Abstract
Virulence of the fungal pathogen Aspergillus fumigatus is in part based on the saprophytic lifestyle that this mold has evolved. A crucial function for saprophytism resides in secreted proteases that allow assimilation of proteinaceous substrates. The impact of extracellular proteolytic activities on the pathogenesis of aspergillosis, however, remains controversial. In order to address this issue, characterization of a conserved regulatory factor, PrtT, that acts on expression of secreted proteases was pursued. Expression of PrtT appears to be regulated posttranscriptionally, and the existence of an mRNA leader sequence implies translational control via eIF2α kinase signaling. Phenotypic classification of a prtTΔ deletion mutant revealed that expression of several major extracellular proteases is PrtT dependent, resulting in the inability to utilize protein as a nutritional source. Certain genes encoding secreted proteases are not regulated by PrtT. Most strikingly, the deletant strain is not attenuated in virulence when tested in a leukopenic mouse model, which makes a strong case for reconsidering any impact of secreted proteases in pulmonary aspergillosis.
Diseases caused by the fungal pathogens of the genus Aspergillus represent a growing and clinically relevant threat in distinct medical settings (31). Aspergilloses have to be considered a frequent complication during immunological therapies or disorders, such as allogeneic stem cell transplantation and chronic granulomatous disease, respectively, and recent evaluations demonstrate the increasingly severe impact of this fungal infection on the health economy (52). Among its various types, so-called invasive forms of aspergillosis are most severe and often life-threatening, especially for the immunocompromised individual. Due to the commonly airborne route of transmission, the pulmonary system represents the prime infection site in a susceptible host, and after successful germination of the infectious propagules, colonization of the infected tissue and eventually hematogenous dissemination may occur. By far the most cases of aspergillosis are caused by the saprophyte A. fumigatus; however, other species of this genus are able to infect and harm individual patients (53).
Besides defined host factors (10), several fungal attributes are considered virulence determinants in the host-pathogen system that results in aspergillosis (25). Based on recently gained insights into the cellular armamentarium that is expressed by members of this genus, the hypothesis that exclusive virulence factors contribute specifically to the virulome of A. fumigatus has been challenged. According to this, it is likely rather that the saprophytic lifestyle exhibited by aspergilli forms the basis for colonizing a susceptible host, which is simply unable to clear the infection (51). Saprophytism is in part characterized by an absorptive nutritional mode, which is based on the extracellular degradation of the surrounding polymeric matrix and successive uptake of oligomeric breakdown products. For the latter, specialized transport systems have evolved, whereas for the former, a plethora of hydrolytic enzymes are expressed and secreted into the environmental vicinity. Among those, proteases appear to play a prominent role, which is indicated by the huge array of proteolytic activities that are presumably encoded by the A. fumigatus genome to comprise more than 100 proteases (33, 36). Three major proteases have been described to be secreted during infection by this fungus: an alkaline serine protease, Alp; a metalloprotease, Mep; and an aspartic protease, Pep (32). Their production is triggered by the presence of proteinaceous substances in the growth medium, whereas free amino acids or oligopeptides repress their expression. Each subclass of proteolytic activity appears to be encoded not by one particular locus but by a family of genes, which hampers a conclusive assessment of their role in the virulence of A. fumigatus. There are several points that would argue in favor of, as well as against, an involvement of extracellular proteolytic activities in the development of invasive aspergillosis: expression during infection could be demonstrated for several proteases (29, 35, 39, 40), and an elastinolytic activity expressed by A. fumigatus had been attributed the ability to cause invasive aspergillosis (24). Also, the alkaline serine protease was able to induce structural changes in the actin cytoskeleton of cultured lung pneumocytes (22), and recently, a nutritional role for breakdown products of the proteinaceous matrix was postulated (14). Yet, in a singular case report the absence of elastinolysis of vessel walls during invasive aspergillosis was evident (9), and, more importantly, virulence attenuation could not be revealed for any A. fumigatus mutant with deletions of single or multiple genes encoding extracellular proteolytic activities (15, 34, 42, 50).
Knowledge about the regulatory means of Aspergillus to trigger expression and secretion of proteases is scarce. Early reports indicated that the presence of protein as the sole source of nitrogen strongly induces expression of the major secreted A. fumigatus proteases, whereas free amino acids or small peptides repress their formation. Also, the presence of a primary N source such as ammonium results in very low levels of extracellular proteolytic activity, which implies that expression of secreted proteases is under the control of the nitrogen catabolite repression system. A more detailed analysis of proteinase secretion by A. fumigatus in the presence of complex substrates such as gelatin, Matrigel, serum, bovine serum albumin (BSA), or even pneumocytes revealed a multifaceted pattern of expression for the major activities (11). Notably, extracellular proteolysis, at least serine proteinase activity, is maintained in the presence of low (0.1%) concentrations of the protein hydrosylate peptone (4). Also, in the model ascomycete A. nidulans an influence of the nitrogen source on proteinase secretion was documented, as well as that of the carbon source and the related catabolite repression system. There, the p53-like transcription factor XprG appears to play a general role in regulating the expression of extracellular proteases in response to starvation (20, 21).
In the biotechnological workhorse A. niger, however, a unique and specific regulator of extracellular proteolytic activity was recently identified in a forward genetics approach by random mutagenesis and screening on growth plates containing casein milk. The aim was to identify isolates that would exhibit reduced protein degradation during production of heterologous proteins (30). Among these prt strains, one putatively regulatory mutant became of interest, and the corresponding prtT gene was identified to encode a Zn2Cys6 protein (37). Further analyses of disruptants of A. niger and A. oryzae validated a conserved function of PrtT in regulating the major protease genes in these two species of Aspergillus. Cross-species genome comparison revealed the interesting fact that prtT was present in several aspergilli but absent in the genome of A. nidulans, where most likely an evolutionary gene deletion event had occurred.
To address the role of secreted proteases in virulence of the pathogenic species A. fumigatus on a broad scale, we here describe our efforts to scrutinize the cellular function of its corresponding prtT gene. A comprehensive characterization of a validated deletion mutant was carried out to reveal that the A. fumigatus prtT gene product is a transcriptional regulator of extracellular proteolytic activities that, however, is not essential for virulence of this opportunistic fungal pathogen.
MATERIALS AND METHODS
Strains, media, and growth conditions.
For general cloning procedures the bacterial strain Escherichia coli DH5α was used (56), which was cultivated in LB (1% Bacto-tryptone, 0.5% yeast extract, 1% NaCl, pH 7.5) medium. Fungal strains used in this study are listed in Table 1. Growth of Aspergillus fumigatus was carried out at 37°C on minimal medium prepared as described by Käfer (18) and supplemented with appropriate sources of carbon and nitrogen as specified. Concentrations of protein-containing supplements were 0.4% for casein, 0.1% for peptone, and 0.4% for BSA. Antibiotic concentrations were 100 μg/ml for ampicillin, 20 μg/ml for phleomycin, and 0.1 μg/ml for pyrithiamine.
