flagellar motility and glycogen levels. Our findings suggest that CsrA governs virulence factor expression in an A/E pathogen by regulating mRNAs encoding translocators, effectors, or transcription factors.
Enteropathogenic Escherichia coli (EPEC) is a major etiologic agent of infantile diarrhea in developing countries, causing the deaths of several hundred thousand children per year (13, 71). EPEC and the related bacterium enterohemorrhagic E. coli (EHEC) cause attaching and effacing (A/E) lesions that are characterized by disruption of the intestinal microvilli and reorganization of the cytoskeleton in infected cells to form actin-filled membrane protrusions, termed “pedestals,” that emanate beneath bacteria attached to the cell surface (48, 69).
The locus of enterocyte effacement (LEE) of EPEC is a pathogenicity island that is necessary and sufficient for the formation of pedestals (63). It consists of five major polycistronic operons, including LEE1, LEE2, LEE3, LEE5, and LEE4; the bicistronic operon grlRA; and several monocistronic genes (21, 26, 67). The LEE1, LEE2, and LEE3 operons encode the architectural components of a type III secretion system (TTSS), whereas LEE4 encodes the translocator exporter SepL; the secreted translocators EspA, EspB, and EspD; the chaperones CesD2 and L0017; the needle complex-forming component of the TTSS EscF; and the effector protein EspF. SepL, along with SepD, forms a molecular switch that coordinates the hierarchical secretion of EspA, EspB, and EspD over effectors in response to calcium and other environmental signals (20, 22). EspA is secreted to form a hollow filamentous organelle that connects the protruding EscF needle of the bacterial TTSS to the host cell membrane (17, 50, 82). EspB and EspD are translocated through this filamentous TTSS and integrated into the host cell membrane, where they form a pore that allows effector molecules to be injected directly into the host cytosol (39, 96). The LEE5 operon encodes the effector Tir, its ligand, the adhesin intimin, and the Tir chaperone CesT (67). Tir is translocated into the host cytosol via the TTSS and subsequently integrated into the host cell membrane, where it serves as a receptor for intimin, which is present on the outer bacterial membrane (44). The interaction of Tir and intimin results in firm attachment of the bacterium to the infected cell. Tir recruits cellular factors, such as tyrosine kinases, Nck, and N-WASP, that activate the Arp2/3 complex and initiate actin polymerization beneath the attached bacteria (9, 32, 42, 93), culminating with the formation of pedestals.
Coordinated spatiotemporal expression from the LEE is critical for pedestal formation by EPEC. Such a mode of regulation is achieved by the presence of a plethora of transcription factors such as Ler, PerC, GrlA, GrlR, QseA, IHF, Fis, and H-NS (21, 29, 30, 67, 83, 94) in response to diverse environmental conditions, including pH, osmolarity, Fe(NO3)3, Ca2+, temperature, quorum sensing, and HCO3− (1, 43, 85-90, 94). Most of these transcription factors affect the expression from the LEE by activating the transcription of ler, which in turn activates transcription from grlRA, LEE2, LEE3, LEE5, escD, and LEE4 (see Fig. 9) (7, 11, 21, 26, 35, 65, 67).
FIG. 9.
Model of LEE regulation by csrA. When expressed in monocopy, binding of CsrA to the LEE4 operon encoding sepL espADB and escF transcript results in their increased steady-state transcript levels. However, activation of escD occurs indirectly via an intermediate regulator(s). For simplicity, the effect is shown to occur via an activator (X). In contrast to the csrA mutant, overexpression of csrA globally represses the transcription from the LEE. This is achieved in part by CsrA binding to the leader segment and resulting in reduced grlRA transcript and consequent GrlA protein levels. Reduced GrlA protein levels lead to reduced Ler protein levels, which in turn result in reduced transcription from the other LEE-encoded operons. csrA also promotes motility by upregulating the flhDC transcript levels in EPEC and represses glycogen biosynthesis. Dotted arrows represent positive genetic circuits that have been demonstrated previously, thick transparent arrows represent the transcription-activating gene product encoded in the LEE1 and grlRA operons, whereas thick filled arrows with shard ends (activated circuits) and thick filled arrows with blunted ends (repressed circuits) represent novel genetic circuits and/or phenotypes of EPEC identified in this paper.
Whereas our understanding of the mechanisms of transcriptional regulation of the LEE is extensive, information about posttranscriptional and posttranslational regulation is more limited. Posttranscriptional control has been suggested in the negative regulation of espADB mRNA in EHEC strains that secrete high levels of EspA. However, the mechanistic basis for this phenomenon has not been established (78). In terms of posttranscriptional and posttranslational regulation, the detailed molecular mechanisms for only the endoribonuclease RNase E (60) and the protease ClpXP have been elucidated (40). Whereas in EHEC RNase E generates the sepL and espADB transcripts by splicing at the C-terminal end of sepL in the precursor sepL espADB transcript (60), the protease ClpXP positively regulates the LEE posttranslationally by affecting the expression of RpoS and GrlR (40). The ribosome binding GTPase BipA and the noncoding RNA DsrA have also been shown to upregulate the expression of the LEE in EPEC and EHEC, respectively, although the detailed molecular mechanisms involved in this regulation have yet to be elucidated (31, 53).
We have identified csrA as a posttranscriptional regulator of EPEC necessary for paralyzing and killing the nematode Caenorhabditis elegans (S. Bhatt and D. Kalman, unpublished data). Previous studies have demonstrated that genes involved in nematode pathogenesis may also facilitate pedestal formation on mammalian cells (3, 66). This latter observation prompted us to investigate the possible role of csrA in mammalian pathogenesis.
CsrA and its orthologue RsmA are homodimeric RNA binding proteins (4, 33, 36, 59, 77) that regulate gene expression posttranscriptionally by binding to sites containing the AGGA/ANGGA motif in the leader segment of transcripts and altering their stability and/or translation (5, 6, 23, 59, 62, 97, 98). Transcripts such as flhDC, whose expression is activated by CsrA, have a putative CsrA binding site(s) located distantly from the Shine-Dalgarno sequence. Binding of CsrA enhances their stability and leads to an increase in the steady-state levels of such transcripts (79, 98). For transcripts that are negatively regulated, CsrA binds to sequences that overlap or lie in close proximity to the Shine-Dalgarno sequence, thereby preventing the 30S ribosomal subunit from binding to the transcript and inhibiting translation and/or facilitating mRNA decay (5, 6, 24, 97).
The carbon storage regulation (Csr) system also includes the noncoding small RNAs CsrB and CsrC, which bind tightly to and sequester 9-10 and 4-5 CsrA dimers, respectively, thereby preventing CsrA from exerting its effect on the mRNAs in its regulon (58, 99). The expression of CsrB and CsrC is activated by CsrA through the BarA-UvrY two-component regulatory system (74, 92, 99). The last component of the Csr system is CsrD, a protein containing vestigial GGDEF and EAL domains, which targets CsrB and CsrC for degradation by the endoribonuclease RNase E, thereby increasing intracellular CsrA activity (91). These negative feedback loops of the Csr system demonstrate that CsrA activity is finely tuned and suggest that the Csr system functions as a homeostatic circuit (91, 92). The role of CsrA and RsmA in regulating virulence factors and host interactions of mammalian and plant pathogens is well established in Salmonella enterica serovar Typhimurium, Erwinia carotovora, Pseudomonas aeruginosa, Helicobacter pylori, Legionella pneumophila, and Yersinia pseudotuberculosis (2, 8, 10, 16, 27, 37, 47, 56, 75).
In this report, we provide evidence that in EPEC, a functional csrA allele is necessary for the formation of pedestals and for membrane depolarization of epithelial cells. CsrA exerts its effect by binding to the leader segment of the sepL espADB mRNA and enhancing the steady-state transcript and protein levels. In contrast to the csrA mutant phenotype, modest overexpression of csrA globally repressed transcription from the LEE operons by binding to and repressing the expression of the global regulator of the LEE activator GrlA. Furthermore, we also provide evidence that CsrA-mediated effects appear through at least one other regulatory factor. Lastly, we show that csrA also regulates other virulence-associated traits such as motility and glycogen biosynthesis. Our results extend the role of csrA as a regulator of virulence of the LEE genes of EPEC and likely other A/E pathogens, which also possess highly conserved csrA orthologs and putative CsrA binding sites in the leader segment of LEE genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
Bacteria were grown in Luria-Bertani (LB) broth or Dulbecco modified Eagle medium (DMEM) containing phenol red (for immunofluorescence microscopy) or lacking glutamine and phenol red (for Western blotting and real-time quantitative reverse transcription [qRT]-PCR) with appropriate antibiotic supplements when needed. The antibiotics used were streptomycin (100 μg/ml), chloramphenicol (25 or 50 μg/ml), kanamycin (50 μg/ml), tetracycline (15 μg/ml), and ampicillin (50 or 100 μg/ml).
