Abstract
We previously identified a gene cluster, epa (for enterocococcal polysaccharide antigen), involved in polysaccharide biosynthesis of Enterococcus faecalis and showed that disruption of epaB and epaE resulted in attenuation in translocation, biofilm formation, resistance to polymorphonuclear leukocyte (PMN) killing, and virulence in a mouse peritonitis model. Using five additional mutant disruptions in the 26-kb region between orfde2 and OG1RF_0163, we defined the epa locus as the area from epaA to epaR. Disruption of epaA, epaM, and epaN, like prior disruption of epaB and epaE, resulted in alteration in Epa polysaccharide content, more round cells versus oval cells with OG1RF, decreased biofilm formation, attenuation in a mouse peritonitis model, and resistance to lysis by the phage NPV-1 (known to lyse OG1RF), while mutants disrupted in orfde2 and OG1RF_163 (the epa locus flanking genes) behaved like OG1RF in those assays. Analysis of the purified Epa polysaccharide from OG1RF revealed the presence of rhamnose, glucose, galactose, GalNAc, and GlcNAc in this polysaccharide, while carbohydrate preparation from the epaB mutant did not contain rhamnose, suggesting that one or more of the glycosyl transferases encoded by the epaBCD operon are necessary to transfer rhamnose to the polysaccharide. In conclusion, the epa genes, uniformly present in E. faecalis strains and involved in biosynthesis of polysaccharide in OG1RF, are also important for OG1RF shape determination, biofilm formation, and NPV-1 replication/lysis, as well as for E. faecalis virulence in a mouse peritonitis model.
Enterococci are among the major causes of endocarditis and nosocomial infections, with Enterococcus faecalis the most commonly identified species (7, 22). The antibiotic resistance of enterococci not only causes difficulty in treatment of enterococcal infections but also appears to promote their survival in hospitalized, antibiotic-exposed individuals, helping to explain their important role in the nosocomial milieu.
In order to understand the mechanisms of enterococcal pathogenesis, which may lead to development of alternative approaches to prevent and/or treat enterococcal infections, we and other investigators have identified several enterococcal factors important for virulence, including proteins and polysaccharides (11). Polysaccharides are composed of repeating units of oligosaccharides, are important components of bacterial cell walls, and can be associated with bacterial surfaces through linkage to the cell membrane, peptidoglycan, or by other unknown mechanisms. Polysaccharides also play important roles in bacterial pathogenesis. They have been shown to be important for adherence to and invasion of host tissues (10, 19), for resistance to host defense systems such as phagocytosis (1, 2, 8, 12, 24, 30), and for induction of host inflammatory responses (14, 25). Vaccine potential of surface polysaccharides has been shown for different bacteria, such as pneumococci (20), and antibodies against capsular carbohydrates have been shown to promote polymorphonuclear leukocyte (PMN)-mediated killing of E. faecalis and Enterococcus faecium and to protect mice against E. faecalis infection (16-18, 24). These capsular carbohydrates of E. faecalis have been shown to be type specific, and the type-specific genes are present only in some populations of enterococci (16-18).
We previously identified an Escherichia coli recombinant clone, TX5159, containing 43-kb cloned DNA from E. faecalis OG1RF, that reacted with sera from four patients with E. faecalis endocarditis but not with rabbit serum raised against surface protein extracts of E. faecalis (31, 32). Sequence analysis of the clone revealed similarity to genes involved in polysaccharide biosynthesis in other organisms, and the ladder-like material expressed in E. coli was sensitive to periodate but resistant to proteinase K treatment, indicating that these genes are involved in polysaccharide biosynthesis (32). The genes identified in this clone were named “orfde1” through “orfde16” (Table 1), and the gene cluster was named “epa” (for enterococcal polysaccharide antigen) (32). Although the start and the end of the epa gene cluster were not determined in E. faecalis, our previous sequence analysis of the cosmid clone revealed putative promoters upstream of orfde1, -2, -3, -4, -5-6, -6, -11, and -16 and our data with reverse transcription-PCR (RT-PCR) and transposon insertions in the E. coli clone indicated that orfde6 (epaE) was cotranscribed with orfde7 to -10 but was transcribed independently from the upstream gene, orfde4 (epaB), and from the downstream gene, orfde11 (epaL) (32, 33). Our previous results also suggested that orfde4 (epaB) and orfde6 (epaE), but not orfde1 and orfde2, are required for polysaccharide biosynthesis (32) and comparison of the orfde4 (epaB) and orfde6 (epaE) disruption mutants with wild-type OG1RF showed that these mutants are deficient in translocation across polarized human enterocyte-like T84 cells (34) and biofilm formation (21), are more susceptible to PMN-mediated killing (26), and are attenuated in a mouse peritonitis model (33). The role of the other genes in polysaccharide biosynthesis has not been previously analyzed. We have also observed that antibody (using patient serum) eluted from the polysaccharide material produced by the E. coli clone did not react with an E. faecalis OG1RF extract in Western blots (33). However, use of whole serum from the same patient detected polysaccharides (by Western blots) in E. faecalis OG1RF but not in the orfde4 (epaB) and orfde6 (epaE) disruption mutants (26), suggesting that the polysaccharide expressed by OG1RF may be different structurally from that expressed by the E. coli clone TX5159 containing the OG1RF epa genes. The structure and location of Epa have not been reported, although other investigators have suggested that Epa may be buried inside the cell wall (13).
TABLE 1.