TABLE 1.
Fungal strains and plasmid constructs
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| ATCC 46645 | Aspergillus fumigatus wild-type strain | 13 |
| AfS61 | prtTΔ deletion mutant (prtT::loxP-Phleor-loxP) | This study |
| AfS62 | Reconstituted strain (prtT<ptrA>) | This study |
| Plasmids | ||
| pJET1.2 | Positive selection vector, pUC19 derivative (bla, multiple-cloning site in Eco47IR) | Fermentas |
| pSK275 | Plasmid containing ptrA cassette conferring resistance to pyrithiamine | 27 |
| pSK341 | Aspergillus marker cassette conferring resistance to phleomycin (pgpdA::ble/tk::trpCt) | 26 |
| pSK462 | Replacement cassette for deletion of prtT in A. fumigatus | This study |
| pSK463 | Reconstitution cassette containing prtT gene coupled to ptrA resistance cassette | This study |
Transformation procedures.
E. coli cells were transformed after treatment with calcium-manganese (12). For A. fumigatus, a procedure of polyethylene glycol-mediated protoplast fusion was followed (38).
Manipulation of nucleic acids and plasmid constructions.
Standard protocols of recombinant DNA technology were carried out (45). Phusion high-fidelity DNA polymerase was generally used in PCRs, and essential cloning steps were verified by sequencing at GATC Biotech. Sequence analyses were performed with the Lasergene Biocomputing software package from DNAStar. Fungal genomic DNA was prepared following the protocol of Kolar et al. (23), and Southern analyses were carried out as described previously (48, 49). Probes for nonradioactive hybridizations were generated and detected using the Gene Images AlkPhos direct labeling and detection system from GE Healthcare. Autoradiographs were produced by exposing washed membranes to Kodak X-Omat films. DNA extraction from homogenized lung tissue was performed using the Qiagen DNeasy blood and tissue kit. Samples of total RNA were isolated with the TRIzol reagent from Invitrogen and used in quantitative real-time PCRs (qRT-PCRs) (see below) after DNase treatment and reverse transcription using the SuperScript III first-strand synthesis SuperMix from Invitrogen.
Plasmids constructed during the course of this study are listed and briefly described in Table 1, and Table 2 lists the sequences of relevant oligonucleotides. In detail, plasmids were constructed as follows. Generation of a gene replacement cassette for deletion of the prtT gene was executed according to the method of Kämper (19) by amplifying flanking regions of the target locus with the primer pairs AB17/20 and AB19/18 to yield a 5′ flank of 2.3 kb and a 3′ flank of 1.65 kb, respectively. After SfiI digestion, fragments were ligated to the SfiI-flanked phleomycin resistance cassette of pSK341, and the resulting replacement cassette was directly inserted in the cloning vector pJET1.2. From the resulting plasmid pSK462, a 6.8-kb replacement module was released by PmeI digestion prior to transformation of A. fumigatus. A reconstitution cassette was generated by insertion of the pyrithiamine resistance gene ptrA amplified from pSK275 with primers Sv551 and Sv556 into the blunted EcoRI site downstream of the prtT coding sequence, which had been inserted as a 6.7-kb PCR amplicon with primers AB17/18 in pJET1.2. The complementing fragment was isolated from this plasmid, pSK463, after a PmeI digest.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| AB17 | 5′-TAT AGT TTA AAC ATT AGG TAA ATG CAT TCT AC-3′ |
| AB18 | 5′-TAT AGT TTA AAC AAG AAT TGA CTG ATA TGC TA-3′ |
| AB19 | 5′-TAT AGG CCA TCT AGG CCT ATT GTC AAT CAT ACA TGG AAT T-3′ |
| AB20 | 5′-TAT AGG CCT GAG TGG CCG GCG GTT GAG TCT TAA ATT TTC-3′ |
| AB28 | 5′-CCA ATG ACA GCA GTG TGT CC-3′ |
| AB29 | 5′-TCC CCG TCC AAG CTG TAG C-3′ |
| AB30 | 5′-CAA ACT CAC GAG GAG ATG AAG-3′ |
| AB31 | 5′-GCA CTG AGG GGT GAT GAC C-3′ |
| AB34 | 5′-TGC CTA CGT TGT CGA CAG TG-3′ |
| AB35 | 5′-TTT GAG GCA TCA CTG TTC TCG-3′ |
| AB36 | 5′-TGA CTC GGA ATA CCT GAC CC-3′ |
| AB37 | 5′-CCA ACT GGG ACT TGA CAG TG-3′ |
| AB38 | 5′-ACT TTC CGC GTG GTC GAC G-3′ |
| AB39 | 5′-GTC GGT CTC AAC TCT CCA TG-3′ |
| AB51 | 5′-GTT CCA GCG AGA TGA GGC G-3′ |
| Sv551 | 5′-GGG AGA TCT GAC AGA CGG GCA ATT G-3′ |
| Sv556 | 5′-GAT GAG ATC TTG CAT CTT TGT TTG TAT TAT AC-3′ |
| Sv576 | 5′-GTC ATT TGT CGG CTG CTG ATC C-3′ |
| Sv589 | 5′-ACA GAG GCC CAT CGA TAT CGC TGC-3′ |
| TubFw | 5′-GGA CGT TAC CTC ACC TGC TC-3′ |
| TubRev | 5′-CAC GCT TGA ACA ACT CCT GA-3′ |
| RTDpp4Fw | 5′-TGT CGA CGC ACA GCA CAT CG-3′ |
| RTDpp4Rev | 5′-CCG GAT CGC ACT GGC ATT GT-3′ |
| qPCRf | 5′-GAT ACC GTC GTA GTC TTA-3′ |
| qPCRr | 5′-TGT CTG GAC CTG GTG AGT-3′ |
qRT-PCR.
qRT-PCRs were performed on a Bio-Rad MyiQ iCycler using the iQ SYBR green Supermix. Sequences of priming oligonucleotides are given in Table 2; gene-specific combinations are AB34/35 (alp1), AB36/37 (pep1), AB38/39 (mep), AB30/31 (sedB), TubFw/TubRev (AFUA_1G10910, encoding β-tubulin), RTDpp4Fw/RTDpp4REv (dppIV), and AB28/29 (dppV). Per reaction mixture, 0.3 μl of cDNA and 1 pmol of each primer were used to perform 40 temperature cycles as follows: 3 min of polymerase activation at 95°C, 15 s at 95°C for initial denaturation, 15 s at 60°C for annealing, and 25 s at 72°C for elongation. Fluorescence was determined at each elongation step. Melting curves were plotted to confirm the specificity of each reaction, and results were quantified according to the comparative threshold cycle method (1).
Proteolysis tests.
Two assays were executed for determination of proteolytic activities, a qualitative assay based on clearance of unprocessed X-ray film material (8) and a quantitative one measuring hydrolysis of dye-coupled collagen (7, 17).
Virulence model of pulmonary aspergillosis.