To determine growth rates, bacterial cultures were grown overnight at 37°C at 250 rpm in LB broth or in LB broth supplemented with the appropriate antibiotic. Overnight cultures were diluted to a starting optical density (OD) of approximately 0.01 in DMEM containing glucose (4.5 g/liter) but lacking phenol red and glutamine. The cultures were grown standing at 37°C in a 5% CO2 incubator, and growth was measured every hour and intermittently at half-hour intervals over a period of 24 h. The growth curve of each strain was determined at least twice with different experimental samples by assaying each sample in duplicate. Growth curves are representative of one such experiment. The strains, plasmids, and oligonucleotides used are listed in Tables 1 and 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Reference or source |
|---|---|---|
| Strains | ||
| EPEC | Wild-type EPEC 2348/69 serotype O127:H6 Strr | June Scott |
| EPEC csrA | EPEC 2348/69 csrA::Cmr Strr Cmr | This study |
| EPEC csrA lacZ csrA+ | EPEC 2348/69 csrA::CmrlacZ::csrA Strr Cmr | This study |
| EPEC grlA 3XFLAG | EPEC 2348/69 Φ(grlA-3XFLAG) Kanr Strr | This study |
| EPEC csrA grlA 3XFLAG | EPEC 2348/69 csrA::Cmr Φ(grlA-3XFLAG) Cmr Kanr Strr | This study |
| EPEC csrA lacZ csrA+grlA 3XFLAG | EPEC 2348/69 csrA::CmrlacZ::csrA Φ(grlA-3XFLAG) Cmr Kanr Strr | This study |
| EPEC(pBR322) | EPEC 2348/69 transformed with empty vector pBR322 Tetr Ampr Strr | This study |
| EPEC(pCRA16) | EPEC 2348/69 transformed with pCRA16 Tetr Strr | This study |
| EPEC grlA 3XFLAG(pBR322) | EPEC 2348/69 Φ(grlA-3XFLAG) containing pBR322 Kanr Tetr Ampr Strr | This study |
| EPEC grlA 3XFLAG(pCRA16) | EPEC 2348/69 Φ(grlA-3XFLAG) containing pCRA16 Kanr Tetr Strr | This study |
| EPEC Δler | EPEC 2348/69 ler deletion | Jay Mellies |
| EPEC Δtir | EPEC 2348/69 tir deletion | B. Brett Finlay |
| EPEC ΔespA | EPEC 2348/69 espA deletion | James B. Kaper |
| EPEC ΔespB | EPEC 2348/69 espB deletion | James B. Kaper |
| EPEC ΔespD | EPEC 2348/69 espD deletion | James B. Kaper |
| S17-1λpir | E. coli pir lysogen of S17-1 (thi pro hsdR hsdM+recA RP4-Tc::Mu-Km::Tn7) Tpr Strr | Phil Rather |
| DH5α | supE44 ΔlacU169 φ80dlacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bettina Bommarius |
| Plasmids | ||
| pTP223 | gam, bet, and exo genes of phage λ cloned under control of lac promoter; Tetr | Kenan Murphy |
| pBR322 | Cloning vector; Tetr Ampr | 85 |
| pCRA16 | csrA gene from K-12 cloned into blunt-ended VspI site of bla of pBR322; Tetr | 97 |
| pJRlacZIns | Suicide vector; Ampr | Vanessa Sperandio |
| pJRcsrA6 | csrA gene from EPEC under its regulatory sequences cloned into EcoRV site of lacZ in pJRLacZIns; Ampr | This study |
Tetr, tetracycline resistance; Cmr, chloramphenicol resistance; Strr, streptomycin resistance; Ampr, ampicillin resistance; Tpr, trimethoprim resistance.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence |
|---|---|
| Knockout/cloning primers | |
| 5′csrAP2(Wanner) | TAAATGCCCCGAAGGAAGTTTCTGTTCACCGTGAAGAGATCTACCAGCGTACATATGAATATCCTCCTTA |
| 3′csrAP1(Wanner) | GAGACGCGGAAAGATTAGTAACTGGACTGCTGGGATTTTTCAGCCTGGAGTGTAGGCTGGAGCTGCTTC |
| c1a | TTATACGCAAGGCGACAAGG |
| c2a | GATCTTCCGTCACAGGTAGG |
| k1a | CAGCCGATTGTCTGTTGTGCCC |
| 5′csrA | GGATAATGCCGGGATACAGAGAGAC |
| 3′csrA[EPEC] | ATTTTGAGGGTGCGTCTCACCG |
| 5′csrA-EcoRV | GCGGCCGATATCAATTGCAATAATATAAGCGTCAGGCAATGc |
| 3′csrA-EcoRV | GCGGCCGATATCGTCAAACAATTTTTCCCACACTTTTATCG |
| 5′grlA-3XFLAG | TCTAATATCTGGAACGAAATGATCTTGAGGCGGAAAAAGGAGAGTGACTACAAAGACCATGACGG |
| 3′grlA-3XFLAG | GAGAAAAAGGCTTACCCTGGAAAACAAAACCCTTAAATATAGCTTCATATGAATATCCTCCTTAG |
| RT-qRT-PCR primers | |
| 5′rrsB | CTTACGACCAGGGCTACACAC |
| 3′rrsB | CGGACTACGACGCACTTTATG |
| 5′ler | GCAGTTCTACAGCAGGAAGCA |
| 3′ler | CGAGCGAGTCCATCATCAG |
| 5′flhD | TGCATACCTCCGAGTTGCTG |
| 3′flhD | GCGTGTTGAGAGCATGATGC |
| 5′tirb | GCAGAAGACGCTTCTCTGAATA |
| 3′tirb | CCCAACTTCAGCATATGGATTA |
| 5′espAb | GCTGCAATTCTCATGTTTGC |
| 3′espAb | GGGCAGTGGTTGACTCCTTA |
| 5′grlR | TTAGCAATGAAGACTCCTGTGG |
| 3′grlR | AGAGAGAACCCCCTGATACAC |
| 5′grlA | AGGCGGTTCCGATAGAAAGT |
| 3′grlA | GCCTCAAGATCATTTCGTTCC |
| 5′escJ | CCAAAGAAATGGACAAAAGTGG |
| 3′escJ | GCTGGGTGGGAAAATAACCT |
| 5′sepL | GAAAGAAGAGGAAGGCACGAC |
| 3′sepL | CAAACATCGCCAAAGTAGGA |
| 5′escD | CACGCCCTATGAAGCAGATAA |
| 3′escD | CAACGCAAAAGTAGCACCAA |
| 5′espD | GCTGCTACGGCTACTTCAGG |
| 3′espD | GCTGTGGTTCTGTTCCCTCT |
| 5′espB | TAGGCTCTTTTGCTGCCATT |
| 3′espB | TTCGCCAGTGCTTTAGTTGA |
| 5′escF | ACAAATGGGTGAAGTAGGTAAAACG |
| 3′escF | GAACCGCAAACTGCAACTCTAAC |
| In vitro transcription primers | |
| 5′-T7p-grlR-EPEC | TAATACGACTCACTATAGGGCATTGCAATCTGGAGAAAAAG |
| 3′-T7p-grlR-EPEC | CAGTGATCATATTTCCATTTT |
| 5′-T7p-escD-EPEC | TAATACGACTCACTATAGGGGATGTAAGTTCACCATATTTT |
| 3′-T7p-escD-EPEC | CGGAAGTTGTAATTCCCGATT |
| 5′-T7p-sepL-EPEC | TAATACGACTCACTATAGGGGTCTAAGAATAGAGTAGAAAG |
| 3′-T7p-sepL-EPEC | CATTAGCCATTGGAAACTCAC |
| phoB-T7 (E. coli K-12) | TAATACGACTCACTATAGGGGCATTAATGATCGCAACCTATTTATTACAACAGGGCAAATCATG |
| GCphoB-T7 (E. coli K-12) | CATGATTTGCCCTGTTGTAATAAATAGGTTGCGATCATTAATGCCCCTATAGTGAGTCGTATTA |
csrA was disrupted by the one-step gene inactivation method described by Datsenko and Wanner and modified for EPEC by Murphy and Campellone (18, 70). Briefly, plasmid pKD3 was used as the template to amplify a chloramphenicol (cat) resistance cassette flanked on its 5′ side with 51 nucleotides (101 to 151 nucleotides downstream of the translation initiation codon of the csrA open reading frame [ORF]) and on its 3′ side with 49 nucleotides (152 to 186 nucleotides downstream of the translation initiation codon of the csrA ORF and 14 nucleotides downstream of the translation termination codon) by using primers 5′csrAP2(Wanner) and 3′csrAP1(Wanner). The PCR product was gel purified, and 250 to 500 ng of the product was electroporated into EPEC expressing the lambda red recombinase proteins Gam, Exo, and Bet from plasmid pTP223 under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (76). Expression of the lambda red recombinase system promotes the site-specific substitution of the wild-type csrA allele with the mutant csrA::Cmr PCR product because of the presence of identical flanking sequences in the PCR product. Recombinants were selected on LB broth plates containing chloramphenicol. grlA was tagged in the chromosome with a 3XFLAG tag in a similar manner by using pKD4 as the template (40, 70, 95). The csrA disruption was verified by locus-specific PCR with primer pair 5′csrA and c2 or 3′csrA(EPEC) and c1, whereas grlA tagging was verified by using the primer pair 5′grlA(qRT-PCR) and k1 (18). PCR products of the expected size were generated and verified by sequencing.
Construction of pCRA16 has been described previously (91). Briefly, a BamHI-EcoRI fragment of approximately 500 bp containing the K-12 csrA gene under the control of its putative promoter(s) was restricted from pCSR10 (80), end filled with the Klenow fragment of DNA polymerase I, and cloned into the blunt VspI site of the bla gene of pBR322 to generate pCRA16. The csrA ORF is oriented in the same direction as the bla gene (91).
EPEC csrA lacZ csrA+ is the csrA disruptant expressing the wild-type csrA allele under the control of its putative promoter(s). csrA was amplified from EPEC by colony PCR with primers 5′csrA-EcoRV and 3′csrA-EcoRV. The 685-bp PCR product contains the intergenic sequence spanning the region from the last 11 nucleotides of the csrA upstream gene alaS to the −35 region of the csrA downstream gene serV and includes csrA under the control of its native cis regulatory elements (as determined by qRT-PCR and Western blotting for LEE-encoding genes regulated by CsrA). The PCR product was gel purified, treated with EcoRV, and cloned into the EcoRV site of the lacZ gene in plasmid pJRlacZins (51) to generate pJRcsrA6. The cloned csrA gene is oriented opposite to the direction of transcription of the lacZ gene. The csrA sequence in pJRcsrA6 was verified by sequencing with the same set of primers. pJRcsrA6 contains the R6Kγ origin of replication and can only replicate in π protein-expressing strains such as S17-1λpir (84). This plasmid was transferred into the csrA mutant of EPEC by conjugation as described previously (51). Integrants were selected by plating dilutions onto LB broth plates supplemented with chloramphenicol, ampicillin, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml), and IPTG (5 mM) and incubated overnight at 37°C. Transconjugants that arose on these plates were confirmed to be EPEC derivatives and contained both the wild-type and csrA mutant alleles. One such colony was selectively purified, and serial dilutions were plated onto LB broth plates devoid of NaCl but supplemented with 5% sucrose to counterselect for loss of the sacB gene carried on the plasmid backbone. One such colony that arose was confirmed to be ampicillin sensitive, chloramphenicol resistant, sucrose resistant (sacB mutant), and white when plated onto LB broth plates containing X-Gal and IPTG (lacZ mutant) and contained both the wild-type and mutant csrA alleles as determined by PCR and was used for all subsequent experiments.
Cell culture and immunofluorescence microscopy.