Annotation of cosmid, OG1RF ID, and V583 ID
| Prior cosmid gene namea | OG1RF ID | V583 ID | % Similarityb | Definition |
|---|---|---|---|---|
| orfde1 | orfde1 | EF2200 | 100 | Methionine aminopeptidase |
| orfde2 | orfde2 | EF2199 | 100 | Probable RNase BN |
| orfde3 | epaA | EF2198 | 100 | Glycosyl transferase, group 4 family protein |
| orfde4 | epaB | EF2197 | 100 | Glycosyl transferase, group 2 family protein |
| orfde5 | epaC | EF2196 | 98 | Glycosyl transferase, group 2 family protein |
| orfde5-6 | epaD | EF2195 | 100 | Glycosyl transferase, group 2 family protein |
| orfde6 | epaE | EF2194 | 100 | Glucose-1-phosphate thymidylyltransferase |
| orfde7 | epaF | EF2193 | 100 | dTDP-4-dehydrorhamnose 3,5-epimerase |
| orfde8 | epaG | EF2192 | 99 | dTDP-glucose 4,6-dehydratase |
| orfde9 | epaH | EF2191 | 99 | dTDP-4-dehydrorhamnose reductase |
| orfde10 | epaI | EF2190 | 99 | Glycosyl transferase, group 2 family protein |
| Not identified | epaJ | EF2189 | 97 | Hypothetical membrane protein |
| Not identified | epaK | EF2184 | 100 | Hypothetical membrane protein |
| orfde11 | epaL | EF2183 | 100 | ABC transporter, permease protein |
| orfde12 | epaMd | EF2182 | 100 | ABC transporter, ATP-binding protein |
| orfde13 | ||||
| orfde14 | epaNe | EF2181 | 99 | Glycosyl transferase, group 2 family protein |
| orfde15 | ||||
| orfde16 | epaO | EF2180 | 99 | Glycosyl transferase, group 2 family protein |
| epaP | EF2179 | 100 | Hypothetical membrane protein | |
| epaQ | EF2178 | 99 | Hypothetical membrane protein | |
| epaR | EF2177 | 100 | Sugar transferase | |
| OG1RF_0164c | EF2176 | Glycosyl transferase, group 2 family protein | ||
| EF2175f | LicD-related protein | |||
| OG1RF_0163 | EF2174 | 81 | Glycosyl transferase, group 25 family protein |
Percentage of similarity between OG1RF and its homologue in V583.
As designated by Bourgogne et al. (4).
orfde12 and orfde13 form the upstream and downstream portions of epaM.
orfde14 and orfde15 form the upstream and downstream portions of epaN.
EF2175 is a pseudogene in OG1RF.
In the present study, we examined the limits of the epa gene cluster in OG1RF, the composition of Epa polysaccharides, and some of the functions associated with the polysaccharide gene cluster.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in the study are listed in Table 2. The epaB (formerly, orfde4) and epaE (formerly, orfde6) disruption mutants TX5179 and TX5180, as well as the epaBCD-complemented strain TX5179.1, were constructed previously (33, 34). Cosmid, OG1RF, and V583 identification (ID) tags are shown in Table 1. A transposon insertion mutant in the OG1RF homologue of EF2174 (OG1RF_0163) (4) was kindly provided by Danielle Garsin (9). Escherichia coli strains were grown in LB medium (Difco Laboratories, Detroit, MI) with appropriate antibiotics, while E. faecalis strains were grown routinely in brain heart infusion (BHI) (Difco) or Todd Hewitt (TH) (Difco) medium with appropriate antibiotics, unless otherwise indicated.
TABLE 2.
Strains used in this study
| Strain (alternative name) | Characteristicsa | Source or reference |
|---|---|---|
| OG1RF (TX4002) | Well-characterized E. faecalis strain; Fusr Rifr | 23 |
| TX10113 | orfde2 (EF2199) disruption mutant, disruption starts at bp +616 of the 908-bp orfde2 gene; Kanr-src | This study |
| TX10114 | epaA (formerly orfde3) disruption mutant, disruption starts at bp +1111 of the 1,136-bp epaA gene; Kanr | This study |
| TX5179 | epaB (formerly orfde4) disruption mutant, disruption starts at bp +243 of the 788-bp epaB gene; Kanr | 33 |
| TX5180 | epaE (formerly orfde6) disruption mutant, disruption starts at bp +252 of the 866-bp epaE gene; Kanr | 33 |
| TX5391 | epaM disruption mutant, disruption starts at bp +1,000 of the 1,217-bp epaM gene; Kanr | This study |
| TX5436 | epaN disruption mutant, disruption starts at bp +2222 of the 3,143-bp epaN gene; Kanr | This study |
| TX5179.1 | epaB-complemented strain, in which the plasmid contains 3,198 bp that includes epaB, epaC, and 682/713 bp of epaD; Eryr Kanr | 34 |
| DAGF29 | Tn917 transposon insertion at bp +1245 from the OG1RF_0163/EF2174 ATG in OG1RF; Eryr | 9 |
Chl, chloramphenicol; Ery, erythromycin; Fus, fusidic acid; Kan, kanamycin; Rif, rifampin.
Construction of disruption mutants, growth, stability, and morphology assays.
The orfde2, epaA, epaM, and epaN disruption mutants of E. faecalis OG1RF were constructed using intragenic fragments (primers are listed in Table S1 in the supplemental material) and the suicide vector pTEX4577 by methods described previously (28). Briefly, an internal fragment was amplified from OG1RF genomic DNA and cloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA). The fragments were then released from the vector by EcoRI and cloned into pTEX4577. The resulting constructs were electroporated into E. faecalis OG1RF, and the mutants were selected with 2,000 μg of kanamycin per ml. Growth of wild-type OG1RF and the mutants was examined by growth in BHI, in Trypticase soy broth (Difco) with 0.25% glucose (TSBG; medium used for biofilm), and in BHI plus 40% horse serum (BHIS) and measurement of the optical density at 600 nm (OD600) at different time points up to 14 h. CFU were determined at 0, 3, 6, and 24 h on BHI agar. Stability of the mutants was examined by growth in BHI broth without antibiotics for 24 h and counting the number of CFU on BHI and BHI-kanamycin (2,000 μg/ml) plates. Bacterial morphology after 24 h of growth in BHI, BHIS, TSBG, and in TH + 1% glucose was observed under a phase-contrast microscope. Statistical analysis was performed using a nonparametric t test and analysis of variance for multiple comparisons.
qRT-PCR.