Outbred male mice (strain CD1, 20 to 28 g; Charles Rivers Breeders) were used for animal experiments. Immunosuppression was carried out by subcutaneous injection of 112 mg/kg hydrocortisone acetate and intraperitoneal injection of 150 mg/kg cyclophosphamide following a sequential protocol as described previously (47). Bacterial infections were prevented by adding 1 g/liter tetracycline and 64 mg/liter ciproxicin to the drinking water. Inocula of 1 × 104 conidiospores in 40 μl of saline were prepared by harvesting spores from 5-day-old slants of solid medium followed by filtration through Miracloth (Calbiochem) and washing with saline. Mice were anesthetized by inhalation of isofluorane and infected by intranasal instillation. The weights of infected mice were monitored for 4 days twice daily before animals were culled to isolate their lungs. In order to assess fungal burdens as a quantitative virulence criterion, qPCRs were performed on equivalent amounts (100 ng) of genomic DNA that had been extracted from equal amounts (250 mg) of homogenized lung tissue (5, 46) using several references for genomic DNA concentration and the oligonucleotides qPCRf and qPCRr to amplify the 18S rRNA locus from the A. fumigatus genome.
RESULTS
The Aspergillus fumigatus genome encodes an ortholog of PrtT.
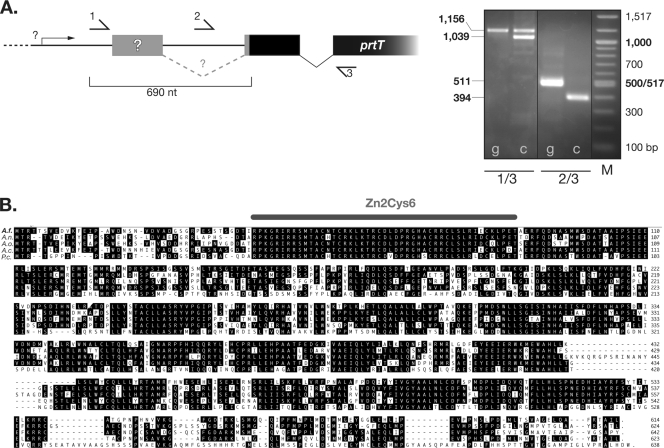
In order to identify the orthologous gene of the A. niger prtT locus, we interrogated the A. fumigatus genome sequence at the CADRE database (28). A BLAST search retrieved an automatically annotated gene entry tagged with the locus identifier AFUA_4G10120. Its deduced gene product displays a high degree of conservation to the PrtT regulatory proteins of A. niger, A. oryzae, and several others, with similarities ranging from 51% to 81%. The automatic annotation predicts a gene product of 696 residues with an unconserved N-terminal part based on the presence of an unusually long (386-nucleotide [nt]) postulated intronic sequence. To map this region of the prtT transcript, diagnostic PCRs from reverse-transcribed mRNA with several specific primer combinations covering the predicted 5′ region were conducted (Fig. 1A). The presence and size of the resulting amplicons indicate the absence of any splicing events for the first predicted intron but hint at the existence of a long leader region of at least 690 nt preceding the translational start site on the prtT transcript. Accordingly, we assume that one of the distal ATG triplets serves as the actual translational start codon to result in a PrtT protein closely related to orthologous ones from other Aspergillus species (Fig. 1B). Based on similarity, the A. fumigatus PrtT is likely to comprise 613 amino acids. Efforts to map the exact transcriptional start sites by 5′ rapid amplification of cDNA ends failed, presumably due to insufficient expression of this putative transcription factor-encoding gene.
FIG. 1.
A. fumigatus prtT encodes a conserved putative transcription factor of the Zn2Cys6 type. (A) The existence of the first annotated intron was examined by diagnostic PCRs covering the 5′ region to indicate that the prtT coding sequence is preceded by an unusually long leader sequence. The putative N terminus based on the putative intron is depicted in gray and the actual coding sequence in black. Priming oligonucleotides used are Sv589 (1), Sv576 (2), and AB51 (3), and calculated fragment sizes amplified from genomic DNA (g) or cDNA (c) are indicated in the left margin. A 100-bp DNA marker (M) was used as a size standard. Results were consistent between strain ATCC 46645 and an unrelated A. fumigatus isolate (D141 [data not shown]). The larger amplicon with primers Sv589 and AB51 (1/3) from cDNA is most likely due to contamination with genomic DNA. (B) Global alignment of the deduced A. fumigatus PrtT sequence with fungal orthologs from A. niger (A.n.; accession number XP_001402055), A. oryzae (A.o.; BAF74781), A. clavatus (A.c.; XP_001271888), and Penicillium crysogenum (P.c.; CAP98521). Consensus residues are boxed in black, and the gray bar indicates the position of the highly conserved Zn2Cys6 DNA binding domain.
The deduced amino acid sequence from prtT contains a DNA binding domain of the C6 zinc finger domain class, which is highly conserved among its orthologous gene products (Fig. 1B). Besides this, no particular domains of specific function could be identified by various database searches.
PrtT is required for protein utilization and degradation.
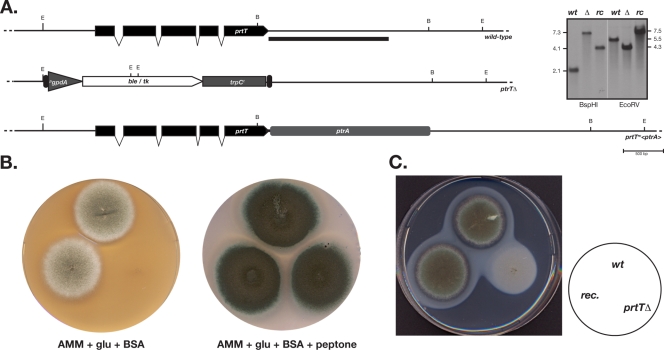
In order to gain insight into the cellular function of the prtT gene product, we constructed a full deletion strain by homologous gene replacement with a suitable selectable marker cassette (Fig. 2A). Southern analysis of several isolates confirmed the replacement event, and one representative was picked for further analysis (see below). Based on this deletant, AfS61, a reconstituted strain, AfS62, was generated by introducing the prtT gene linked to a selectable marker at the resident genomic position, which could be confirmed by Southern analysis. The resulting set of strains was eventually subjected to phenotypic inspection on various types of growth media. The most strikingly phenotype was evident on plates supplemented with the protein BSA as the sole source of nitrogen (Fig. 2B). On this type of medium, a clear growth retardation could be observed, resulting in the near absence of mycelial expansion. Only after prolonged incubation of the growth plates was a very sparse hyphal mesh produced by strain AfS61, whereas proper growth was present for the wild-type progenitor and the reconstituted strain AfS62. Predigested protein, however, supported growth, as could be demonstrated by growing the prtTΔ strain on minimal medium in the presence of 0.1% peptone as a nitrogen source. As a first indication of which cellular function this growth phenotype is based on, strains were propagated on minimal medium containing casein. In a wild-type situation, acidification results in protein precipitation around the growing colony to result in a turbid zone; upon prolonged growth, this precipitate zone clears up due to hydrolytic degradation by the abundantly secreted proteases. Mycelia of the prtT deletant strain did not display any clearance around or beneath the hyphal mat on casein plates, in contrast to the wild-type strain or the reconstituted strain (Fig. 2C). Accordingly, although casein is a different substrate, the growth defect of AfS61 in the presence of BSA might be due to the strain's inability to break down this polymeric substrate.