3T3 cells were maintained and passaged under standard culture conditions in DMEM supplemented with 10% fetal bovine serum and penicillin and streptomycin. Pedestal formation by EPEC and its isogenic derivatives on 3T3 cells was performed as described previously (3).
Preparation of cell lysates, trichloroacetic acid precipitation, and Western blotting.
Synthesis of EspA and Tir and secretion of EspA, EspB, EspD, and Tir were analyzed by Western blotting. Bacterial cultures were grown under standing conditions at 37°C in a 5% CO2 incubator to an OD of 0.2 to 0.3, 0.5, or 1.0 and pelleted by centrifugation at 3,000 rpm. The proteins in the supernatant were precipitated at −20°C by the addition of trichloroacetic acid to a final concentration of 10%. The precipitated proteins were pelleted and washed with acetone and centrifuged. The pellets were air dried to ensure complete evaporation of the acetone prior to suspending them in 100 μl of a 1:1 volumetric ratio of 2× Laemmli buffer containing ß-mercaptoethanol (54) and 1 M Tris-HCl (pH 6.8). The suspensions were boiled for 10 min prior to 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. To ensure equal loading, the volume of the sample loaded was normalized to the OD of the cultures when harvested.
To determine protein levels in the cell lysate, the cell pellets were suspended in 200 μl of 50 mM Tris-HCl (pH ∼7.5) and sonicated to disrupt the cell membrane. Cell debris was pelleted by centrifugation, and the supernatant was transferred to a fresh Eppendorf tube. Protein concentrations were determined by the Lowry assay (61) as recommended by the manufacturer (Bio-Rad), after which the lysates were mixed with an equal volume of 2× Laemmli buffer containing ß-mercaptoethanol. The lysates were boiled at 95°C for 10 min. Equal amounts of protein were subjected to 12% SDS-polyacrylamide gel electrophoresis. Proteins were electroblotted onto a polyvinylidene difluoride membrane and probed with one of the following antibodies: mouse monoclonal anti-FLAG M2 (F-3165; 10 μg/ml), rabbit anti-Ler (1:2500), rabbit anti-EspA (1:50,000), rabbit anti-EspB (1:10,000), rat anti-EspD (1:500), or rabbit anti-Tir (1:50,000). The anti-FLAG antibody was purchased from Sigma. Each experiment was repeated at least three times with similar results each time.
RNA isolation.
RNA isolation was performed by using the MasterPure RNA purification kit from EPICENTRE as recommended by the manufacturer, with the exception that the DNase I treatment was performed for 45 min and then an additional 5 μl of DNase I was added to the samples and the reaction was allowed to continue for another 45 min prior to inactivation of the DNase I. The DNase I-treated RNA was subjected to phenol-chloroform extraction, followed by isopropanol precipitation, and the procedure was repeated. Precipitated RNA was resuspended in 50 μl of TE buffer (10 mM Tris-HCl [pH 7.8],1 mM EDTA), and 1 μl of ScriptGuard RNase inhibitor was added to the RNA samples.
Real-time qRT-PCR.
The iScript One-Step real-time qRT-PCR kit with SYBR green (Bio-Rad) was used to determine the transcript levels of genes as recommended by the manufacturer, with some modifications. Each reaction mixture had a total volume of 25 μl. The reaction mixture consisted of 1× SYBR green RT-PCR mix (12.5 μl of 2× reaction buffer containing 0.4 mM dATP, dCTP, dGTP, and dTTP; magnesium ions; iTaq DNA polymerase, 20 nM fluorescein; SYBR green I dye; and stabilizers), a primer pair (0.75 μl at a final concentration of 300 nM), nuclease-free water, iScript reverse transcriptase (0.5 μl), and DNase I-treated RNA (50 ng). The reaction conditions were (i) 53°C for 20 min, (ii) 95°C for 10 min, (iii) 40 cycles of 95°C for 0.5 min and 60°C for 1 min, and (iv) 4°C for ∞. Melting curve analysis were done to ensure the specificity of the generated product. The melting curve conditions were (i) 95°C for 1 min, (ii) 55°C for 1 min, and (iii) 40 cycles of 55°C for 10 s with an increment of 0.5°C. Control reactions were conducted to ensure the absence of genomic DNA contamination in purified RNA and that the observed fluorescence was not the result of primer dimer formation. Real-time qRT-PCR was performed in duplicate or triplicate with multiple experimental samples. The cycle threshold method 2−ΔΔCt was used to quantify the transcript levels after normalizing them to the housekeeping gene rrsB. The relative transcript level for EPEC or EPEC(pBR322) in the first sample of the first experiment was defined as 1, and all other values were determined relative to this. The unpaired Student t test was used to assay for the statistical significance of differences. A P value of <0.01 was considered statistically significant for real-time qRT-PCR-based analysis.
In vitro transcription and RNA electrophoretic mobility shift assay (EMSA).
The MEGAshortscript kit from Ambion was used for in vitro transcription as recommended by the manufacturer. Briefly, a PCR product containing the T7 promoter at its 5′ end and the entire leader segment encompassing the putative CsrA binding sites of grlR, sepL, and escD was generated. Transcripts were run on a 5% polyacrylamide gel, the band corresponding to the correct-size transcript was excised, and the gel fragment was suspended in 250 μl of elution buffer (0.5 M ammonium acetate, 1 mM EDTA, 0.2% SDS) along with 2 μl of Superase-in (Ambion) and eluted overnight at 4°C and then at room temperature for 30 min. The eluate was subjected to phenol-chloroform extraction and ethanol precipitation and suspended in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) buffer. Five hundred nanograms of the RNA was run on a gel to verify the size and integrity of the RNA. Thirty to 40 pmol of the RNA was dephosphorylated, and the RNA was purified by another round of phenol-chloroform extraction and ethanol precipitation. The purified RNA was suspended in 10 μl of nuclease-free water. RNA was radioactively labeled and suspended in 10 μl of TE buffer. Because the csrA ORF and the CsrA protein are identical between EPEC and K-12, C-terminally His-tagged CsrA, purified from E. coli K-12, was used for the binding reactions. Purification of recombinant His-tagged CsrA has been described elsewhere (68). His-tagged CsrA was suspended in protein dilution buffer (10 mM Tris-HCl [pH 7.5], 2 mM dithiothreitol, 10% glycerol). RNA was denatured at 85°C and slowly cooled to 25°C over 25 min in a Bio-Rad ICycler prior to addition to the binding reaction mixture. The components of the binding reaction mixture included 50 pM RNA in TE, 1× binding buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM KCl), 3.25 ng/μl total yeast RNA, 20 mM dithiothreitol, 7.5% glycerol, 0.01 μl of Superase-in (Ambion), and 2 μl of different concentrations of His-tagged CsrA in a total volume of 10 μl. The reaction mixtures were incubated at 37°C for 30 min, after which the samples were loaded onto a 10% native polyacrylamide gel. The gel was exposed to a PhosphorImager plate overnight, and the bands were detected with a PhosphorImager. ImageQuaNT (Molecular Dynamics) was used to perform densitometric analysis. The apparent equilibrium dissociation constant (KD) for CsrA was determined from two independent experiments with the equation Y = {Ymax × [(x/KD)n]}/{1 + [(x/KD)n]}. For competition-based gel shift experiments, unlabeled specific and nonspecific (phoB) transcripts were added at final concentrations of 500 pM, 5 nM, and 50 nM, which represent 10-, 100-, and 1,000-fold excesses relative to the labeled transcript.
Transepithelial resistance (TER) assay.
The resistance of cells in response to wild-type and mutant EPEC strains was monitored in real time with the electric cell substrate impedance sensing (ECIS) 1600R device (Applied BioPhysics, Troy, NY). Caco2-BBE cells were seeded into ECIS 8W1E electrodes (5 × 105 cells/400 μl/electrode) and kept at 37°C in 5% CO2 and 90% humidity. When the cells were confluent, they were washed and changed to serum-free, antibiotic-free DMEM. Cell resistance was measured at a frequency of 500 Hz and a voltage of 1 V. Details of the operation, equivalent resistance-capacitance circuit, and modification of the ECIS system can be found in the manufacturer's instructions. Bacterial strains were grown overnight in LB broth and transferred to serum-free and antibiotic-free DMEM, and confluent Caco2-BBE monolayers were infected at a multiplicity of infection of 10. TER was measured continuously postinfection. TER assays were conducted at least three times with separate experimental samples with each sample assayed in triplicate per experiment. The depicted results are representative of one such experiment.
Motility assay.
Cultures were grown to an OD at 600 nm (OD600) of 1.0, and 1 μl of the culture was stabbed onto 0.3% 0.5× DMEM containing glucose (2.25 g/liter) and sodium pyruvate (0.5 mM) but lacking phenol red and glutamine. The stabbed agar plates were incubated upright overnight at 37°C, and the diameter of the “diffusion halo” was measured 30 h later to determine the extent of motility of the bacterium. Motility assays were performed on three separate occasions.
Glycogen biosynthesis assay.
A loopful of a bacterial culture grown to an OD600 of 1.0 was streaked onto Kornberg medium (1.1% K2HPO4, 0.85% KH2PO4, 0.6% yeast extract, 2% glucose, 1.5% agar) (83), after which the plates were incubated overnight at 37°C. The plates were then exposed to iodine crystals to determine the extent of glycogen biosynthesis in the different bacterial strains. Iodine forms complexes with glycogen, as a result of which strains producing glycogen appear blackish-brown whereas strains that do not synthesize glycogen do not exhibit any color change after exposure to iodine. Glycogen biosynthesis assays were performed on three separate occasions. The pictures shown are representative of one such experiment.
RESULTS
csrA regulates adherence and pedestal formation on mammalian cells.
Disruption of csrA does not affect the growth of EPEC, as evident from similar doubling times for the wild type (48.04 ± 0.8 min) and the csrA mutant (41.15 ± 9.2 min) (Fig. 1A, B, and C). Because csrA was necessary for virulence against C. elegans (data not shown, unpublished data), we next assessed whether disruption of csrA affected virulence in mammalian cells. To do this, we assessed the capacity of EPEC to adhere to and form pedestals on mammalian 3T3 cells (49) and to disrupt the resistance across polarized epithelial cells (12, 34). Although cells were infected with the same number of bacteria, the csrA mutant exhibited reduced adherence compared to that of EPEC or the single-copy complemented strain (n = 4; Fig. 1D, compare 4′,6-diamidino-2-phenylindole [DAPI] staining in column 2, row 3 [csrA mutant], with column 1, row 3 [EPEC], or column 3, row 3 [csrA lacZ csrA+]). Furthermore, whereas clustering of the bacteria was evident for the wild-type strain and the complemented strain (yellow arrows), the csrA mutant primarily appeared as solitary isolated cells.