RNA was extracted using RNAwiz (Ambion, Austin, TX) from cells collected at the late log growth phase from BHI broth as described previously (5). Five micrograms of RNA was treated twice with DNA-free (Ambion, Austin, TX) according to the supplier's protocol. Quantitative RT-PCR (qRT-PCR) on cDNA was performed using SYBR green (Applied Biosystems, Foster City, CA) and an XFRTG cycler (Applied Biosystems) as previously published (6). The primers are listed in Table S1 in the supplemental material, and their location is illustrated by an asterisk in Fig. 1B. The amount of transcript obtained for each of the primer sets was normalized with the amount of gyrB transcripts. Statistical analysis was performed using a nonparametric t test.
FIG. 1.
OG1RF and its derivatives. (A) Illustration of the epa gene cluster. In white are the genes belonging to the epa locus. (B) Disruptions in orfde2, epaA, epaB, epaE, epaM, and epaN (using pTEX4577) are denoted by an inverted triangle, while disruption of OG1RF_163 was made using Tn917. The name of the strain is in boldface, and the disrupted gene is indicated in parentheses. The arrows above the genes represent the transcripts determined by qRT-PCR. The asterisks indicate the locations of the qRT-PCR primers for each gene. (C) Growth curve of OG1RF and its derivatives in TSBG (biofilm medium). (D) Growth curve of OG1RF and its derivatives in BHIS. These experiments (C and D) were performed at least twice, and results from one representative experiment are shown.
Extraction of crude carbohydrates from E. faecalis and analysis.
E. faecalis OG1RF and its derivatives were inoculated into 200 ml of TH broth with 1.0% glucose and grown without shaking for 24 h at 37°C. Cells were harvested and resuspended in 1.5 ml phosphate-buffered saline (PBS), followed by addition of 100 μl of mutanolysin (5 U/μl) and 250 μl of lysozyme (40 mg/ml). After 1 h of incubation at 37°C, 20 μl of MgSO4 (1 M), 4 μl of CaCl2 (0.5 M), 20 μl of RNase (10 mg/ml), and 20 μl of DNase I (10 mg/ml) were added and the mixture was incubated for an additional 2 h at 37°C, followed by addition of 40 μl of proteinase K (25 mg/ml) and overnight incubation at 56°C. All enzymes were obtained from Sigma (St. Louis, MO). Cell debris was removed by centrifugation, supernatant was recovered and extracted with CIP (chloroform-isoamyl alcohol-phenol at 24:1:25 [vol/vol/vol]), and polysaccharides were precipitated by ethanol (final concentration, 80%). After centrifugation, the pellet was washed with 75% ethanol, air-dried, and dissolved in 200 μl of distilled water (dH2O). To separate polysaccharides, 10-μl samples were run on a 0.8% agarose gel and stained with Stains-All (Sigma) overnight in the dark and destained with light. A Western blot was performed as described previously (31).
Purification and compositional analysis of polysaccharides.
A crude extract of carbohydrates prepared (as described above) from 20 liters of E. faecalis OG1RF was used for column purification. The crude extract (about 15 ml) was first purified using BioGel P-100 (Bio-Rad, Hercules, CA) and 3 bed volumes of 50 mM Tris-HCl (pH 8.0). The collected fractions were assayed by a colorimetric method for carbohydrate quantification. Briefly, 60 μl of sample was added into a well of a 96-well microtiter plate, followed by addition of 30 μl of 5% phenol and 140 μl of 95.5% H2SO4. The plate was incubated at room temperature for 20 min, and the OD490 was measured. The approximate amount of carbohydrates in a sample was calculated by comparing the OD490 of the sample versus that of a series of glucose standards (10, 5, 2.5, 1.25, and 0.625 mg/ml). The fractions were pooled and further purified using the Macroprep DEAE support (Bio-Rad) and a NaCl gradient (0 to 1 M in 50 mM Tris-HCl). The pooled fractions were assayed with an enterococcal patient serum using dot blots, dialyzed with dH2O, and kept at −80°C. The pooled (prepurified) fractions were further purified at the Complex Carbohydrate Research Center, University of Georgia, by Superose 6 size exclusion column (elution buffer, 10 mM ammonium formate [pH 5.0]) or normal-phase chromatography. For normal-phase chromatography, polysaccharide was loaded onto an Avicel PH101 microcrystalline cellulose column which had been preequilibrated in 50% ethanol followed by elution with an ethanol gradient (50 to 0%) buffered to pH 5.65 with triethylammonium acetate. A crude extract of carbohydrates from TX5179 (epaB) was also sent to the Complex Carbohydrate Research Center, where it was purified by Superose 6 size exclusion column. Compositional and 31P-nuclear magnetic resonance analyses were performed by the Complex Carbohydrate Research Center, University of Georgia.
Biofilm assay.
Biofilm formation by OG1RF and derivatives was determined as described previously with cells grown in TSBG (21). After 24 h of growth and before being processed for biofilm formation, OD600 was determined (growth OD). Biofilm formation was assessed at OD570 after processing and crystal violet staining. The experiments were performed twice, and eight wells per strain were used in each experiment. One-way analysis of variance with Bonferroni's post test was used to compare OG1RF and derivatives.
Phage infection assays.
Phage NPV-1 stock (29) was prepared using E. faecalis OG1RF. To compare the phage sensitivity of OG1RF and its derivatives, E. faecalis strains were grown in BHI broth with appropriate antibiotics at 37°C overnight. Cells were washed and resuspended in 10 mM MgSO4 (adjusted to an OD550 of ≈1 for all strains). Ca. 100 NPV-1 phage (in a 100-μl volume) were mixed with 100 μl of OD-adjusted bacterial cells and incubated at room temperature for 20 min, followed by addition of 4 ml BHI soft agar (0.7%), and then poured onto a BHI agar plate. Plaques were counted after overnight incubation at 37°C. For the adsorption test, 100 μl of bacterial cells (prepared as described above) and 1,000 NPV-1 phage in a 100-μl volume were mixed, the mixture was centrifuged after 1, 5, or 10 min of incubation at room temperature, and the supernatant was taken and the titer of phage was determined using OG1RF.
Electron microscopy.