FIG. 2.
The prtT gene supports protein assimilation by A. fumigatus. (A) Deletion and reconstitution of the prtT locus. The genomic situation at the target locus is shown schematically for the wild-type progenitor ATCC 46645, the deletant AfS61, and the reconstituted strain AfS62. On the right side confirmative Southern analyses after digestion of genomic DNA with BspHI and EcoRV and hybridization with a 3′-specific probe (black bar) are displayed. (B) Growth tests on minimal medium supplemented with BSA with or without peptone as a nitrogen source. Media were point inoculated with equal amounts of spores and incubated at 37°C to reveal a severe growth retardation of the prtTΔ strain when undigested protein serves as the nitrogen source, a phenotype that is completely reversed when hydrolyzed protein such as peptone is added. (C) Growth on casein-containing medium shows the absence of a clearance halo around the growing edge of the AfS61 (prtTΔ) colony and therefore deficient degradation of precipitated protein. The inoculation legend presented on the right is valid for panels B and C.
A prtT deletant is strongly impaired in secreting proteolytic activities.
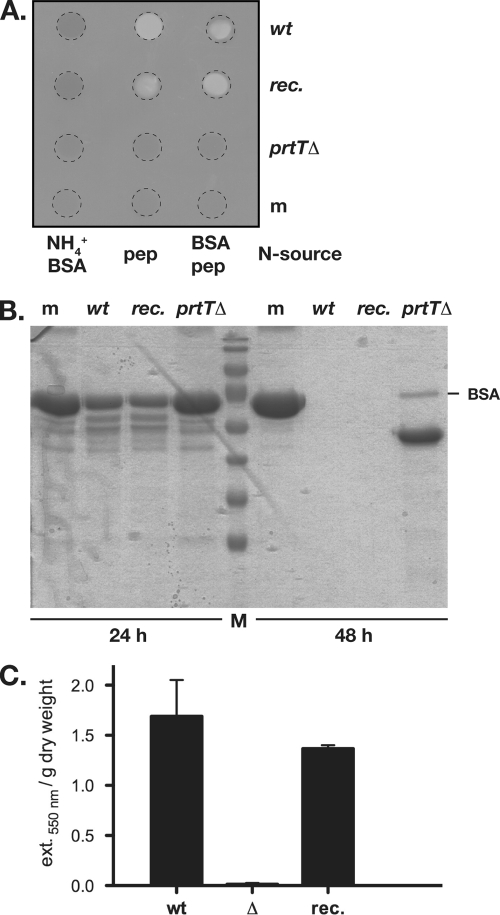
Based on the hypothesis that the prtT deletion mutant is impaired in expressing extracellular proteases, a degradation assay was performed with this strain when grown in liquid culture in the presence of BSA as well as small amounts of peptone (4). The latter substrate supports growth of the mutant but does not suppress secretion of extracellular proteases in the wild type (data not shown). Accordingly, these culture conditions allow proper data evaluation due to comparable growth of the analyzed strains. Supernatants from the BSA-peptone cultures and from cultures supplemented with alternative sources of nitrogen were subjected to a crude assay for proteolytic activity by spotting aliquots on sheets of unprocessed X-ray films. A clearing zone, which is based on gelatin hydrolysis in the light-sensitive layer, could be observed only for strains expressing PrtT (Fig. 3A). From these assays it was evident also that the presence of the primary nitrogen source ammonium suppresses expression of extracellular proteolytic activities. To assess degradation of the protein substrate in the course of fungal growth, supernatants from the BSA-peptone cultures were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and proteins visualized by Coomassie blue staining (Fig. 3B). After 24 h, degradation of BSA was evident for the wild-type strain as well as the reconstituted isolate, whereas the prtTΔ strain was apparently unable to degrade this substrate. Upon prolonged cultivation, complete hydrolysis of the growth substrate could be observed for the strains carrying the prtT gene; the prtT deletant, however, was able to break down BSA to a limited extent, resulting in a distinct degradation product of approximately 50 kDa in the culture supernatant. Shift experiments with pregrown mycelia of AfS61 transferred to BSA-containing but peptone-lacking medium did not result in this degradation pattern (data not shown), indicating that the expression of the accountable proteolytic activity is linked to fungal growth. To quantify the actual proteolytic capacities from the culture supernatants, colorimetric assays with the unspecific substrate azocoll were carried out. In this assay, almost no detectable activity was evident in supernatants from the prtTΔ strain AfS61, in sharp contrast to the wild-type progenitor ATCC 46645 or the reconstituted strain AfS62 (Fig. 3C). In summary, these data indicate that the presence of prtT is vital for the expression of extracellular proteases by A. fumigatus, which enables this saprophyte to assimilate proteinaceous growth substrates.
FIG. 3.
PrtT is required for expression of extracellular proteolytic activities. (A) Supernatants from cultures supplemented with various sources of nitrogen were spotted on an unprocessed X-ray film to test for the presence of proteolytic activities. In the presence of the primary N source ammonium, expression of secreted proteases that would hydrolyze the gelatin-containing light-sensitive layer is absent, but not in the presence of protein or the proteolysis product peptone. In contrast to the wild-type isolate (wt) and the reconstituted strain (rec.) the prtTΔ mutant apparently is impaired in producing extracellular proteases. Drop zones are indicated by dashed circles; untreated medium (m) was included in the spot tests. (B) Visualization of protein degradation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis after 24 and 48 h of cultivation supports the near-complete absence of extracellular proteases in the culture supernatants containing BSA and peptone when inoculated with the prtTΔ deletion strain AfS61. (C) Quantitative assessment of extracellular proteolytic activities secreted by the wild-type isolate, the prtTΔ deletion strain and the reconstituted strain when grown in the presence of BSA and peptone reveals the almost-complete absence of extracellular proteases in the prtTΔ culture supernatant. Error bars indicate standard deviations.
PrtT regulates transcription of major secreted proteases.