FIG. 1.
Inactivation of csrA does not affect EPEC growth but greatly diminishes adherence, pedestal formation, and disruption of TER. (A and B) Bacterial cultures grown overnight were diluted in DMEM (lacking phenol red) to a starting OD600 of ∼0.01, and the growth of the strains was measured hourly and intermittently at half-hour intervals. Shown are the growth curves of the strains on logarithmic (A) and linear (B) scales. EPEC and its isogenic csrA mutant grow at similar rates. Furthermore, EPEC(pBR322) and EPEC(pCRA16) also grow at the same rate to an OD of ∼0.25, after which the multicopy csrA expressor grows at a slower rate. (C) The doubling times of EPEC, the csrA mutant, EPEC(pBR322), and EPEC(pCRA16) were determined from at least two independent experiments with each strain assayed in duplicate per experiment. The values represent the mean ± standard deviation from all of the experiments. (a) The doubling time of EPEC(pCRA16) was determined in the early log phase (OD600, <0.25) prior to the first biphasic lag (lower black arrow) displayed by the strain. (D) 3T3 cells were infected for 5 h with equal numbers of bacteria. Cells were then fixed with formaldehyde and permeabilized with Triton X-100. Pedestal formation was visualized by immunofluorescence microscopy by using antibodies against Tir and pY. Fluorescein isothiocyanate-phalloidin was used to detect filamentous actin beneath the adherent bacteria. DAPI was used to detect adherent bacteria and the cellular nucleus. Adherence was diminished in the csrA mutant, as evident from the relatively low number of DAPI-stained mutant bacteria compared to those of the wild-type or the complemented strain (compare column 2, row 3 (csrA mutant), with column 1, row 3 (EPEC), or column 3, row 3 (csrA lacZ csrA+). Yellow arrows represent clustering of bacteria that was routinely observed in the wild-type and complemented strains but not in the csrA mutant. The ability to form pedestals was also reduced in the csrA mutant, as manifested by diminished Tir, pY, and polymerized actin staining. Pedestal assays were performed on three separate occasions with independent bacterial cultures, and similar results were obtained in each experiment. Images are representative of one such experiment. Scale bars represent 20 μm. (E) The extent of pedestal formation was quantified by counting a total of 100 cells from at least 10 different frames, and the percentage of cells with 10 or more pedestals per cell was determined. The values and error bars represent the standard deviation of the mean from one such experiment. ** denotes a P value of <0.001. FITC, fluorescein isothiocyanate.
Using fluorescence microscopy in conjunction with antibodies against phosphotyrosine (pY) and Tir and fluorescein isothiocyanate-phalloidin to visualize polymerized actin, we next assessed the formation of actin pedestals. In cells infected with EPEC, >80% of the cells displayed >10 pedestals per cell (Fig. 1E), and Tir and pY staining was evident directly beneath attached bacteria and at the tips of actin pedestals (Fig. 1D, column 1). By contrast, with the csrA mutant, the percentage of cells displaying pedestals was reduced by ∼35-fold (Fig. 1E), as was the number of pedestals per cell (Fig. 1D). Accordingly, pY and Tir staining was diminished in cells infected with the csrA mutant compared to that in cells infected with wild-type EPEC (Fig. 1D, column 2). Complementation of the mutant with csrA in a single copy restored adherence, pedestal formation, and localization of pY and Tir to the levels seen with wild-type EPEC (Fig. 1D, column 3, and E).
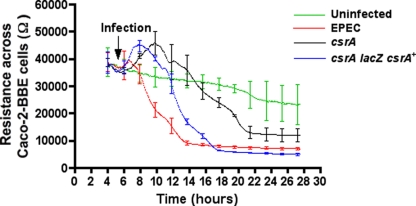
Disruption of TER across polarized Caco-2BBe cells depends on csrA.
Previous reports have shown that adherence, pedestal formation, and the effectors EspF and Map mediate the capacity of EPEC to disrupt TER across polarized Caco-2BBe cells (12, 19, 64). Because mutation of csrA diminishes the ability of EPEC to adhere and form pedestals, we next examined its effects on TER. For EPEC, the time required to disrupt the TER to 50% of the value at the point of infection (t1/2) was ∼5.5 h, whereas the csrA mutant required approximately 2.5 times as long (t1/2 of ∼14.3 h; Fig. 2). By 9.5 h postinfection, EPEC decreased the resistance across the cells by over 76% (more than fourfold), whereas the csrA mutant reduced the TER by less than 15% during the same period. The complemented strain regained the ability to disrupt the TER with significantly faster kinetics than those of the csrA mutant (t1/2 of ∼7.9 h), and by 9.5 h it had decreased the TER by over 63% (Fig. 2). Interestingly, even after prolonged infection by the csrA mutant, there was still some residual endpoint resistance (∼12,000 Ω) that was reproducibly higher than that observed in cells infected with EPEC (7,000 Ω) or with the complemented strain (5,000 Ω) (Fig. 2). Taken together, these data suggest that csrA not only regulates the time of onset but also the kinetics and extent of membrane depolarization of mammalian cells in vitro.
FIG. 2.
csrA is necessary for disruption of TER across Caco-2BBe cells. Caco-2BBe cells were seeded into ECIS 8W1E electrodes (5 × 105 cells/400 μl/electrode) and kept at 37°C in a 5% CO2 incubator with 90% humidity. Bacterial cultures grown overnight were used to infect the cells at a multiplicity of infection of 10. TER was measured continuously postinfection. Disruption of csrA delayed the onset and reduced the rate and extent of membrane depolarization. Each experiment was performed on three separate occasions, with triplicate samples being assayed in every experiment. Error bars represent standard deviations of the mean values obtained at the designated time points from one such experiment. The start of infection of the cells by the bacterial strains is indicated by the arrow.
csrA is necessary for synthesis and secretion of translocators but only affects the secretion of the effector Tir.
Mutations in espA, espB, espD, or tir negate the ability of EPEC and Citrobacter rodentium to form pedestals on tissue culture cells in vitro or to colonize mice, respectively (14, 21, 28, 44-46, 50, 55). Disruption of csrA reduced the secreted (Fig. 3A), as well as the bacterium-associated (Fig. 3B), levels of EspA, EspB, and EspD. Complementation of the csrA mutant with csrA expressed in a single copy under the control of its putative promoter(s) restored both the secreted (Fig. 3A) and the bacterium-associated (Fig. 3B) amounts of EspA, EspB, and EspD to nearly wild-type levels.
FIG. 3.
Secretion of EspA, EspB, EspD, and Tir is diminished in the csrA mutant. (A and B) EPEC, the csrA mutant, and the csrA lacZ csrA+ monocopy complemented strain were grown in DMEM lacking phenol red at 37°C in a 5% CO2 incubator to an OD of ∼1.0. Western blotting for secreted (A) and EPEC-associated (B) EspA, EspB, EspD, and Tir was performed with polyclonal antibodies as described in Materials and Methods. Whereas both the secreted and bacterium-associated levels of EspA, EspB, and EspD were reduced, only the secretion of Tir was reduced in the csrA mutant. Western blotting experiments were conducted on at least three separate occasions with similar results. The image shown is a representation of one such experiment.
Two prominent bands corresponding to Tir were visible in the secreted fraction of the wild type and the complemented strain (Fig. 3A, lanes 1 and 3) but absent in the tir mutant (data not shown). Whereas the slower-migrating band displayed a modest reduction, the faster-migrating band was absent in the csrA mutant (Fig. 3A, compare lane 1 with lane 2). Our results are consistent with previous observations identifying at least two variants of Tir in EPEC, of which one migrates at 80 kDa (upper band) and the other, likely representing a truncated form, migrates at 70 kDa (lower band) (25, 26). Both bands were routinely observed in the secreted fraction (Fig. 3A), but only the 80-kDa band was consistently observed in cell lysates (Fig. 3B). In contrast to the secretion of Tir, the bacterium-associated level of Tir was not substantially affected by the disruption of csrA (Fig. 3B). Overall, these data suggest that disruption of csrA reduces the secretion of Tir without appreciably affecting the amount of Tir protein associated with the bacterium or its synthesis (see below).
csrA regulates the transcript levels of the architectural components of the TTSS.
Because the bacterium-associated levels of EspA, EspB, and EspD were reduced in the csrA mutant, we next explored the possibility that reduced levels of protein were due to reduced transcript levels. Real-time qRT-PCR for espA, espD, and espB demonstrated that transcript levels for these genes were reduced by approximately 16-, 28-, and 14-fold, respectively, in the csrA mutant (Fig. 4A). Complementation with csrA in a single copy restored the transcript levels to those seen in the parental strain (Fig. 4A). Thus, reduced steady-state protein levels of the translocators in the mutant can be accounted for by reduced transcript levels. Interestingly, whereas espA and espB showed similar n-fold reductions in transcript levels, espD exhibited a 1.75-fold greater reduction (Fig. 4A).
FIG. 4.
sepL, espA, espD, espB, escD, and escF, but not tir, transcript levels are reduced in the csrA mutant. (A and B) RNA was isolated from bacterial cultures grown to an OD of ∼1.0, and real-time qRT-PCR was performed as described in Materials and Methods. Shown are the relative transcript levels of sepL, espA, espD, espB (A), tir, escD, and escF (B) in csrA mutant and csrA lacZ csrA+ strains normalized to those of wild-type EPEC. Results are means ± standard deviations from triplicate experiments, with each sample being assayed in duplicate per experiment. The unpaired Student t test was used to assay for the statistical significance of differences between the csrA mutant and EPEC or the csrA lacZ csrA+ strain. A P value of <0.01 was considered statistically significant. ** denotes a P value of <0.001, whereas * denotes a P value of <0.005.