E. faecalis OG1RF and the orfde4 (epaB) mutant were grown overnight in BHI broth at 37°C. In the first set of experiments, the cells were processed directly for electron microscopy (3). In a second set of experiments, 500 μl of the cultures was collected and resuspended in 500 μl of fresh BHI broth, followed by mixing with phage NPV-1 (bacterium/phage ratio of 1:2). After 10, 30, or 45 min of incubation at 37°C, the bacterial cells were collected, resuspended in 500 μl of 2.5% glutaradehyde, and incubated at room temperature for 20 min. After centrifugation, the cell pellets were kept at 4°C before processing for electron microscopy (3). At least 10 different fields were examined, and representative fields were selected.
Mouse peritonitis model.
In vivo testing was performed with the mouse peritonitis model, and the 50% lethal dose and Kaplan-Meier survival curves were determined and log rank analysis was performed as described previously (27).
RESULTS
Organization of the epa cluster and effect of disruption of epa genes on growth and cell morphology.
Based on the results to be discussed below and on the OG1RF genome sequence (4), the genes previously designated as “orfde3” to “orfde16” (derived from the cosmid sequence) (31, 32) were renamed here as “epaA” to “epaO” (Fig. 1A and Table 1). In addition, three genes downstream of epaO are part of an operon with epaLMNO (see below); thus, these three genes were designated “epaP,” “epaQ,” and “epaR,” the last one encoding a sugar transferase. We also identified two short ORFs (epaJ and epaK) between epaI (orfde10) and epaL (orfde11) that encode hypothetical membrane proteins that were not identified in the previously described cosmid clone of this region. Finally, epaM corresponds to the area previously annotated in the cosmid as “orfde12” and “orfde13” (forming the upstream and downstream portions of epaM) and epaN corresponds to the area including both orfde14 and orfde15, bringing the total number of known genes in the epa locus in OG1RF to 18 (Fig. 1A). An updated annotation of those genes is included in Table 1. Disruption of orfde2, epaA, epaM, and epaN resulted in OG1RF derivatives TX10113, TX10114, TX5391, and TX5436, respectively. (All of the disruptions are illustrated in Fig. 1B and listed in Table 2.) We confirmed the polar effect of our disruption mutants on the downstream genes by qRT-PCR. Indeed, the genes within each of the epaBCD, epaEFGHIJ, and epaLMNOPQ loci were found to be cotranscribed (data not shown). In conclusion, with the disruption of five genes, we affected the production of 16 out of the 18 proteins encoded by the epa locus.
The epa mutants, the orfde2 mutant, and DAGF29 (transposon insertion in OG1RF_0163) as well as OG1RF were tested for growth and phenotype in BHI (routine broth), BHIS (BHI-40% serum), and TSBG (biofilm medium). In TSBG, three mutants (epaB, epaE, and epaN) grew as well as wild-type OG1RF, while the epaA and epaM mutants showed slower exponential growth phase (Fig. 1C) and did not reach the same OD in the stationary phase as OG1RF. In BHIS, only the epaM mutant was affected, with a lag phase 1 h longer than that of the other strains (Fig. 1D). The epaB, epaE, and epaM mutants presented growth patterns and survival rates in the stationary phase comparable to those of wild-type OG1RF. Finally, in BHI, the epaE and epaN mutants grew as well as OG1RF during the exponential growth phase, while the epaB mutant showed only a slightly lower OD600 in the stationary phase (10%; P = 0.0286) (data not shown). The epaA and epaM mutants generated lower OD600s than wild-type OG1RF, with a slower exponential growth phase and a lower survival rate in the stationary phase (1 log lower CFU than OG1RF at 24 h; P < 0.05) (data not shown). However, if the data are corrected for multiple comparisons, the significance disappears. The orfde2 and DAFG29 mutants behaved like wild-type OG1RF under all conditions tested.
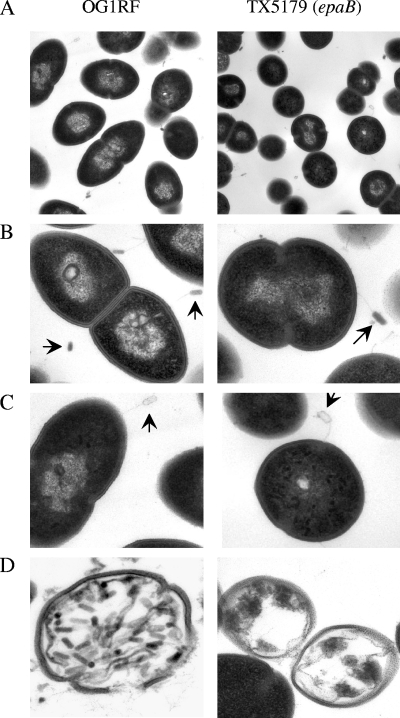
By phase-contrast microscopy with cells grown overnight in TH broth with 1.0% glucose, we observed oval cells present mostly as diplococci for OG1RF and the orfde2 and DAGF29 mutants and round, mainly single cells (shorter than OG1RF cells) for the epaA, epaB, epaE, epaM, and epaN mutants (Fig. 2A). Interestingly, the epaB mutant complemented with the epaBCD-containing plasmid (where 229/238 amino acids [aa] of EpaD should be translated) showed a mixed population with oval and round cells, indicating that the truncated version of EpaD may be partially functional or that the stoichiometry of the enzymes/intermediaries in the pathways controlled by the products of EpaB and EpaC was not fully restored. The different shapes of wild-type OG1RF and the epaB mutant were further seen in electron microscopy (Fig. 2B).
FIG. 2.
Morphology of OG1RF and the epaB mutant cells by phase-contrast microscopy (A) and thin-section electron microcopy (B). Disruptions are denoted by an inverted triangle.
Effect of disruption of epa genes on polysaccharide content.