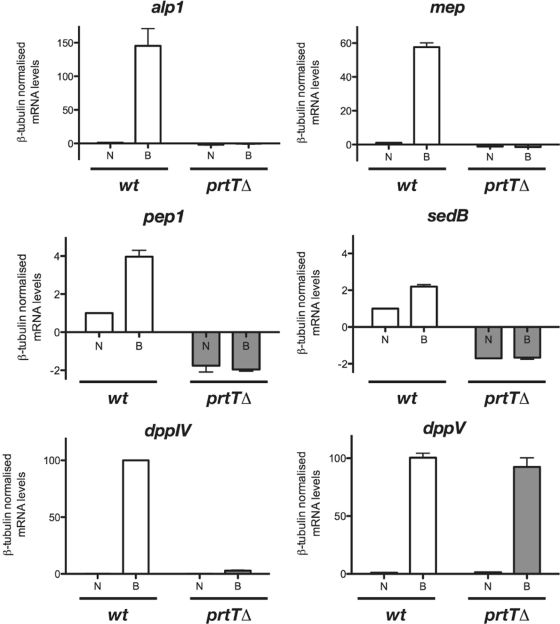
As it was evident that the prtT gene product would regulate the expression of extracellular proteolytic activities, we monitored steady-state transcript levels of several selected genes encoding major secreted proteases of A. fumigatus, in particular alp1 (AFUA_84G11800), mep (AFUA_8G07080), pep1 (AFUA_5G13300), sedB/tppA (AFUA_4G03490), dppIV (AFUA_4G09320), and dppV (AFUA_2G09030), which encode a serine alkaline protease (16), a metalloprotease (15), aspergillopepsin (43), a sedolisin (41), and two dipeptidyl-peptidases (2, 3), respectively (Fig. 4). Upon a shift from minimal medium supplemented with primary sources of carbon and nitrogen to medium containing BSA as the sole N source, expression of any of the major extracellular protease-encoding transcripts alp1, mep, and pep1 increased in the wild-type isolate, and this transcriptional induction was absent in strain AfS61 carrying the prtT deletion. Transcript levels of the sedolisin-encoding gene sedB did not increase significantly upon a shift to BSA-containing medium, whereas transcription of the two genes encoding extracellular dipeptidyl-peptidases, dppIV and dppV, was strongly induced when BSA was present. This regulatory pattern was unaltered in the prtTΔ genetic background for the dppV gene. For dppIV, however, a much smaller range of induction was evident, resulting in basal levels of transcription. This clearly demonstrates that induced expression of some of the major secreted proteases is regulated on the transcriptional level by the PrtT regulator; however, this regulation is not entirely comprehensive, and the expression pattern for some extracellular proteases might not depend exclusively on PrtT, as demonstrated for the dipeptidyl-peptidase-encoding genes dppIV and dppV.
FIG. 4.
PrtT regulates transcriptional induction of several secreted proteases. Steady-state levels of transcripts from several protease-encoding genes were monitored by qRT-PCR after a shift from minimal medium containing NH4+ (N) as a nitrogen source to BSA (B)-containing cultures for 2 h. Expression rates were normalized to mRNA levels of the β-tubulin-encoding gene (AFUA_1G10910) and set arbitrarily to 1 for the wild-type strain grown in minimal medium, except for expression of dppIV, which could not be detected in either strain grown in NH4+-containing medium; in this case, the transcript level of the BSA-grown wild-type strain was set arbitrarily to 100. Error bars indicate standard deviations.
The prtT gene product is not a virulence determinant in leukopenic mice.
In order to assess any contribution of the PrtT regulator to A. fumigatus virulence, the corresponding deletion mutant AfS61 and its wild-type progenitor ATCC 46645 were tested in a murine model of pulmonary aspergillosis. From small groups (n = 5) of leukopenic animals that had been infected with low doses of fungal spores, similar weight loss profiles were determined for the prtTΔ deletion strain and the wild-type isolate (Fig. 5A). These data correlate with disease progression and were essentially very similar for either A. fumigatus strain. Moreover, lungs of infected animals were isolated at 4 days postinfection to extract genomic DNA from the homogenized tissue. From these samples the fractions of fungal DNA, corresponding to the fungal burden in the respective organ, were determined by quantitative PCR amplifying the 18S ribosomal DNA locus (Fig. 5B). No significant differences between the cohorts of infected animals were evident, which indicates that fungal growth is unaltered for the prtTΔ strain. Although fungal burden is not directly indicative of virulence, fungal growth is a recognized pathogenicity trait (44). Accordingly, it is likely that regulated expression of the major extracellular proteases is not a virulence determinant of A. fumigatus in the course of pulmonary aspergillosis, at least not in the disease model used.
FIG. 5.
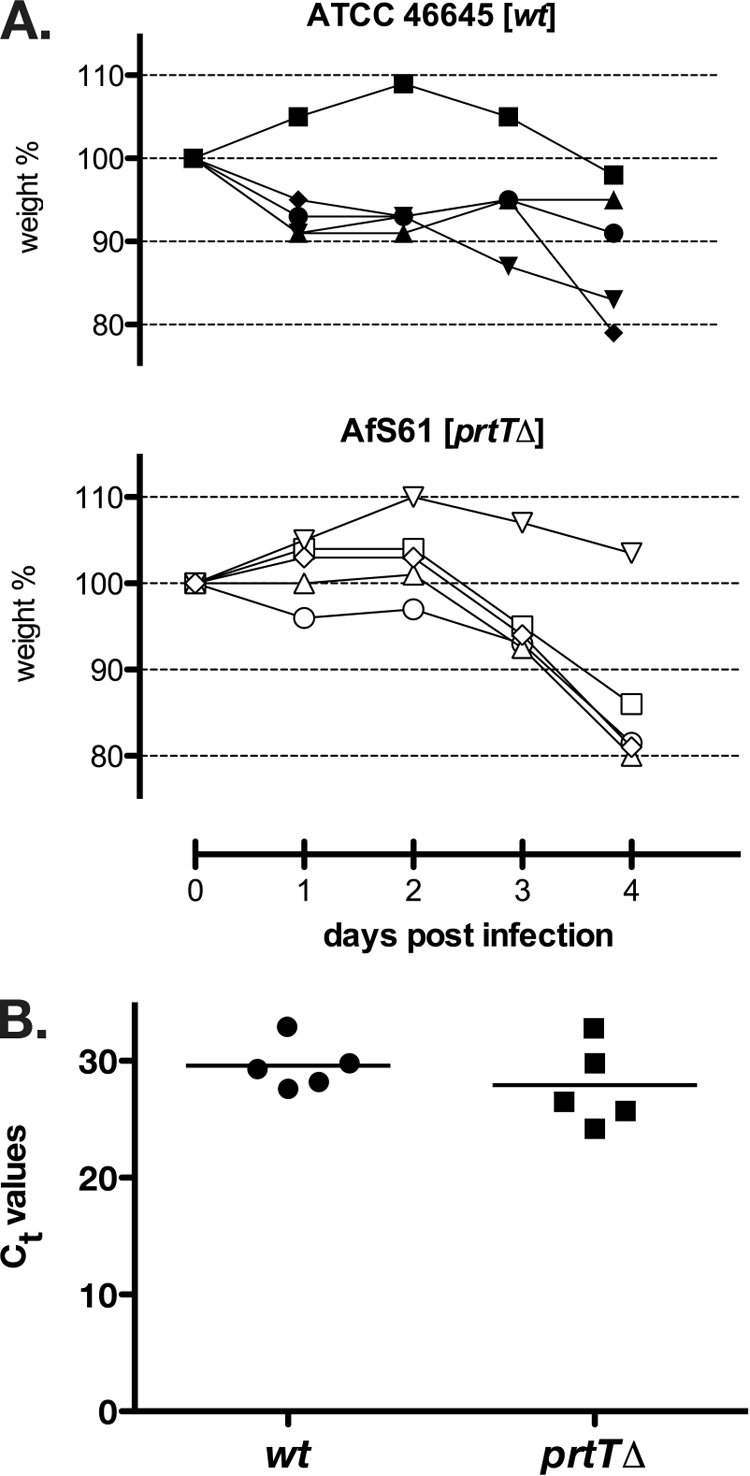
The prtTΔ deletant strain grows at rates comparable to those for the wild-type reference in the lungs of leukopenic mice. Shown are weight loss data for infected animals (A) and threshold cycle (Ct) values from qPCRs performed on DNA samples extracted from corresponding lung tissues (B). Similar disease progressions and quantities of fungal burden can be deduced from these data, which indicates that the deletion mutant AfS61 is not impaired in virulence in this kind of disease model.