A σ70 promoter for the LEE4 operon is located upstream of sepL, which is the first gene in the operon (67). In EPEC and EHEC, sepL, espA, espD, and espB are transcribed as a single polycistronic transcript (60, 67). In EHEC, this polycistronic transcript is processed at the C terminus of sepL to yield two transcripts containing sepL and espADB, respectively (60), whereas for EPEC this phenomenon has yet to be demonstrated. SepL facilitates the secretion of translocators (20, 52, 72) without affecting their synthesis. Because CsrA binds to the 5′ leader segment of transcripts and affects gene expression, we scanned the RNA sequence in the leader segment of the sepL transcript for putative CsrA binding sites. Two such sites were identified (Fig. 5A and B), suggesting that CsrA might bind to and affect the sepL espADB transcript levels. Consistent with this prediction, sepL transcript levels were reduced by ∼17.2-fold in the csrA mutant (Fig. 4A). Together, these data suggest that csrA not only regulates the synthesis of EspA, EspB, and EspD but may directly affect their export by regulating the levels of SepL. Consistent with data showing similar levels of bacterium-associated Tir in the wild type and the csrA mutant, no significant differences in the tir transcript levels in these strains were observed (Fig. 4B). Thus, reduced secretion of Tir in the mutant is likely due to an effect of csrA on a regulator or a component of the TTSS.
FIG. 5.
CsrA binds to the leader segment of the LEE4 operon. (A) Cartoon depicting the location of putative CsrA binding sites in the leader segment of the LEE4 transcript. The thick black arrows depict the genetic organization of sepL, espA, espD, espB, cesD2, and escF. Two putative CsrA binding sites identified upstream of sepL are depicted by white rectangles. The vertical line with an arrow at the apex indicates the transcription start site of the LEE4 operon in EPEC. (B) The primary sequence in the leader segment of the sepL transcript of EPEC, EHEC, and C. rodentium was scanned to determine the presence of putative CsrA binding sites by comparing the sequence to the SELEX (selected evolution of ligands by exponential enrichment)-determined CsrA consensus sequence (A/GUACAA/GGGAUGU) (23). The σ70 transcriptional start site for sepL is highlighted in gray. The predicted Shine-Dalgarno sequence for the transcript is in bold and italicized. The predicted CsrA binding sites are in boldface. Note that each predicted CsrA binding site contains the highly conserved ANGGA motif (boldface and underlined) present in known CsrA-regulated transcripts (4, 62). The highlighted AUG codon represents the translation initiation codon. References for the transcriptional start site and/or the predicted Shine-Dalgarno sequence are shown on the right. (C) RNA EMSA was performed with purified His-tagged CsrA and 32P-labeled sepL leader RNA as described in Materials and Methods. CsrA bound to the leader segment of sepL at a concentration of 10 nM (S1). Higher-molecular-weight ribonucleoprotein complexes were observed with increasing CsrA concentrations of ≥80 nM (S2). U1 and U2 represent the unbound transcript, whereas S1 and S2 represent shifted ribonucleoprotein complexes. (D) Binding to the sepL transcript was specific because unlabeled sepL but not the noncompetitor RNA, phoB, successfully competed with the labeled transcript. CsrA and sepL or phoB RNA concentrations are indicated at the bottom of each lane. (E) The calculated apparent equilibrium binding constant of CsrA for sepL RNA was 23 ± 1.7 nM. Error bars represent standard errors of the means determined from two independent experiments. (F and G) The steady-state protein levels of Ler in EPEC, the csrA mutant, and the csrA lacZ csrA+ strain (F) or chromosomally 3XFLAG-tagged GrlA in EPEC grlA 3XFLAG, csrA mutant grlA 3XFLAG, and csrA lacZ csrA+ grlA 3XFLAG (G) were determined by Western blotting. Ler and 3XFLAG-tagged GrlA protein levels were unaffected in the csrA disruptant.
EscF forms the needle complex of the TTSS that connects the EscC outer membrane protein of the TTSS to the EspA filament. Inactivation of escF diminishes the secretion of EspA, EspB, and EspD and pedestal formation by EPEC (100). escF is present on the same polycistronic transcript as sepL espADB (67), suggesting that its transcript levels might also be reduced in the csrA mutant. However, the transcript levels of escF did not show the same extent of reduction as sepL espADB and were reduced by only ∼5.5-fold in the csrA mutant (Fig. 4B). It is possible that besides being present on the longer transcript, escF is also transcribed from an additional promoter(s) in the intergenic region between espB and escF and this transcript is not subject to CsrA-mediated regulation.
Besides escF, we also assayed for the transcript levels of escD. EscD is homologous to the Yersinia YscD protein, both of which are predicted to be inner membrane proteins and components of the TTSS necessary for the secretion of translocators and effectors (73). escD is located between the eae gene of the LEE5 operon and the sepL gene of the LEE4 operon (67). escD and LEE4 have overlapping divergent promoters, and their transcriptional initiation nucleotides are adjacent to one another, albeit on opposite strands (67). We found that escD transcript levels were reduced ∼2.5-fold in the csrA mutant (Fig. 4B) and restored when the mutant was complemented with csrA in single copy. Overall, these data suggest that reduced synthesis of EscF and EscD in the csrA mutant may, in part, be responsible for reduced secretion of Tir and the translocators by affecting assembly of the TTSS. Expression from the LEE1 and LEE2 operons, which encode other components of the TTSS, was not significantly affected in the csrA mutant (data not shown).
Purified CsrA binds to the leader segment of the sepL espADB, but not the escD, transcript.
Two putative CsrA binding sites were identified in the 5′ leader segment of sepL (Fig. 5A and B), suggesting that CsrA might regulate sepL espADB mRNA levels by directly binding to the transcript. Binding of CsrA to the leader segment of the sepL transcript was detectable at a protein concentration of 10 nM (Fig. 5C), with an apparent equilibrium binding constant of 23 ± 1.7 nM (Fig. 5E). No unbound transcript was evident at a CsrA concentration of 80 nM (Fig. 5C), and multiple shifted ribonucleoprotein complexes were evident at concentrations of ≥160 nM (Fig. 5C). The binding of CsrA to the sepL transcript was specific, as shown by competition assays with unlabeled sepL and the nonspecific phoB RNA, to which CsrA does not bind (Fig. 5D). The binding data and real-time qRT-PCR results suggest that increased steady-state levels of sepL, espA, espD, espB, and escF in the wild-type strain are the result of direct interaction between CsrA and the polycistronic transcript. CsrA failed to bind to the escD transcript (data not shown), suggesting that csrA-mediated activation of escD occurred through an intermediate regulator.
The transcription factor GrlA activates the transcription of ler (7, 21, 38), which in turn upregulates the steady-state transcript levels of escD (21, 26, 67). To determine whether the effect of csrA on escD and LEE4 may also be occurring through grlA and ler, we assayed for the expression of these genes. Protein levels of Ler and FLAG-tagged GrlA were unchanged in the csrA mutant (Fig. 5F and G), suggesting that the effects of csrA on escD and LEE4 occurred independently of Ler and GrlA.
Overexpression of csrA globally represses expression from the LEE.
Previous studies with csrA suggest that fine tuning of the activity of CsrA is critical for the virulence of S. enterica serovar Typhimurium (2). This observation prompted us to assess the effect of increasing the activity of CsrA in EPEC. The noncoding small RNAs CsrB and CsrC bind to and sequester CsrA, thereby inhibiting its activity. These RNAs are strongly regulated in vivo, and deletion of both csrB and csrC increases CsrA activity in the cell, a condition that would be more relevant for in vivo interpretation (92, 99). Because CsrB and CsrC exhibit compensatory regulation, loss of either RNA leads to an increase in the remaining RNA (99). Thus, single deletions have weak or negligible effects on CsrA-regulated genes and processes. In this regard, isogenic csrB or csrC single mutants show no difference in the expression of Tir (data not shown), and we were unable to construct a csrB csrC double mutant of EPEC, for unknown reasons. Thus, high activity of CsrA was achieved by increasing the levels of CsrA via expression of csrA from a plasmid. Multicopy expression of csrA resulted in a modest ∼1.5-fold ± 0.1-fold increase in CsrA protein levels above that of the empty-vector-containing strain (Fig. 6A). Overexpression of CsrA reduced the transcript levels of escJ, tir, and espA in the LEE2, LEE5, and LEE4 operons and the monocistronic gene escD by 76-, 43-, 13-, and 45-fold, respectively (Fig. 6B). Consistent with the observed reduction in transcript levels of tir and espA, the overall steady-state protein levels were also reduced (data not shown). Reduced tir transcript levels in the overexpressor, but not the csrA mutant (compare Fig. 6B with 4B), suggest that at some level of regulation, disruption and overexpression of csrA affect Tir levels via distinct pathways.
FIG. 6.
Expression of csrA from a multicopy plasmid globally represses expression from the LEE via GrlA. (A) Cultures of EPEC(pBR322) and EPEC(pCRA16) grown overnight were diluted in DMEM and allowed to grow to an OD of ∼1.0. CsrA protein levels were elevated by ∼1.5 ± 0.1 in the overexpressor [EPEC(pCRA16)] compared to those in the empty-vector-containing strain [EPEC(pBR322)]. (B and C) RNA was isolated from EPEC(pBR322) and EPEC(pCRA16) grown to an OD of ∼1.0, and real-time qRT-PCRs for escJ (LEE2), tir (LEE5), espA (LEE4), and escD (B) and ler (LEE1), grlR, and grlA (C) transcripts were performed as described above. Multicopy expression of csrA repressed the transcript levels from all of the LEE operons tested. Error bars represent the standard deviation of the mean from two separate experiments, with triplicate samples assayed every experiment. The unpaired Student t test was used to assay for the statistical significance of differences between EPEC(pBR322) and EPEC(pCRA16). A P value of <0.01 was considered statistically significant. ** denotes a P value of <0.001. (D) Western blotting for the steady-state protein levels of Ler and 3XFLAG-tagged GrlA was performed with cell lysates of EPEC(pBR322) and EPEC(pCRA16) grown to an OD of ∼0.5. Multicopy expression of csrA repressed the steady-state protein levels of Ler and GrlA-3XFLAG. NS refers to a cross-reacting nonspecific band. (E) The steady-state Ler levels were reduced in the overexpressor, even when both strains were growing at the same rate (OD600 of ∼0.2). NS refers to a cross-reacting nonspecific band. EPEC(pBR322) and EPEC(pCRA16) grew at comparable rates to an OD600 of ∼0.25, after which the overexpressor grew at a slightly slower rate (Fig. 1A, B, and C).