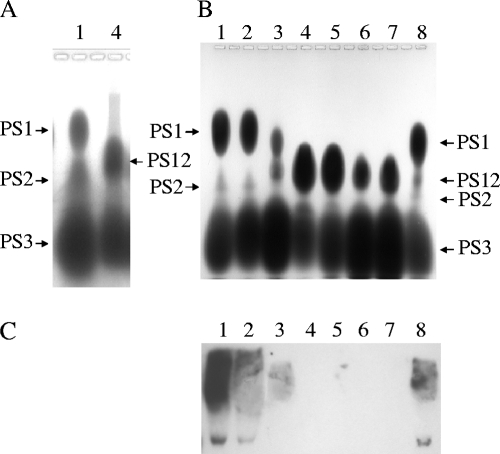
Our initial comparison of the polysaccharide contents of OG1RF and the epaB mutant showed three polysaccharide bands with OG1RF extracts (named PS1, PS2, and PS3) and two polysaccharide bands (PS12 and PS3) for the epaB mutants (Fig. 3A); PS12 was so named because it ran in a position between PS1 and PS2 and was not present in OG1RF (Fig. 3A). We then compared the polysaccharide contents of all of the mutants and found the following. (i) PS1 was completely missing in the epaB, epaE, epaM, and epaN mutants (Fig. 3B, lanes 4, 5, 6, and 7) and was reduced in the epaA mutant (Fig. 3B, lane 3), while PS1 production was restored in the epaB-complemented strain (lane 8). (ii) Depending on the extracts, a variable amount of PS2 (in some extracts, a very small amount) was detected in wild-type OG1RF (lane 1) and the epaB-complemented strain. (iii) The new polysaccharide band, PS12, was seen in the epaA (in small amounts), epaB, epaE, epaM, and epaN mutants and was also detected in residual amounts in the epaB-complemented strain. (iv) The orfde2 mutant (lane 2) showed a polysaccharide pattern similar to that of wild-type OG1RF (Fig. 3B).
FIG. 3.
Polysaccharides of OG1RF and its derivatives. (A and B) Stains-All-stained polysaccharide extracts in 0.8% agarose gel; (C) Western blots with polysaccharide extracts and patient serum. Lane 1, OG1RF; lane 2, TX10113 (the orfde2 mutant); lane 3, TX10114 (the epaA mutant); lane 4, TX5179 (the epaB mutant); lane 5, TX5180 (the epaE mutant); lane 6, TX5391 (the epaM mutant); lane 7, TX5436 (the epaN mutant); lane 8, TX5179.1 (the epaB-complemented strain).
Serum from a patient with E. faecalis endocarditis (S0013) has previously been shown to react with polysaccharide extracts of wild-type OG1RF but not with those of the epaB and the epaE mutants (26). In the present study, S0013 reacted with polysaccharide extracts of wild-type OG1RF and the orfde2 and epaA mutants, while no reaction was detected for polysaccharide extracts of the epaB, epaE, epaM, and epaN mutants. The reactive pattern was restored for the epaB-complemented strain (Fig. 3C).
Composition of OG1RF polysaccharides.
Polysaccharides were purified from OG1RF (grown in TH broth with 1% glucose without shaking for 24 h at 37°C), and one purified fraction reacted with S0031 and migrated the same distance in the gel as PS1; thus, it was named “Epa” due to its absence in epa mutants and its reaction with patient serum. Compositional analysis revealed the presence of glucose, rhamnose, N-acetylglucosamine (glcNAc), N-acetyl galactosamine (galNAc), and galactose in this material. No phosphate was detected in this material. Rhamnose was not found in the polysaccharide of the epaB mutant, but instead mannose was present; glucose, galNAc, galactose, and glcNAc were also found in the polysaccharide of the epaB mutant. Both polysaccharides from OG1RF and the epaB mutant were observed to have a molecular mass of ∼31 kDa by size exclusion.
Effect of disruption of epa genes on biofilm formation.
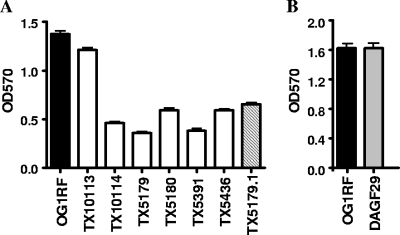
The epaA, epaB, epaE, epaM, and epaN mutants showed significant reduction in biofilm formation (OD570) compared to wild-type OG1RF (at least 50%; P < 0.001), while biofilm produced by the orfde2 and DAGF29 mutants was comparable to that of OG1RF (Fig. 4A and B). Interestingly, although the growth ODs (OD600) of the epaA and epaM mutants under biofilm conditions (static environment) were significantly lower (0.293 and 0.283, respectively; P < 0.001, as in batch cultures) than that of OG1RF (0.352), which may have contributed to their lower biofilm OD, the growth OD600s of the epaB and epaE mutants were significantly higher (0.419 and 0.409, respectively; P < 0.001) while still generating less biofilm. On the other hand, the epaB-complemented strain showed a 1.5- to 2-fold increase in biofilm formation compared to the epaB mutant (P < 0.001), although not equal to that of wild-type OG1RF (Fig. 4A). This result was expected since the epaB-complemented strain is not fully restored, as seen by microscopy with the mixed-shape population.
FIG. 4.
Biofilm formation by OG1RF and its derivatives. (A) TX10113, TX10114, TX5179, TX5180, TX5391, TX5436, and TX5179.1 are the orfde2, epaA, epaB, epaE, epaM, and epaN mutants and the epaB-complemented strain, respectively. (B) DAGF29 is the OG1RF_0163 transposon insertion mutant (9).
Effect of disruption of epa genes on phage sensitivity.
E. faecalis OG1RF has previously been found to be sensitive to a phage called NPV-1 (29). We had planned to use this phage to eliminate extracellular E. faecalis wild-type OG1RF and mutants for phagocytosis and intracellular survival studies, and so we tested phage sensitivity of OG1RF and its derivatives. We observed clear plaques when growing OG1RF with NPV-1. However, testing for phage sensitivity with the epa mutants showed that NPV-1 (≥1:2 dilution) did not form plaques with the epaB, epaE, epaM, or epaN mutants, while reduction in PFU was shown with the epaA mutant compared to wild-type OG1RF (Fig. 5); some lysis was observed with undiluted phage. The epaB-complemented strain produced slightly lower PFU than wild-type OG1RF, and the orfde2 and DAGF29 mutants produced similar PFU to wild-type OG1RF (Fig. 5). To determine whether the resistance of the epa mutants to NPV-1 was due to deficiency in phage adsorption, wild-type OG1RF and the epaA and the epaB mutants were compared, and no significant difference was detected for these strains (data not shown).
