Abstract
The role of secreted proteases in the virulence of the pathogenic fungus Aspergillus fumigatus remains controversial. Recently, the Aspergillus niger transcription factor PrtT was shown to control the expression of multiple secreted proteases. In this work, the gene which encodes the PrtT homolog in A. fumigatus was cloned and its function analyzed using a deletion mutant strain. Deletion of A. fumigatus prtT resulted in the loss of secreted protease activity. The expression of six secreted proteases (ALP, MEP, Dpp4, CpdS, AFUA_2G17330, and AFUA_7G06220) was markedly reduced. Culture filtrates from the prtT deletion strain exhibited reduced killing of lung epithelial cells and lysis of erythrocytes. However, the prtT deletion strain did not exhibit altered virulence in lung-infected mice. These results suggest that PrtT is not a significant virulence factor in A. fumigatus.
Fungi belonging to the genus Aspergillus are important opportunistic pathogens of immunocompromised patients. Aspergillus fumigatus is the main causative agent of aspergillosis. In the last 15 years there has been a sharp upsurge in the incidence and severity of fungal infections caused by these organisms. This has been attributed to more aggressive cytotoxic chemotherapy, an increase in the number of bone marrow and organ transplant recipients, and the emergence of AIDS (18).
Evidence accumulating over the last decade suggests that A. fumigatus utilizes multiple virulence factors to infect and colonize its host, including toxins and proteases, protective pigments and antioxidants, thermotolerance, and small spore size (26).
Proteases have been implicated as virulence factors in viral (9), bacterial (8, 38), and fungal (22, 23a) pathogenesis. The following evidence implicates secreted A. fumigatus proteases in virulence: (i) secreted A. fumigatus proteases induce proinflammatory cytokine release in infected macrophages and epithelial cells, thereby alerting the immune system (14); (ii) infected lung epithelial cells also undergo protease-dependent changes to the actin cytoskeleton, leading to cell peeling and death (15, 36); (iii) proteases are secreted in vivo during infection, and protease-specific antisera show labeling of mycelium in the lungs of patients and experimentally infected animals (22); (iv) loss of elastase protease activity in mutagenized strains of A. fumigatus has been correlated with decreased virulence in vivo (16); and (v) mice intratracheally injected with purified ALP1 protease showed a marked degree of lower respiratory tract destruction (10).
However, deletion analyses of selected proteases (ALP1, MEP, PEP1, MEP, and ALP1) have failed to conclusively demonstrate a significant role in virulence in animal models (22). This is probably due to the large number of proteases secreted by A. fumigatus and the functional redundancy among them. Its genome encodes approximately 111 proteases and 26 nonpeptidase homologs (MEROPS peptidase database for A. fumigatus, http://merops.sanger.ac.uk/), of which 47 have a signal sequence directing them to the endoplasmic reticulum and possible extracellular secretion (23a). It is not technically feasible to delete more than a few these genes at a time. Instead, we selected and designed an alternative approach: to delete the A. fumigatus gene prtT (AFUA_4G10120), which encodes a putative transcription factor controlling the expression of multiple secreted proteases. Previous studies of Aspergillus niger, an important industrial producer of citric acid and recombinant proteins, have shown that random mutagenesis and subsequent inactivation of the prtT locus result in a mutant mold strain with strongly reduced protease activity. The prtT gene in A. niger was subsequently cloned by complementation and shown to encode a zinc finger-containing putative transcription factor (27, 37). We identified a single A. fumigatus homolog of A. niger prtT (AFUA_4G10120) by BlastP sequence analysis.
In this report we describe the effects of prtT deletion on growth, protease expression, and virulence in A. fumigatus. Our approach analyzes one of the molecular mechanisms regulating protease expression in a pathogenic filamentous fungus, a field that remains largely unexplored.
MATERIALS AND METHODS
Strains and culture conditions.
A. fumigatus strain AF293, originally isolated at autopsy from a patient with invasive pulmonary aspergillosis, was used throughout this study. For continuous growth, the different A. fumigatus strains were grown on YAG medium, which consists of 0.5% (wt/vol) yeast extract, 1% (wt/vol) glucose, and 10 mM MgCl2, supplemented with trace elements, vitamins, and 1.5% (wt/vol) agar when needed (1). Skim milk (SM) medium, used in the protease assays, consisted of 1% (wt/vol) glucose, 1% (wt/vol) SM (Difco, Livonia, MI), 0.1% (wt/vol) Casamino Acids (Difco), 7 mM KCl, 2 mM MgSO4, 50 mM Na2HPO4-NaH2PO4 buffer)pH 5.3), and 0.05% (wt/vol) Triton X-100, supplemented with vitamins, trace elements, and 1.5% agar when needed (19). For starch, bovine serum albumin (BSA), and collagen plates, the SM was replaced by 1% (wt/vol) starch (Difco), 1% (wt/vol) BSA (Amresco, Solon, OH), or 0.2% (wt/vol) collagen (Sigma-Aldrich, St. Louis, MO), respectively. No Casamino Acids were added to the BSA- and collagen-containing plates. Conidia were harvested in 0.2% (vol/vol) Tween 80, resuspended in double-distilled water, and counted with a hemocytometer. Escherichia coli strain DH10B (Invitrogen, Carlsbad, CA) was used for cloning.
Construction and verification of the A. fumigatus prtT disruption mutant.