In EPEC, grlR and grlA are on the same bicistronic transcript because a PCR product was generated in a real-time qRT-PCR with a primer specific for grlR and another specific for grlA (data not shown). The grlRA operon encodes a repressor, GrlR, and an activator, GrlA (21). GrlA activates the expression from the LEE by upregulating the transcription of ler (7, 21), whereas binding of GrlR to GrlA has been proposed to prevent such activation (15, 38, 41). Ler, in turn, upregulates the transcription from LEE2, LEE3, LEE5, LEE4, and escD (26, 67). Because we observed reduced transcript levels of genes under the control of the ler regulon, we assessed the expression of ler, grlR, and grlA in the csrA-overexpressing strain. The transcript levels of the three genes were reduced by 7-, 14-, and 29-fold, respectively, in the overexpressor, as determined by real-time qRT-PCR (Fig. 6C). The corresponding protein levels of Ler and 3XFLAG-tagged GrlA were also substantially reduced (Fig. 6D), suggesting that overexpression of csrA globally shuts off the transcription from the LEE by repressing the expression of grlA. We were unable to assay for GrlR because no antibody was available, and we were unable to epitope tag grlR on the chromosome. Notably, no Ler protein was detectable in the csrA overexpressor, even when ler was expressed from a multicopy plasmid, despite the fact that complementation of EPEC Δler with the same plasmid resulted in Ler protein levels that were ∼17-fold higher than those of EPEC containing a single chromosomal copy of ler (data not shown).
We could not attribute the observed differences in gene expression from the LEE simply to the subtle alteration in growth exhibited by the csrA-overexpressing strain. While barely observable on a logarithmic scale (Fig. 1A), in a linear plot of growth, the csrA-overexpressing strain exhibited biphasic growth (black arrows in Fig. 1B), which resulted in a mild but detectable growth defect (Fig. 1B). This growth defect was evident only after the strain had grown past the early log phase beyond an OD600 of ∼0.25 as both strains grew at similar rates in the early log phase, with doubling times of ∼51.61 ± 8.3 and ∼54.70 ± 6.4 min for EPEC(pBR322) and EPEC(pCRA16), respectively (Fig. 1C). Steady-state levels of Ler protein were reduced in the csrA overexpressor (Fig. 6E), even when both strains were growing at the same rate (OD600, ∼0.2).
CsrA binds to the 5′ leader segment of the grlRA transcript.
Three predicted CsrA binding sites were identified in the leader segment of the grlRA transcript (Fig. 7A and B). Transcripts that are repressed by CsrA often have binding sites that overlap the Shine-Dalgarno sequence or are located nearby (5, 97). One of the predicted CsrA binding sites in grlRA is located one codon downstream of the translation initiation codon in EPEC (Fig. 7B). Bioinformatic analysis, together with the observation that grlR and grlA transcript levels were reduced in the csrA-overexpressing strain (Fig. 6C and D), suggested that CsrA, when present at high levels, might repress these genes by binding to the leader segment of the grlRA transcript. Binding of CsrA to the grlRA leader segment was evident at a CsrA concentration of 2.5 nM; at 80 nM, all of the transcript was present in the bound form (Fig. 7C). Multiple shifted ribonucleoprotein species were evident at CsrA concentrations as low as 20 nM (Fig. 7C, S1 and S2). On further increasing the CsrA concentration (≥320 nM), the transcript was evident in supershifted CsrA-RNA complexes S3 and S4 (Fig. 7C). Binding to the grlRA leader was specific because unlabeled grlRA, but not phoB, RNA competed with the labeled transcript (Fig. 7D). The apparent equilibrium binding constant of CsrA for grlRA was 6 ± 0.8 nM (Fig. 7E). Taken together, these data suggest that overexpression of CsrA globally represses transcription from the LEE in part by binding to the grlRA transcript and repressing the expression of GrlA.
FIG. 7.
Purified CsrA binds to the leader segment of the grlRA transcript. (A) Cartoon depicting the locations of putative CsrA binding sites in the leader segment of the grlRA transcript. The thick black arrows depict the genetic organization of grlR and grlA, respectively, in the grlRA operon. The three white rectangles in the leader segment and the 5′ region of the grlR ORF represent the predicted sites to which CsrA dimers (bound black balls) might bind. The vertical line with an arrow at the apex indicates the transcription start site. (B) The primary sequence of the leader segment of the grlRA transcript of EPEC, EHEC, and C. rodentium was scanned to determine the presence of putative CsrA binding sites. The σ70 transcriptional start site of the grlRA operon is highlighted in gray. The predicted Shine-Dalgarno sequence of the transcript is in bold and italicized. The predicted CsrA binding sites are in boldface. Note that each predicted CsrA binding site contains the highly conserved ANGGA motif (boldface and underlined) (4, 62). The highlighted trinucleotide AUG represents the translation initiation codon. References for the transcriptional start sites are shown on the right. (C) RNA EMSA with purified His-tagged CsrA and 32P-labeled grlRA leader RNA was performed as described above. The binding of CsrA to the leader segment of the grlRA transcript began to occur at a concentration of 2.5 nM. Higher-molecular-weight ribonucleoprotein complexes were observed beginning at a concentrations of 20 nM (S2, S3, and S4). CsrA and grlRA RNA concentrations are indicated at the bottom of each lane. U1 represents the unbound transcript, whereas S1, S2, S3, and S4 represent shifted ribonucleoprotein complexes. (D) Binding to grlRA was specific because high concentrations of unlabeled grlRA RNA, but not of the noncompetitor phoB RNA, successfully competed with the labeled transcripts. (E) The apparent equilibrium binding constant of CsrA for the grlRA transcript was 6 nM. The error bars represent the standard errors of the means determined from two independent experiments.
csrA regulates motility and glycogen biosynthesis in EPEC.
In nonpathogenic E. coli K-12, CsrA activates flagellar biogenesis by directly binding to the untranslated leader segment of the flhDC transcript and increasing its half-life (98). Similarly, inactivation of csrA rendered EPEC nonmotile, whereas complementation of the mutant with csrA restored motility (Fig. 8A). High levels of CsrA appeared to have a dose-dependent effect on motility, as the overexpressor was hypermotile compared to the empty-vector-containing strain (Fig. 8A). Consistent with the observed reduction in motility, flhD transcript levels exhibited an approximately sixfold reduction in the csrA mutant and the transcript levels were restored in the complemented strain (Fig. 8B). Besides motility, csrA also regulated glycogen biosynthesis in EPEC, with a csrA mutation resulting in elevated glycogen levels (Fig. 8C). Glycogen biosynthesis was repressed in a dose-dependent manner as multicopy presence of csrA in EPEC [EPEC(pCRA16)] did not elicit any color change on exposure to iodine in comparison to EPEC containing the empty vector [EPEC(pBR322)] (Fig. 8C).
FIG. 8.
csrA activates motility and represses glycogen biosynthesis in EPEC. (A) For motility assays, cultures grown overnight were diluted in LB broth or in LB broth supplemented with the appropriate antibiotic and allowed to grow to an OD of 1.0. One microliter of the culture was stabbed onto 0.3% 0.5× DMEM agar plates and incubated for 30 h at 37°C. Motility assays were performed in triplicate, and the diameter of the bacterial halo was measured as an indicator of bacterial motility. The unpaired Student t test was used to assay for the statistical significance of differences between the csrA mutant and EPEC or the csrA lacZ csrA+ strain or between EPEC(pBR322) and EPEC(pCRA16). A P value of <0.01 was considered statistically significant. ** denotes a P value of <0.001, and * denotes a P value of <0.01. (B) Real-time qRT-PCR for flhD (master regulator of flagella) was performed as described above. flhD transcript levels were substantially reduced in the csrA mutant and restored when the mutant was complemented with csrA in monocopy. ** denotes a P value of <0.001, whereas * denotes a P value of <0.005. (C) To measure glycogen biosynthesis, bacterial cultures were grown to an OD600 of ∼1.0, streaked onto Kornberg plates, and incubated at 37°C overnight, after which the plates were exposed to iodine vapor. Glycogen levels were elevated in the csrA mutant compared to those in the wild type (EPEC) or the complemented strain (csrA lacZ csrA+), as evident from increased black-brown color formation in the mutant. csrA repressed glycogen biosynthesis in a dose-dependent manner because multicopy expression of csrA in the wild-type background [EPEC(pCRA16)] did not lead to a color change, in contrast to the brown color observed for the empty-vector-containing strain [EPEC(pBR322)], which contains a single copy of csrA.
DISCUSSION
We have shown that csrA regulates gene expression from the LEE pathogenicity island of EPEC and is necessary for pedestal formation and disruption of TER, two in vitro correlates of pathogenesis. Whereas disruption and multicopy expression of csrA lead to a reduction in the levels of the LEE4-encoded transcripts, the mechanistic basis differs. The two predicted CsrA binding sites observed in the leader segment of the sepL transcript do not overlap the predicted Shine-Dalgarno sequence of sepL and are located 17 and 45 nucleotides upstream. Our results suggest that binding of CsrA to the leader segment of the sepL espADB transcript results in increased steady-state transcript levels in EPEC. In contrast, multicopy expression of csrA decreases transcription from the LEE operons, including LEE4, by repressing the expression of the transcriptional activator GrlA (Fig. 9).
csrA disruption and overexpression also differentially regulate the expression of Tir. Whereas tir transcript and bacterium-associated Tir protein levels are unaffected in the csrA mutant, reduced secretion of Tir in the csrA mutant is partly due to reduced synthesis of EscD and EscF. In the multicopy expressor, by contrast, tir transcript levels are greatly diminished, resulting in reduced Tir synthesis.
It is noteworthy that csrA, when expressed in a single copy, upregulates the expression of genes that form the architectural components of the TTSS (e.g., escD and escF), translocators that form a filamentous conduit between the EscF needle of the TTSS and the host cytosol (e.g., espA, espB, and espD) and the “gatekeeper switch” sepL. SepL and SepD interact (20, 72) to facilitate the hierarchical secretion of translocators over the effectors in response to changes in calcium and possibly other environmental signals (20), perhaps as a means of ensuring the formation of a functional secretion apparatus and subsequent delivery of effectors directly into the host cytosol rather than the medium. EPEC with a mutation in sepL is insensitive to the presence or absence of calcium and displays dysregulated secretion of effectors and translocators (20). Thus, SepL may be a component of a sensor apparatus that detects a reduced intracellular calcium concentration in the host cell and then promotes the release of effectors. Our results suggest that csrA is immediately proximal to sepL in a regulatory cascade, and thus it might play a role in coordinating this hierarchical secretion of translocators over effectors by posttranscriptionally regulating the expression of sepL in response to different environmental stimuli. Future experiments aimed at addressing whether csrA transcript or CsrA protein levels are affected by environmental factors known to regulate the expression and/or secretion of the proteins encoded by LEE4 genes will shed light on the possible involvement of CsrA in such a hierarchical secretion mechanism.