DISCUSSION
Pathogenicity of the ubiquitous saprophyte A. fumigatus has been subjected to extensive scrutiny for the last few years, since this filamentous fungus appears to lack bona fide virulence factors. In order to address the open question about the contribution of secreted proteolytic activities to the fungal virulome, we directed our efforts toward a conserved regulator of extracellular protease expression, the prtT gene product. Characterization of a defined deletion mutant revealed that PrtT is indeed required for transcription of several genes encoding secreted proteases, which in turn enable A. fumigatus to assimilate proteinaceous substrates. However, the absence of regulation of expression of extracellular proteases appears not to influence virulence. It is evident that by our in vitro characterization of the prtTΔ mutant we address the saprophytic capacity of A. fumigatus only in part. Yet, the data on PrtT-dependent expression of extracellular proteolytic activities give insights into fungal characteristics that contribute to saprophytism.
In our analysis of the automatically annotated prtT gene, we became aware of an unusually long leader sequence preceding the coding region that is translated into the conserved regulatory protein. Diagnostic PCRs indicate that this leader is at least 690 nt in length, with no splicing events being evident. For the A. niger ortholog of prtT, a comparable situation was described, with the widest-reaching stretch upstream of the translational start codon being 800 nt long (37). Leader stretches are common among genes that are modulated in their expression by posttranscriptional mechanisms, for instance, translational regulation via small upstream open reading frames. Sequence analysis of the postulated leader region indeed indicates the presence of various AUG triplets followed by in-frame stop codons. Accordingly, a regulatory mechanism acting on PrtT expression via eIF2α kinase signaling seems likely (54). In this scenario, nutritional stress such as the presence of protein as a secondary source of nitrogen would trigger phosphorylation of the translation initiation factor subunit eIF2α and relaxation of the inhibitory functions of upstream open reading frames in the prtT mRNA leader region to result in proper translation of the actual coding sequence. In line with this is the fact that steady-state levels of the prtT transcript do not change upon a shift from the primary N source ammonium to BSA (data not shown), a situation in which extracellular proteases become abundantly expressed. Yet, additional regulatory mechanisms, such as phosphorylation or regulated translocation, may modulate PrtT activity as well.
The observation that expression of extracellular proteolytic activity is abolished by the presence of ammonium accounts for an influence of the wide-domain regulatory system of nitrogen catabolite repression, which is essentially mediated by the GATA-type transcription factor AreA (6, 55). Regulation of expression by PrtT appears to be more specific, as it acts on transcription of a subset of secreted proteases, among them the major activities Alp, Mep, and Pep. The fact that these genes are targets of PrtT-mediated regulation is properly illustrated by the phenotypic behavior of the corresponding deletant: growth on protein as the sole source of nitrogen and carbon became markedly reduced as degradation of this substrate was considerably impaired. Several other protease-encoding genes might not be affected by PrtT, as is supported by the unaltered expression of dppV or sedB, and also basal levels of protease expression might still be present in the prtTΔ deletion background. This is corroborated by low transcript levels for the dipeptidyl-peptidase-encoding gene dppIV in the prtTΔ deletant and the observation that prolonged propagation of this strain in BSA-containing medium results in limited but distinct degradation of this protein (Fig. 3B). This specific pattern may result from the action of a specific endoprotease expressed in small amounts or bound to the fungal cell wall. Future studies might explore the PrtT-directed transcriptome and its associated proteome or secretome to reveal the depth of the PrtT regulon. Furthermore, the mechanism of regulating PrtT activity remains to be clarified, as well as the signal transduction cascade that mediates the degradative response of A. fumigatus when challenged with proteinaceous substrates.
The main finding of this study lies in the observation that an A. fumigatus strain with the prtT gene deleted is as virulent as its progenitor in a murine model of pulmonary aspergillosis testing leukopenic animals. From the observed phenotypes displayed by the deletant, it is obvious that PrtT contributes to protein degradation and therefore most likely to the saprophytic lifestyle of A. fumigatus, though it appears not to be essential for pathogenicity in the aforementioned animal model. This kind of aspergillosis model, which is devoid of phagocytic cells of the innate immune system, is appropriate when testing for growth or fitness of an A. fumigatus mutant at the site of infection, two qualities strongly related to fungal virulence (44). We chose not to employ a standard model following progression of deaths among cohorts of infected animals but instead quantified fungal burden as a read-out for virulence, based on the consideration that this would reveal even subtle differences more consistently. Data from the corresponding qRT-PCRs strongly indicate that the prtTΔ strain is not attenuated, a conclusion that is supported by the weight loss profiles of the infected animals in the course of the infection experiments. Yet, as regulation of extracellular protease expression by PrtT possibly is not comprehensive, we cannot rule out that secreted proteases other than the PrtT-affected ones contribute to substrate tapping or invasion during aspergillosis. Moreover, it might well be that extracellular proteases are involved in interacting with the primary lines of immune defense, an aspect that is not covered by our study. Virulence tests in the model using cortisone-treated animals did not, however, reveal any attenuation in virulence for a prtTΔ mutant (Nir Osherov, personal communication). In summary, these findings resemble the insights gained from various protease deletion mutants (15, 34, 42) and do not support a role of secreted proteases in pulmonary aspergillosis. The impact of cell wall-linked proteolytic activities, however, remains to be addressed.
One noteworthy aspect of the prtT gene lies in the fact that it is absent in the less pathogenic species A. nidulans. Quantification of extracellular proteolytic activity from a wild-type strain of A. nidulans revealed levels similar to the ones expressed by the A. fumigatus prtTΔ deletant (our unpublished observation). This indicates that A. nidulans has not evolved compensatory means, and it may be reasoned that the prtT gene loss event is evolutionarily recent. It is tempting to speculate that the associated deletion event in A. nidulans might have affected its virulence potential. Our data do not support such a hypothesis, and based on the multifactorial nature of Aspergillus virulence, it has to be assumed that other additional variations make the difference between the saprophyte A. nidulans and the pathogen A. fumigatus.