FIG. 5.
Phage sensitivity of OG1RF and its derivatives. NPV-1 phage was used to infect TX10113, TX10114, TX5179, TX5180, TX5391, TX5436, and TX5179.1, which are the orfde2, epaA, epaB, epaE, epaM, and epaN mutants and the epaB-complemented strain, respectively. DAGF29 is the OG1RF_0163 transposon insertion mutant (9). PFU are shown.
Electron microscopy of OG1RF and the epaB mutant grown with NPV-1 phage showed that phage particles, with filled or empty heads and a long tail, were attached to both wild-type OG1RF and the epaB mutant cells after 10 min (Fig. 6A and B). At 45 min, after looking at many fields under the microscope, there was evidence of bacterial lysis by phage particles with OG1RF cells, but not with the epaB mutant (Fig. 6C).
FIG. 6.
Electron microscopy of OG1RF and the epaB mutant infected by NPV-1 phage. (A) Cell morphology in the presence of phage; (B) attachment of phage (full head) to bacteria; (C) attachment of phage (empty head) to bacteria. In panel D, OG1RF cells are filled with phage particles and show early lysis, whereas no phage was seen inside TX5179. The arrows point to the phage.
Effect of disruption of epa genes on virulence.
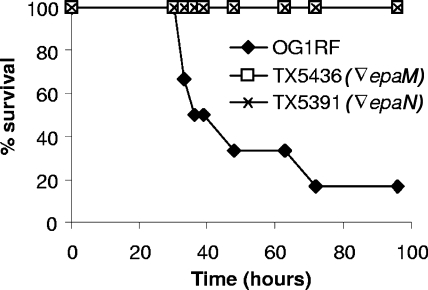
Previously, we have shown that disruptions in epaB (orfde4) and epaE (orfde6) caused significant attenuation in the mouse peritonitis model (33). In the present study, we examined the epaM and epaN mutants in this model and found that the two strains were significantly attenuated (Fig. 7) (P = 0.0051 and 0.0051 by log rank for the epaM and epaN mutants versus OG1RF, respectively). Since epaM had a lower survival rate than OG1RF and epaN in BHI broth once the culture reached the stationary phase, the accuracy of the inocula was verified by CFU determination. The results presented in Fig. 7 were obtained after inoculation of 3.5 × 108 cells of the epaM mutant versus 3.4 × 108 cells of the OG1RF wild type and 3.4 × 108 cells of the epaN mutant versus 3.4 × 108 cells of the OG1RF wild type.
FIG. 7.
Survival curves of OG1RF and its derivatives in the mouse peritonitis model. Six mice per group were used, and P = 0.0051 for OG1RF versus TX5391 (the epaM mutant) or TX5436 (the epaN mutant). The inocula for OG1RF, TX5391, and TX5436 were 3.4 × 108, 3.5 × 108, and 3.4 × 108 CFU, respectively. Disruptions are denoted by an inverted triangle.
DISCUSSION
Biosynthesis of polysaccharides generally involves synthesis of nucleotide sugar precursors in the cytoplasm, formation and polymerization of repeating units, and export to the cell surface. The E. faecalis epa gene cluster contains genes encoding enzymes and transporters potentially involved in all of these processes. For example, the predicted EpaA protein shows similarity to undecaprenyl-phosphate α-N-acetylglucosaminyltransferase (a member of the glycosyl transferase group 4 family), while the products of epaB, -C, -D, -I, -N, and -O and OG1RF_0164 (4) show similarity to the glycosyl transferase group 2 family. The predicted OG1RF_0163 (4) protein shows similarity to glycosyl transferase group 25, the predicted product of epaR is a sugar transferase, epaEFG and -H are similar to genes involved in dTDP-rhamnose biosynthesis pathway of various organisms, and epaL and -M encode proteins with similarity to components of ABC transport systems that might be involved with exporting polysaccharide components (32).
To determine the limits of the epa genes of OG1RF, we constructed disruption mutants and compared the polysaccharide contents of the epaA, epaB, epaE, epaM, and epaN mutants with that of wild-type OG1RF. Our results demonstrated that the previously described genes orfde3 to -16 are important for biosynthesis of Epa by OG1RF and, based on our observations, we have renamed these genes “epaA” to “epaO,” while EF2179, EF2178, and EF2177, cotranscribed with epaO, were named “epaP,” “epaQ,” and “epaR.” In all, it appears that the epa gene cluster is comprised of 18 genes flanked by orfde2 and OG1RF_0164, with the expression of 16 of these 18 affected by our disruptions in epaA, epaB, epaE, and epaN. A disruption mutant of OG1RF_0163 behaved as OG1RF in the assays tested (biofilm formation, phage sensitivity, and cell shape). Similarly, neither orfde1 nor orfde2 is required for Epa production, consistent with our previous observation that, in E. coli with the cloned epa gene cluster, transposon insertions in orfde1 and orfde2 did not affect polysaccharide antigen production (32). At the same time, we found that disruption of the epa genes (epaA, -B, -E, -M, and -N) resulted in a new polysaccharide, possibly due to accumulation of some intermediary products resulting from loss of epa-encoded function(s). It is also interesting to note that we were unsuccessful in obtaining an epaO insertion mutant, although epaM and epaN insertion mutants were obtained. One possibility is that an intermediary substrate generated by epa genes is toxic to the cell in the absence of one or several of the proteins encoded by epaOPQR. Another possibility is that some epa-encoded functions are complemented by other genes of OG1RF, allowing mutants to be made, while others (e.g., one of the EpaOPQR functions) are not. Such possibilities could also explain why some mutants show a growth defect while others do not.