We have demonstrated that multicopy expression of csrA globally represses expression from the LEE locus, likely via GrlA. A grlA mutation might be expected to act epistatically with respect to a grlR mutation, because GrlR affects expression from the LEE locus by binding to and inhibiting GrlA activity (15, 38). Hence, a reduction in GrlR and GrlA should lead to overall transcriptional repression from lee1, resulting in reduced levels of the master regulator Ler. Consistent with this idea, ler transcript and Ler protein levels, as well as transcription from all of the LEE operons tested, were reduced in the CsrA overexpressor.
It is intriguing that the affinity of CsrA for the grlRA transcript is higher than that for the sepL transcript, especially as the repression of grlRA by CsrA appears to occur only at high CsrA concentrations. A likely explanation is that although CsrA initially binds to the grlRA transcript at low concentrations (KD, ∼6 nM), it does not produce physiologically relevant changes in gene expression, as evidenced by unchanged GrlA and Ler protein levels in the csrA mutant and the wild-type strain. However, at higher CsrA concentrations, higher-order complexes that affect gene expression may form. Consistent with this idea, multiple CsrA-grlRA ribonucleoprotein species are observed in gel shifts at higher CsrA concentrations (40 nM) (Fig. 7C, band S2). It is unlikely that S2 represents an artifact resulting from nonspecific aggregates of CsrA because whereas almost all of the grlRA leader transcript is evident in the supershifted species S2 at 80 nM, the sepL transcript remained in the S1 form. Notably, a concentration of 40 nM is within the range in which CsrA binds to other E. coli mRNAs (5). Our data also raise the possibility of at least one additional CsrA-regulated factor that affects gene expression from the LEE in lieu of the fact that escD transcript levels are reduced in the csrA mutant but purified CsrA does not bind to its leader segment.
CsrA is a pleiotropic regulator in EPEC as it also regulates motility and glycogen metabolism. The flhDC mRNA leader segment contains binding sites for CsrA that mediate its effects on motility in E. coli K-12 (98). These binding sites are conserved in EPEC. Thus, it is likely that in EPEC, as in K-12, flagellar biogenesis and consequent motility are mediated by the direct binding of CsrA to the flhDC leader transcript. However, there may also be differences in the CsrA-mediated regulation of motility between EPEC and K-12. E. coli K-12 is motile under diverse environmental conditions, including LB and LB supplemented with exogenous carbon sources such as acetate and succinate, whereas a glucose concentration of 10 mM represses motility (98). By contrast, EPEC remains motile on 0.5× DMEM, which contains 12.5 mM glucose, whereas an isogenic csrA mutant is nonmotile.
Thus, CsrA represents a novel posttranscriptional regulatory molecule in A/E pathogens that activates and represses the expression of the LEE-encoded genes in a concentration-dependent manner. Based on putatively identified CsrA binding sites in the leader segment of sepL and grlRA of EHEC and C. rodentium (Fig. 5B and 7B), and the conserved mode of regulation of ler by GrlR and GrlA (7, 21), we predict that CsrA functions similarly in these A/E pathogens.
An outstanding question is how EPEC and other A/E pathogens integrate transcriptional and posttranscriptional control of the LEE with motility and metabolism as they transit through the digestive tract. Our results raise the possibility that an important component of this coordinated regulatory system may be CsrA. Thus, regulation of the concentration or activity of CsrA, through transcriptional control or via its binding partners CsrB/C, in response to extracellular signals from other bacteria, or even signals from the host, may have pronounced effects on pathogenesis in vivo. We speculate that it would be advantageous for EPEC to mimic commensal strains, perhaps in a manner akin to the csrA overexpressor, during passage through the upper gastrointestinal tract. Thus, increased expression or activity of CsrA would be expected to repress the expression of virulence factors encoded on the LEE (Fig. 9), thereby minimizing energy expenditures on unneeded transcripts and reducing the possibility of detection of pathogen-specific antigens by the innate immune system. Moreover, increased flagellar motility might facilitate transit to the sites of colonization. As EPEC approaches the small intestine, decreasing CsrA protein levels or activity may prove advantageous. Thus, reducing flagellar motility in coordination with increased transcription and translation of translocators, followed by effectors, would permit the bacteria to efficiently attach and colonize. Using the mutants we have developed, we are currently testing how coordinated regulation of flagellar motility, metabolism, and LEE expression by CsrA contributes to pathogenesis and immune evasion in vivo.
Acknowledgments
We thank Bernie Weiss and Phil Rather for critical evaluation of the manuscript and Guy Benian, Charlie Moran, and the members of the Kalman, Romeo, and Weiss labs for helpful discussions and suggestions. We are grateful to June Scott (Emory University) for providing the wild-type strain EPEC 2348/69, Jim Kaper and Michael Donnenberg (University of Maryland) for strains (EPEC ΔespA, EPEC ΔespB, and EPEC ΔespD), and antibodies (anti-Tir, anti-EspA, and anti-EspB), B. Brett Finlay (University of British Columbia) for EPEC Δtir, Jay Mellies (Reed College) for EPEC Δler, Vanessa Sperandio (UT-Southwestern, Dallas) for pJRLacZIns, Rebekah DeVinney (University of Calgary) for the anti-EspD antibody, Phil Rather (Emory University) for S17-1λpir, Bettina Bommarius (Emory University) for DH5α, and Ilan Rosenshine (The Hebrew University, Jerusalem) for anti-Ler.
This work was supported by NIH grants R01DK074731-01 and R01-A1056067-01 to D.K. and R01-GM059969 to T.R. S.B. is the recipient of National Science Foundation award 0450303 and subaward I-66-606-63 to Emory University.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Abe, H., I. Tatsuno, T. Tobe, A. Okutani, and C. Sasakawa. 2002. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 703500-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 686790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anyanful, A., J. Dolan-Livengood, T. Lewis, S. Sheth, M. N. DeZalia, M. Sherman, L. V. Kalman, G. M. Benian, and D. Kalman. 2005. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol. Microbiol. 57988-1007. [DOI] [PubMed] [Google Scholar]
- 4.Babitzke, P., and T. Romeo. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10156-163. [DOI] [PubMed] [Google Scholar]
- 5.Baker, C. S., L. A. Eory, H. Yakhnin, J. Mercante, T. Romeo, and P. Babitzke. 2007. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol. 1895472-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, C. S., I. Morozov, K. Suzuki, T. Romeo, and P. Babitzke. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 441599-1610. [DOI] [PubMed] [Google Scholar]
- 7.Barba, J., V. H. Bustamante, M. A. Flores-Valdez, W. Deng, B. B. Finlay, and J. L. Puente. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 1877918-7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnard, F. M., M. F. Loughlin, H. P. Fainberg, M. P. Messenger, D. W. Ussery, P. Williams, and P. J. Jenks. 2004. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 5115-32. [DOI] [PubMed] [Google Scholar]
- 9.Bommarius, B., D. Maxwell, A. Swimm, S. Leung, A. Corbett, W. Bornmann, and D. Kalman. 2007. Enteropathogenic Escherichia coli Tir is an SH2/3 ligand that recruits and activates tyrosine kinases required for pedestal formation. Mol. Microbiol. 631748-1768. [DOI] [PubMed] [Google Scholar]
- 10.Burrowes, E., C. Baysse, C. Adams, and F. O'Gara. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152405-418. [DOI] [PubMed] [Google Scholar]
- 11.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39664-678. [DOI] [PubMed] [Google Scholar]
- 12.Canil, C., I. Rosenshine, S. Ruschkowski, M. S. Donnenberg, J. B. Kaper, and B. B. Finlay. 1993. Enteropathogenic Escherichia coli decreases the transepithelial electrical resistance of polarized epithelial monolayers. Infect. Immun. 612755-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H. D., and G. Frankel. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 2983-98. [DOI] [PubMed] [Google Scholar]
- 14.Cleary, J., L. C. Lai, R. K. Shaw, A. Straatman-Iwanowska, M. S. Donnenberg, G. Frankel, and S. Knutton. 2004. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150527-538. [DOI] [PubMed] [Google Scholar]
- 15.Creasey, E. A., R. M. Delahay, S. J. Daniell, and G. Frankel. 2003. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 1492093-2106. [DOI] [PubMed] [Google Scholar]
- 16.Cui, Y., A. Chatterjee, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 1775108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniell, S. J., N. Takahashi, R. Wilson, D. Friedberg, I. Rosenshine, F. P. Booy, R. K. Shaw, S. Knutton, G. Frankel, and S. Aizawa. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3865-871. [DOI] [PubMed] [Google Scholar]
- 18.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean, P., and B. Kenny. 2004. Intestinal barrier dysfunction by enteropathogenic Escherichia coli is mediated by two effector molecules and a bacterial surface protein. Mol. Microbiol. 54665-675. [DOI] [PubMed] [Google Scholar]
- 20.Deng, W., Y. Li, P. R. Hardwidge, E. A. Frey, R. A. Pfuetzner, S. Lee, S. Gruenheid, N. C. Strynakda, J. L. Puente, and B. B. Finlay. 2005. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 732135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 1013597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donnenberg, M. S., J. Yu, and J. B. Kaper. 1993. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J. Bacteriol. 1754670-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubey, A. K., C. S. Baker, T. Romeo, and P. Babitzke. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 111579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubey, A. K., C. S. Baker, K. Suzuki, A. D. Jones, P. Pandit, T. Romeo, and P. Babitzke. 2003. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 1854450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott, S. J., S. W. Hutcheson, M. S. Dubois, J. L. Mellies, L. A. Wainwright, M. Batchelor, G. Frankel, S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 331176-1189. [DOI] [PubMed] [Google Scholar]
- 26.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 686115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forsbach-Birk, V., T. McNealy, S. Chunwei, D. Lynch, and R. Marre. 2004. Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int. J. Med. Microbiol. 29415-25. [DOI] [PubMed] [Google Scholar]
- 28.Foubister, V., I. Rosenshine, M. S. Donnenberg, and B. B. Finlay. 1994. The eaeB gene of enteropathogenic Escherichia coli is necessary for signal transduction in epithelial cells. Infect. Immun. 623038-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34941-952. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41549-559. [DOI] [PubMed] [Google Scholar]