Acknowledgments
We thank all members of the research group, especially Michaela Dümig for technical assistance. We thank colleagues of the institute for discussions and support, and we are indebted to Matthias Brock for providing strain ATCC 46645. Hubertus Haas is acknowledged for discussing an evolutionary aspect.
Financial aid was received from the German Research Foundation via its Priority Program SPP1160 (KR2294/1-3), the Free State of Bavaria, and the University of Würzburg; E.M.B. is supported by the Medical Research Council.
Editor: A. Casadevall
Footnotes
Published ahead of print on 29 June 2009.
REFERENCES
- 1.Aarskog, N. K., and C. A. Vedeler. 2000. Real-time quantitative polymerase chain reaction. A new method that detects both the peripheral myelin protein 22 duplication in Charcot-Marie-Tooth type 1A disease and the peripheral myelin protein 22 deletion in hereditary neuropathy with liability to pressure palsies. Hum. Genet. 107494-498. [DOI] [PubMed] [Google Scholar]
- 2.Beauvais, A., M. Monod, J. P. Debeaupuis, M. Diaquin, H. Kobayashi, and J.-P. Latgé. 1997. Biochemical and antigenic characterization of a new dipeptidyl-peptidase isolated from Aspergillus fumigatus. J. Biol. Chem. 2726238-6244. [DOI] [PubMed] [Google Scholar]
- 3.Beauvais, A., M. Monod, J. Wyniger, J. P. Debeaupuis, E. Grouzmann, N. Brakch, J. Svab, A. G. Hovanessian, and J.-P. Latgé. 1997. Dipeptidyl-peptidase IV secreted by Aspergillus fumigatus, a fungus pathogenic to humans. Infect. Immun. 653042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchara, J. P., G. Larcher, F. Joubaud, P. Penn, G. Tronchin, and D. Chabasse. 1993. Extracellular fibrinogenolytic enzyme of Aspergillus fumigatus: substrate-dependent variations in the proteinase synthesis and characterization of the enzyme. FEMS Immunol. Med. Microbiol. 781-91. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, J. C., G. K. Abruzzo, J. W. Anderson, A. M. Flattery, C. J. Gill, V. B. Pikounis, D. M. Schmatz, P. A. Liberator, and C. M. Douglas. 2001. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob. Agents Chemother. 453474-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caddick, M. X., D. Peters, and A. Platt. 1994. Nitrogen regulation in fungi. Antonie van Leeuwenhoek 65169-177. [DOI] [PubMed] [Google Scholar]
- 7.Chavira, R., Jr., T. J. Burnett, and J. H. Hageman. 1984. Assaying proteinases with azocoll. Anal. Biochem. 136446-450. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, A. L., P. Ying, and V. A. Fischetti. 1991. A method to detect proteinase activity using unprocessed X-ray films. Anal. Biochem. 19320-23. [DOI] [PubMed] [Google Scholar]
- 9.Denning, D. W., P. N. Ward, L. E. Fenelon, and E. W. Benbow. 1992. Lack of vessel wall elastolysis in human invasive pulmonary aspergillosis. Infect. Immun. 605153-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Pauw, B., T. J. Walsh, J. P. Donnelly, D. A. Stevens, J. E. Edwards, T. Calandra, P. G. Pappas, J. Maertens, O. Lortholary, C. A. Kauffman, D. W. Denning, T. F. Patterson, G. Maschmeyer, J. Bille, W. E. Dismukes, R. Herbrecht, W. W. Hope, C. C. Kibbler, B. J. Kullberg, K. A. Marr, P. Munoz, F. C. Odds, J. R. Perfect, A. Restrepo, M. Ruhnke, B. H. Segal, J. D. Sobel, T. C. Sorrell, C. Viscoli, J. R. Wingard, T. Zaoutis, and J. E. Bennett. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 461813-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gifford, A. H., J. R. Klippenstein, and M. M. Moore. 2002. Serum stimulates growth of and proteinase secretion by Aspergillus fumigatus. Infect. Immun. 7019-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 20463-113. [DOI] [PubMed] [Google Scholar]
- 13.Hearn, V. M., and D. W. Mackenzie. 1980. Mycelial antigens from two strains of Aspergillus fumigatus: an analysis by two-dimensional immunoelectrophoresis. Mykosen 23549-562. [PubMed] [Google Scholar]
- 14.Ibrahim-Granet, O., M. Dubourdeau, J.-P. Latgé, P. Ave, M. Huerre, A. A. Brakhage, and M. Brock. 2008. Methylcitrate synthase from Aspergillus fumigatus is essential for manifestation of invasive aspergillosis. Cell. Microbiol. 10134-148. [DOI] [PubMed] [Google Scholar]
- 15.Jaton-Ogay, K., S. Paris, M. Huerre, M. Quadroni, R. Falchetto, G. Togni, J.-P. Latgé, and M. Monod. 1994. Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol. Microbiol. 14917-928. [DOI] [PubMed] [Google Scholar]
- 16.Jaton-Ogay, K., M. Suter, R. Crameri, R. Falchetto, A. Fatih, and M. Monod. 1992. Nucleotide sequence of a genomic and a cDNA clone encoding an extracellular alkaline protease of Aspergillus fumigatus. FEMS Microbiol. Lett. 71163-168. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, N., N. S. Tan, B. Ho, and J. L. Ding. 2007. Azocoll protease activity assay. Nat. Protoc. doi: 10.1038/nprot.2007.484. [DOI]
- 18.Käfer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1933-131. [DOI] [PubMed] [Google Scholar]
- 19.Kämper, J. 2004. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271103-110. [DOI] [PubMed] [Google Scholar]
- 20.Katz, M. E., K. A. Gray, and B. F. Cheetham. 2006. The Aspergillus nidulans xprG (phoG) gene encodes a putative transcriptional activator involved in the response to nutrient limitation. Fungal Genet. Biol. 43190-199. [DOI] [PubMed] [Google Scholar]
- 21.Katz, M. E., A. Masoumi, S. R. Burrows, C. G. Shirtliff, and B. F. Cheetham. 2000. The Aspergillus nidulans xprF gene encodes a hexokinase-like protein involved in the regulation of extracellular proteases. Genetics 1561559-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kogan, T. V., J. Jadoun, L. Mittelman, K. Hirschberg, and N. Osherov. 2004. Involvement of secreted Aspergillus fumigatus proteases in disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected A549 lung pneumocytes. J. Infect. Dis. 1891965-1973. [DOI] [PubMed] [Google Scholar]
- 23.Kolar, M., P. J. Punt, C. A. van den Hondel, and H. Schwab. 1988. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62127-134. [DOI] [PubMed] [Google Scholar]
- 24.Kothary, M. H., T. Chase, Jr., and J. D. Macmillan. 1984. Correlation of elastase production by some strains of Aspergillus fumigatus with ability to cause pulmonary invasive aspergillosis in mice. Infect. Immun. 43320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krappmann, S. 2007. Pathogenicity determinants and allergens, p. 377-400. In G. H. Goldman and S. A. Osmani (ed.), The aspergilli: genomics, medical aspects, biotechnology, and research methods. CRC Press, Boca Raton, FL.