Compositional analysis of Epa (the band (PS1) from OG1RF that reacted with E. faecalis patient serum and was not seen in the epa mutants) showed rhamnose as one of its major sugar residues. The carbohydrate compositional analysis of the epaB mutant revealed replacement of rhamnose by mannose in PS12, suggesting that the glycosyl transferases encoded by the epaBCD operon contribute to rhamnose transfer to the polysaccharide. Since transcription of at least one glycosyl transferase was affected for each of the epaB, epaE, and epaM mutants, in-frame deletion will be required to define the function of each of the six glycosyl transferases located in this epa cluster. The predicted protein similarities and the lack of wild-type Epa in the epaE, epaM, and epaN mutants suggest that these genes may contribute to biosynthesis (epaE to -I) and transport (epaL and -M) of rhamnose.
Although the PS1 and PS12 polysaccharides ran at different positions in agarose gels, they showed similar molecular sizes in a size exclusion column, suggesting different charges of these polysaccharides. Hsu et al. found previously that a surface-located heteroglycan (∼20 kDa) isolated from clinical strains of E. faecalis was composed of rhamnose, glucose, galactose, mannosamine, and glucosamine (15). The similar composition of the heteroglycan to our Epa polysaccharide (∼31 kDa) suggests possible involvement of epa genes in biosynthesis of this heteroglycan. Hancock et al. also reported similar composition of a cell wall polysaccharide isolated from E. faecalis, strain FA2-2, although the molecular mass of that polysaccharide was 50 kDa (13). The difference in sizes of these polysaccharides may due to differences in methodology and/or different strains used in the studies.
We also found that disruption of epa genes had an effect on E. faecalis cell shape, producing more round cells. In addition, disruptions of epaA and epaM appear to have an additional effect on cell growth, but not disruption of epaB, epaE, and epaN. Our study also found that the epa genes were important for lytic infection of OG1RF by phage NPV-1. Although attachment of this phage to the epa mutants was not obviously affected, it was not clear whether the epa genes influence the entry of the phage DNA into the mutant cells and/or influence phage replication inside the mutant cells; thus, at the present time, it is not clear how epa genes affect the NPV-1 lytic infection cycle.
It was also interesting to find that the epaA mutant, unlike other epa mutants, made a smaller amount of Epa and showed an intermediate phenotype in phage infection: e.g., the epaA mutant, when infected by the phage NPV-1, produced reduced PFU versus wild-type OG1RF, while other epa mutants produced 0 PFU. These results suggest that the truncated EpaA product (missing only the last 8 aa) in the epaA disruption mutant has partial activity or that some function of EpaA may be partially complemented by other factors in E. faecalis. We previously found that in the E. coli recombinant clone with a Tn7 insertion in the 3′ end of epaA, expression of the polysaccharides was still detected, even in the rfe (E. coli homologue of epaA) strain (32); this suggests the EpaA may not be needed by E. coli to synthesize the polysaccharides or, again, that truncated EpaA may be partially active. The intermediate phenotype of the epaA mutant was not seen in biofilm formation, suggesting that EpaA functions differently in these processes.
Previously, we showed that the epa genes were present in 12 of 12 distinct E. faecalis strains by Southern blots with PCR products of orfde4 to -10 (26), suggesting that Epa, unlike the serotype-specific capsular polysaccharide, Cps (13), is a common component of E. faecalis. In the present study, we further confirmed widespread distribution of epa genes in E. faecalis by dot blot hybridization with 92 E. faecalis strains (data not shown). Although the epa genes are widespread, genomic comparison indicates that differences occur in the epa gene cluster and in the surrounding area. In OG1RF, a unique gene cluster is present downstream of the epa gene cluster replacing EF2166 to EF2176 present in V583 (4). This OG1RF unique gene cluster predicts 14 proteins, 9 of which are likely associated with polysaccharide biosynthesis (4). In V583, a small transposon carrying four genes (EF2185 to EF2188) is inserted in the 74 bp between epaJ and epaK, leaving these two small ORFs and their respective promoter areas intact. Preliminary data indicate that neither the OG1RF unique gene cluster nor the transposon detected in V583 is present in two other sequenced E. faecalis strains (personal observation).
Future studies to determine the localization of the Epa components in the outer layers of E. faecalis, as well as the effect of disruption of Epa on expression and/or localization of other E. faecalis virulence factors, may provide insight into the pleiotrophic effect of Epa on translocation, biofilm formation, PMN-mediated killing, and virulence. Such studies should also advance our understanding of the human response to E. faecalis infection, which may potentially help the design of immunotherapeutic agents against E. faecalis infections.
Supplementary Material
Acknowledgments
We thank Patricia Navarro in the Department of Pathology and Laboratory Medicine for providing technical support for electron microscopy and Suresh R. Pai for construction of some of the epa mutants and performing some of the animal experiments.
The present work was supported by grant NIH R37 AI47923 from the Division of Microbiology and Infectious Diseases, NIAID, to B.E.M. The polysaccharide compositional and structural analysis was supported in part by the Department of Energy-funded (DE-FG09-93ER-20097) Center for Plant and Microbial Complex Carbohydrates, University of Georgia.