- 31.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48507-521. [DOI] [PubMed] [Google Scholar]
- 32.Gruenheid, S., R. DeVinney, F. Bladt, D. Goosney, S. Gelkop, G. D. Gish, T. Pawson, and B. B. Finlay. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3856-859. [DOI] [PubMed] [Google Scholar]
- 33.Gutiérrez, P., Y. Li, M. J. Osborne, E. Pomerantseva, Q. Liu, and K. Gehring. 2005. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J. Bacteriol. 1873496-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guttman, J. A., Y. Li, M. E. Wickham, W. Deng, A. W. Vogl, and B. B. Finlay. 2006. Attaching and effacing pathogen-induced tight junction disruption in vivo. Cell. Microbiol. 8634-645. [DOI] [PubMed] [Google Scholar]
- 35.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heeb, S., S. A. Kuehne, M. Bycroft, S. Crivii, M. D. Allen, D. Haas, M. Camara, and P. Williams. 2006. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J. Mol. Biol. 3551026-1036. [DOI] [PubMed] [Google Scholar]
- 37.Heroven, A. K., K. Bohme, M. Rohde, and P. Dersch. 2008. A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 681179-1195. [DOI] [PubMed] [Google Scholar]
- 38.Huang, L. H., and W. J. Syu. 2008. GrlA of enterohemorrhagic Escherichia coli O157:H7 activates LEE1 by binding to the promoter region. J. Microbiol. Immunol. Infect. 419-16. [PubMed] [Google Scholar]
- 39.Ide, T., S. Laarmann, L. Greune, H. Schillers, H. Oberleithner, and M. A. Schmidt. 2001. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3669-679. [DOI] [PubMed] [Google Scholar]
- 40.Iyoda, S., and H. Watanabe. 2005. ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J. Bacteriol. 1874086-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jobichen, C., M. Li, G. Yerushalmi, Y. W. Tan, Y. K. Mok, I. Rosenshine, K. Y. Leung, and J. Sivaraman. 2007. Structure of GrlR and the implication of its EDED motif in mediating the regulation of type III secretion system in EHEC. PLoS Pathog. 3e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalman, D., O. D. Weiner, D. L. Goosney, J. W. Sedat, B. B. Finlay, A. Abo, and J. M. Bishop. 1999. Enteropathogenic E. coli acts through WASP and Arp2/3 complex to form actin pedestals. Nat. Cell Biol. 1389-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kenny, B., A. Abe, M. Stein, and B. B. Finlay. 1997. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect. Immun. 652606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenny, B., R. Devinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91511-520. [DOI] [PubMed] [Google Scholar]
- 45.Kenny, B., and B. B. Finlay. 1995. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc. Natl. Acad. Sci. USA 927991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenny, B., L. C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20313-323. [DOI] [PubMed] [Google Scholar]
- 47.Kerrinnes, T., Z. B. Zelas, W. Streckel, F. Faber, E. Tietze, H. Tschape, and S. Yaron. 2009. CsrA and CsrB are required for the post-transcriptional control of the virulence-associated effector protein AvrA of Salmonella enterica. Int. J. Med. Microbiol. 299333-341. [DOI] [PubMed] [Google Scholar]
- 48.Knutton, S., M. M. Baldini, J. B. Kaper, and A. S. McNeish. 1987. Role of plasmid-encoded adherence factors in adhesion of enteropathogenic Escherichia coli to HEp-2 cells. Infect. Immun. 5578-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 571290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 172166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraiss, A., J. Blass, and J. Reidl. 1998. A suicide plasmid (pJR lacZins) for targeted integration for non-native genes into the chromosome of Escherichia coli. Tech. Tips Online 1TO1249. [Google Scholar]
- 52.Kresse, A. U., F. Beltrametti, A. Muller, F. Ebel, and C. A. Guzman. 2000. Characterization of SepL of enterohemorrhagic Escherichia coli. J. Bacteriol. 1826490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laaberki, M., N. Janabi, E. Oswald, and F. Repoila. 2006. Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157:H7: interplay between Ler, GrlA, HNS and RpoS. Int. J. Med. Microbiol. 296197-210. [DOI] [PubMed] [Google Scholar]
- 54.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 55.Lai, L. C., L. A. Wainwright, K. D. Stone, and M. S. Donnenberg. 1997. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect. Immun. 652211-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawhon, S. D., J. G. Frye, M. Suyemoto, S. Porwollik, M. McClelland, and C. Altier. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 481633-1645. [DOI] [PubMed] [Google Scholar]
- 57.Leverton, L. Q., and J. B. Kaper. 2005. Temporal expression of enteropathogenic Escherichia coli virulence genes in an in vitro model of infection. Infect. Immun. 731034-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu, M. Y., G. Gui, B. Wei, J. F. Preston, 3rd, L. Oakford, U. Yuksel, D. P. Giedroc, and T. Romeo. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 27217502-17510. [DOI] [PubMed] [Google Scholar]
- 59.Liu, M. Y., and T. Romeo. 1997. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 1794639-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lodato, P. B., and J. B. Kaper. 2009. Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 71273-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193265-275. [PubMed] [Google Scholar]
- 62.Lucchetti-Miganeh, C., E. Burrowes, C. Baysse, and G. Ermel. 2008. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 15416-29. [DOI] [PubMed] [Google Scholar]
- 63.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23399-407. [DOI] [PubMed] [Google Scholar]
- 64.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. P. Nougayrede, M. S. Donnenberg, and G. Hecht. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mellies, J. L., A. M. Barron, and A. M. Carmona. 2007. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect. Immun. 754199-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mellies, J. L., A. M. Barron, K. R. Haack, A. S. Korson, and D. A. Oldridge. 2006. The global regulator Ler is necessary for enteropathogenic Escherichia coli colonization of Caenorhabditis elegans. Infect. Immun. 7464-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33296-306. [DOI] [PubMed] [Google Scholar]
- 68.Mercante, J., K. Suzuki, X. Cheng, P. Babitzke, and T. Romeo. 2006. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J. Biol. Chem. 28131832-31842. [DOI] [PubMed] [Google Scholar]
- 69.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 411340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'Connell, C. B., E. A. Creasey, S. Knutton, S. Elliott, L. J. Crowther, W. Luo, M. J. Albert, J. B. Kaper, G. Frankel, and M. S. Donnenberg. 2004. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol. Microbiol. 521613-1625. [DOI] [PubMed] [Google Scholar]
- 73.Ogino, T., R. Ohno, K. Sekiya, A. Kuwae, T. Matsuzawa, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, and A. Abe. 2006. Assembly of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 1882801-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pernestig, A. K., D. Georgellis, T. Romeo, K. Suzuki, H. Tomenius, S. Normark, and O. Melefors. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 185843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 1836676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poteete, A. R., and A. C. Fenton. 1984. Lambda red-dependent growth and recombination of phage P22. Virology 134161-167. [DOI] [PubMed] [Google Scholar]
- 77.Rife, C., R. Schwarzenbacher, D. McMullan, P. Abdubek, E. Ambing, H. Axelrod, T. Biorac, J. M. Canaves, H. J. Chiu, A. M. Deacon, M. DiDonato, M. A. Elsliger, A. Godzik, C. Grittini, S. K. Grzechnik, J. Hale, E. Hampton, G. W. Han, J. Haugen, M. Hornsby, L. Jaroszewski, H. E. Klock, E. Koesema, A. Kreusch, P. Kuhn, S. A. Lesley, M. D. Miller, K. Moy, E. Nigoghossian, J. Paulsen, K. Quijano, R. Reyes, E. Sims, G. Spraggon, R. C. Stevens, H. van den Bedem, J. Velasquez, J. Vincent, A. White, G. Wolf, Q. Xu, K. O. Hodgson, J. Wooley, and I. A. Wilson. 2005. Crystal structure of the global regulatory protein CsrA from Pseudomonas putida at 2.05 Å resolution reveals a new fold. Proteins 61449-453. [DOI] [PubMed] [Google Scholar]
- 78.Roe, A. J., H. Yull, S. W. Naylor, M. J. Woodward, D. G. Smith, and D. L. Gally. 2003. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect. Immun. 715900-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 291321-1330. [DOI] [PubMed] [Google Scholar]
- 80.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1754744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 82.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 9811638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharp, F. C., and V. Sperandio. 2007. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 752432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 85.Sircili, M. P., M. Walters, L. R. Trabulsi, and V. Sperandio. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 722329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 703085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expressionof the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9615196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 1835187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 1008951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43809-821. [DOI] [PubMed] [Google Scholar]
- 91.Suzuki, K., P. Babitzke, S. R. Kushner, and T. Romeo. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 202605-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suzuki, K., X. Wang, T. Weilbacher, A. K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 1845130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swimm, A., B. Bommarius, Y. Li, D. Cheng, P. Reeves, M. Sherman, D. Veach, W. Bornmann, and D. Kalman. 2004. Enteropathogenic Escherichia coli use redundant tyrosine kinases to form actin pedestals. Mol. Biol. Cell 153520-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 1482735-2744. [DOI] [PubMed] [Google Scholar]
- 95.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 9815264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wachter, C., C. Beinke, M. Mattes, and M. A. Schmidt. 1999. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 311695-1707. [DOI] [PubMed] [Google Scholar]
- 97.Wang, X., A. K. Dubey, K. Suzuki, C. S. Baker, P. Babitzke, and T. Romeo. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 561648-1663. [DOI] [PubMed] [Google Scholar]
- 98.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40245-256. [DOI] [PubMed] [Google Scholar]
- 99.Weilbacher, T., K. Suzuki, A. K. Dubey, X. Wang, S. Gudapaty, I. Morozov, C. S. Baker, D. Georgellis, P. Babitzke, and T. Romeo. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48657-670. [DOI] [PubMed] [Google Scholar]
- 100.Wilson, R. K., R. K. Shaw, S. Daniell, S. Knutton, and G. Frankel. 2001. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell. Microbiol. 3753-762. [DOI] [PubMed] [Google Scholar]