- 26.Krappmann, S., Ö. Bayram, and G. H. Braus. 2005. Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot. Cell 41298-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krappmann, S., N. Jung, B. Medic, S. Busch, R. A. Prade, and G. H. Braus. 2006. The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis. Mol. Microbiol. 6176-88. [DOI] [PubMed] [Google Scholar]
- 28.Mabey, J. E., M. J. Anderson, P. F. Giles, C. J. Miller, T. K. Attwood, N. W. Paton, E. Bornberg-Bauer, G. D. Robson, S. G. Oliver, and D. W. Denning. 2004. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res. 32D401-D405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markaryan, A., I. Morozova, H. Yu, and P. E. Kolattukudy. 1994. Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect. Immun. 622149-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattern, I. E., J. M. van Noort, P. van den Berg, D. B. Archer, I. N. Roberts, and C. A. van den Hondel. 1992. Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol. Gen. Genet. 234332-336. [DOI] [PubMed] [Google Scholar]
- 31.McNeil, M. M., S. L. Nash, R. A. Hajjeh, M. A. Phelan, L. A. Conn, B. D. Plikaytis, and D. W. Warnock. 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980-1997. Clin. Infect. Dis. 33641-647. [DOI] [PubMed] [Google Scholar]
- 32.Monod, M., K. Jaton-Ogay, and U. Reichard. 1999. Aspergillus fumigatus-secreted proteases as antigenic molecules and virulence factors. Contrib. Microbiol. 2182-192. [DOI] [PubMed] [Google Scholar]
- 33.Monod, M., O. Jousson, and U. Reichard. 2009. Aspergillus fumigatus secreted proteases, p. 87-106. In J.-P. Latgé and W. J. Steinbach (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC.
- 34.Monod, M., S. Paris, J. Sarfati, K. Jaton-Ogay, P. Ave, and J.-P. Latgé. 1993. Virulence of alkaline protease-deficient mutants of Aspergillus fumigatus. FEMS Microbiol. Lett. 10639-46. [DOI] [PubMed] [Google Scholar]
- 35.Moutaouakil, M., M. Monod, M. C. Prevost, J. P. Bouchara, S. Paris, and J.-P. Latgé. 1993. Identification of the 33-kDa alkaline protease of Aspergillus fumigatus in vitro and in vivo. J. Med. Microbiol. 39393-399. [DOI] [PubMed] [Google Scholar]
- 36.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 4381151-1156. [DOI] [PubMed] [Google Scholar]
- 37.Punt, P. J., F. H. Schuren, J. Lehmbeck, T. Christensen, C. Hjort, and C. A. van den Hondel. 2008. Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet. Biol. 451591-1599. [DOI] [PubMed] [Google Scholar]
- 38.Punt, P. J., and C. A. van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216447-457. [DOI] [PubMed] [Google Scholar]
- 39.Reichard, U., S. Buttner, H. Eiffert, F. Staib, and R. Rüchel. 1990. Purification and characterisation of an extracellular serine proteinase from Aspergillus fumigatus and its detection in tissue. J. Med. Microbiol. 33243-251. [DOI] [PubMed] [Google Scholar]
- 40.Reichard, U., H. Eiffert, and R. Rüchel. 1994. Purification and characterization of an extracellular aspartic proteinase from Aspergillus fumigatus. J. Med. Vet. Mycol. 32427-436. [DOI] [PubMed] [Google Scholar]
- 41.Reichard, U., B. Lechenne, A. R. Asif, F. Streit, E. Grouzmann, O. Jousson, and M. Monod. 2006. Sedolisins, a new class of secreted proteases from Aspergillus fumigatus with endoprotease or tripeptidyl-peptidase activity at acidic pHs. Appl. Environ. Microbiol. 721739-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichard, U., M. Monod, F. Odds, and R. Rüchel. 1997. Virulence of an aspergillopepsin-deficient mutant of Aspergillus fumigatus and evidence for another aspartic proteinase linked to the fungal cell wall. J. Med. Vet. Mycol. 35189-196. [DOI] [PubMed] [Google Scholar]
- 43.Reichard, U., M. Monod, and R. Rüchel. 1995. Molecular cloning and sequencing of the gene encoding an extracellular aspartic proteinase from Aspergillus fumigatus. FEMS Microbiol. Lett. 13069-74. [DOI] [PubMed] [Google Scholar]
- 44.Rhodes, J. C. 2006. Aspergillus fumigatus: growth and virulence. Med. Mycol. 44(Suppl. 1)S77-S81. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Sheppard, D. C., K. A. Marr, D. N. Fredricks, L. Y. Chiang, T. Doedt, and S. G. Filler. 2006. Comparison of three methodologies for the determination of pulmonary fungal burden in experimental murine aspergillosis. Clin. Microbiol. Infect. 12376-380. [DOI] [PubMed] [Google Scholar]
- 47.Smith, J. M., C. M. Tang, S. Van Noorden, and D. W. Holden. 1994. Virulence of Aspergillus fumigatus double mutants lacking restrictocin and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect. Immun. 625247-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Southern, E. 2006. Southern blotting. Nat. Protoc. 1518-525. [DOI] [PubMed] [Google Scholar]
- 49.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98503-517. [DOI] [PubMed] [Google Scholar]
- 50.Tang, C. M., J. Cohen, T. Krausz, S. Van Noorden, and D. W. Holden. 1993. The alkaline protease of Aspergillus fumigatus is not a virulence determinant in two murine models of invasive pulmonary aspergillosis. Infect. Immun. 611650-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tekaia, F., and J. P. Latge. 2005. Aspergillus fumigatus: saprophyte or pathogen? Curr. Opin. Microbiol. 8385-392. [DOI] [PubMed] [Google Scholar]
- 52.Tong, K. B., C. J. Lau, K. Murtagh, A. J. Layton, and R. Seifeldin. 2009. The economic impact of aspergillosis: analysis of hospital expenditures across patient subgroups. Int. J. Infect. Dis. 1324-36. [DOI] [PubMed] [Google Scholar]
- 53.Walsh, T. J., and A. H. Groll. 2001. Non-fumigatus species of Aspergillus: perspectives on emerging pathogens in immunocompromised hosts. Curr. Opin. Investig. Drugs 21366-1367. [PubMed] [Google Scholar]
- 54.Wek, R. C., H. Y. Jiang, and T. G. Anthony. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 347-11. [DOI] [PubMed] [Google Scholar]
- 55.Wilson, R. A., and H. N. Arst, Jr. 1998. Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “streetwise” GATA family of transcription factors. Microbiol. Mol. Biol. Rev. 62586-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 173469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]