Editor: A. Camilli
Footnotes
Published ahead of print on 6 July 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.AlonsoDeVelasco, E., A. F. Verheul, J. Verhoef, and H. Snippe. 1995. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol. Rev. 59591-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arduino, R. C., K. Jacques-Palaz, B. E. Murray, and R. M. Rakita. 1994. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect. Immun. 625587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamford, D. H., and L. Mindich. 1980. Electron microscopy of cells infected with nonsense mutants of bacteriophage phi 6. Virology 107222-228. [DOI] [PubMed] [Google Scholar]
- 4.Bourgogne, A., D. A. Garsin, X. Qin, K. V. Singh, J. Sillanpaa, S. Yerrapragada, Y. Ding, S. Dugan-Rocha, C. Buhay, H. Shen, G. Chen, G. Williams, D. Muzny, A. Maadani, K. A. Fox, J. Gioia, L. Chen, Y. Shang, C. A. Arias, S. R. Nallapareddy, M. Zhao, V. P. Prakash, S. Chowdhury, H. Jiang, R. A. Gibbs, B. E. Murray, S. K. Highlander, and G. M. Weinstock. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgogne, A., S. G. Hilsenbeck, G. M. Dunny, and B. E. Murray. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 1882875-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourgogne, A., K. V. Singh, K. A. Fox, K. J. Pflughoeft, B. E. Murray, and D. A. Garsin. 2007. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J. Bacteriol. 1896490-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez-Guerrero, M. L., C. Verdejo, J. Azofra, and M. de Gorgolas. 1995. Hospital-acquired infectious endocarditis not associated with cardiac surgery: an emerging problem. Clin. Infect. Dis. 2016-23. [DOI] [PubMed] [Google Scholar]
- 8.Garcia, E., and R. Lopez. 1997. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol. Lett. 1491-10. [DOI] [PubMed] [Google Scholar]
- 9.Garsin, D. A., J. Urbach, J. C. Huguet-Tapia, J. E. Peters, and F. M. Ausubel. 2004. Construction of an Enterococcus faecalis Tn917-mediated-gene-disruption library offers insight into Tn917 insertion patterns. J. Bacteriol. 1867280-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson, R. L., M. K. Lee, C. Soderland, E. Y. Chi, and C. E. Rubens. 1993. Group B streptococci invade endothelial cells: type III capsular polysaccharide attenuates invasion. Infect. Immun. 61478-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmore, M., P. Coburn, S. Nallapareddy, and B. Murray. 2002. Enterococcal virulence, p. 301-354. In M. S. Gilmore, D. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 12.Haft, R. F., M. R. Wessels, M. F. Mebane, N. Conaty, and C. E. Rubens. 1996. Characterization of cpsF and its product CMP-N-acetylneuraminic acid synthetase, a group B streptococcal enzyme that can function in K1 capsular polysaccharide biosynthesis in Escherichia coli. Mol. Microbiol. 19555-563. [DOI] [PubMed] [Google Scholar]
- 13.Hancock, L. E., and M. S. Gilmore. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. USA 991574-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, B., S. Poole, and M. Wilson. 1996. Bacterial modulins: a novel class of virulence factors which cause host tissue pathology by inducing cytokine synthesis. Microbiol. Rev. 60316-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu, C. T., A. L. Ganong, B. Reinap, Z. Mourelatos, J. Huebner, and J. Y. Wang. 2006. Immunochemical characterization of polysaccharide antigens from six clinical strains of enterococci. BMC Microbiol. 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebner, J., A. Quaas, W. A. Krueger, D. A. Goldmann, and G. B. Pier. 2000. Prophylactic and therapeutic efficacy of antibodies to a capsular polysaccharide shared among vancomycin-sensitive and -resistant enterococci. Infect. Immun. 684631-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huebner, J., Y. Wang, W. A. Krueger, L. C. Madoff, G. Martirosian, S. Boisot, D. A. Goldmann, D. L. Kasper, A. O. Tzianabos, and G. B. Pier. 1999. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect. Immun. 671213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hufnagel, M., L. E. Hancock, S. Koch, C. Theilacker, M. S. Gilmore, and J. Huebner. 2004. Serological and genetic diversity of capsular polysaccharides in Enterococcus faecalis. J. Clin. Microbiol. 422548-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulse, M. L., S. Smith, E. Y. Chi, A. Pham, and C. E. Rubens. 1993. Effect of type III group B streptococcal capsular polysaccharide on invasion of respiratory epithelial cells. Infect. Immun. 614835-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, C. J., L. H. Lee, C. S. Lu, and A. Wu. 2001. Bacterial polysaccharides as vaccines—immunity and chemical characterization. Adv. Exp. Med. Biol. 491453-471. [DOI] [PubMed] [Google Scholar]
- 21.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 723658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 346-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 1755216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakita, R. M., V. C. Quan, K. Jacques-Palaz, K. V. Singh, R. C. Arduino, M. Mee, and B. E. Murray. 2000. Specific antibody promotes opsonization and PMN-mediated killing of phagocytosis-resistant Enterococcus faecium. FEMS Immunol. Med. Microbiol. 28291-299. [DOI] [PubMed] [Google Scholar]
- 25.Soell, M., M. Diab, G. Haan-Archipoff, A. Beretz, C. Herbelin, B. Poutrel, and J.-P. Klein. 1995. Capsular polysaccharide types 5 and 8 of Staphylococcus aureus bind specifically to human epithelial (KB) cells, endothelial cells, and monocytes and induce release of cytokines. Infect. Immun. 631380-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng, F., K. D. Jacques-Palaz, G. M. Weinstock, and B. E. Murray. 2002. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect. Immun. 702010-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng, F., E. C. Nannini, and B. E. Murray. 2005. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the protein as a possible immunotherapy target. J. Infect. Dis. 191472-480. [DOI] [PubMed] [Google Scholar]
- 28.Teng, F., L. Wang, K. V. Singh, B. E. Murray, and G. M. Weinstock. 2002. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect. Immun. 701991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trotter, K. M., and G. M. Dunny. 1990. Mutants of Enterococcus faecalis deficient as recipients in mating with donors carrying pheromone-inducible plasmids. Plasmid 2457-67. [DOI] [PubMed] [Google Scholar]
- 30.Xu, S., R. D. Arbeit, and J. C. Lee. 1992. Phagocytic killing of encapsulated and microencapsulated Staphylococcus aureus by human polymorphonuclear leukocytes. Infect. Immun. 601358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, Y., L. Jiang, B. E. Murray, and G. M. Weinstock. 1997. Enterococcus faecalis antigens in human infections. Infect. Immun. 654207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu, Y., B. E. Murray, and G. M. Weinstock. 1998. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect. Immun. 664313-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, Y., K. V. Singh, X. Qin, B. E. Murray, and G. M. Weinstock. 2000. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect. Immun. 68815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng, J., F. Teng, G. M. Weinstock, and B. E. Murray. 2004. Translocation of Enterococcus faecalis strains across a monolayer of polarized human enterocyte-like T84 cells. J. Clin. Microbiol. 421149-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.