A 5,120-bp DNA fragment flanking the A. fumigatus prtT gene was generated by PCR, using the Expand high-fidelity PCR system (Roche Diagnostics, Indianapolis, IN) and primers PrtT outer 5′ and PrtT outer 3′, designed to contain an AscI restriction site at their 5′ ends (Table 1). The product of this PCR was cloned into the TA vector pGEM-T-Easy (Promega, Madison WI). A 773-bp fragment, which included 307 bp of the N-terminal prtT open reading frame, was then removed by digestion with XbaI and HindIII and replaced with a hygromycin-selectable marker to produce the pAfPrtT-D plasmid. The hygromycin cassette, containing 5′ and 3′ HindIII and XbaI restriction sites, was generated by PCR amplification using primers Hyg 5′ and Hyg 3′ (Table 1). For transformation, 10 μg of spin-purified AscI-digested pAfPrtT-D plasmid was used. Transformation was performed as previously described by Romano et al. (31). For Southern blot analysis, genomic DNA was extracted from wild-type (WT) A. fumigatus AF293 and five independent transformants (prtTΔ-D1 to -5 strains). Southern hybridization analysis was performed as previously described (11). Briefly, 10-mg fungal genomic DNA samples were digested with XbaI and run on a 1% (wt/vol) agarose gel. The restricted DNA was transferred to a Nytran N nylon membrane (Schleicher & Schuell Bioscience, Keene, NH) and hybridized with an [α-32P]dCTP-radiolabeled AfPrtT 5′ probe at 65°C. The probe (785 bp) was generated by PCR with primers PrtT outer 5′ and PrtT up 3′ (Table 1). The prtT-KI-1 (KI denotes knock-in) control strain was prepared by complementing the prtT-D1 strain with a plasmid containing the pgpdA-phleomycin cassette and the 5,120-bp DNA fragment flanking the above-described A. fumigatus prtT gene. This plasmid was generated by cloning a pgpdA-phleomycin cassette into the pAfPrtT-D plasmid via an NdeI restriction site. The primers used to amplify the pgpdA-phleomycin cassette from pAN-8.1 (28) were pAN-8.1 NdeI-5′ and pAN-8.1 NdeI-3′ (Table 1). Two prtT-KI strains were obtained and verified for prtT mRNA expression by reverse transcriptase PCR (RT-PCR). Both were phenotypically identical to the control WT strain AF293 as assessed by protease activity experiments (see Results).
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′ → 3′) |
|---|---|
| PrtT outer 5′ | ATGGCGCGCCGCACTGATCTCGTCCTATCGGTT |
| PrtT outer 3′ | ATGGCGCGCCCTAAAGTCTCCTACCTACCTCAG |
| Hyg 5′ | ATAAGCTTGTCGACAGAAGATGATAT |
| Hyg 3′ | ATAAGCTTGCTCTCCCTTATGCGACTCCTGCA |
| PrtT up 3′ | AGTGACAGAACAGGCGACAG |
| pAN-8.1 NdeI 5′ | ATCATATGGAATTCCCTTGTATCTCTACACAC |
| pAN-8.1 NdeI 3′ | TGCACCATATGCGGTGTGAAATA |
| gpdA 5′ | TCTCCAACGTTCTTGCACC |
| gpdA 3′ | CCACTCGTTGTCGTACCAGG |
| prtT inner 5′ | GGTGATAGCGAAACAGAAGAGACAAG |
| prtT inner 3′ | ATAGACTCCACTGGTAGTTAGGTGTTCTG |
| AFUA_8G07080 (Mep) 5′ | AACTTTGCCACTCCTCCTGA |
| AFUA_8G07080 (Mep) 3′ | GCCGTGCTTATCGATCAAAT |
| AFUA_4G11800 (Alp1) 5′ | GCATCAATGTCAACCACGTC |
| AFUA_4G11800 (Alp1) 3′ | CATCGAAAGCGTTCTCAACA |
| AFUA_6G00310 (CpdS) 5′ | AGCTGGTTCAAGCACTTCGT |
| AFUA_6G00310 (CpdS) 3′ | TGAAGGGCTTTTTGGACATC |
| AFUA_4G09320 (DppIV) 5′ | AAGGCCTACATTGCCTCAGA |
| AFUA_4G09320 (DppIV) 3′ | CTCCTCGTACAACCTCTTGG |
| AFUA_2G17330 5′ | CACCGAGGCCATTGTAGATT |
| AFUA_2G17330 3′ | TAACCCCAGACGATTTTTGC |
| AFUA_7G06220 5′ | TACTATCGGGGCCAGTTACG |
| AFUA_7G06220 3′ | AGAGAGCTCCTGGGTTGGAT |
| AFUA_4G03790 5′ | ATCCTGGCTCAGTCTGCTGT |
| AFUA_4G03790 3′ | CGAACGTCCTTTTGTTGGAT |
Genomic DNA extraction.
Conidia (107) were added to 25 ml YAG medium and incubated at 37°C for 16 h. After growth, the mycelium was collected with Miracloth (Calbiochem, La Jolla CA), lyophilized for 24 h, and pulverized. An ∼50-μl volume of powder was taken for genomic DNA extraction. To extract genomic DNA from A. fumigatus, we used the Epicentre MasterPure yeast DNA purification kit (Epicenter Biotechnologies, Madison, WI) according to the manufacturer's instructions.
RNA extraction and analysis.
Total RNA was isolated from each strain using the Qiagen RNeasy plant kit (Qiagen, Valencia, CA) following the filamentous fungus protocol. For RT-PCR analysis, 3 μg of total RNA from each sample was digested with DNase (DNA free; Ambion, Austin, TX) according to the manufacturer's instructions. The RNA concentration was assessed in a Thermo Scientific NanoDrop 1000 spectrophotometer, and 1 μg was taken for the RT reaction using AffinityScript multiple-temperature reverse transcriptase (Stratagene, TX). The first-strand cDNA was PCR amplified using gene-specific primers for 300- to 500-bp fragments of the A. fumigatus proteases. gpdA was used as a normalizing control (Table 1).
Analysis of fungal enzymatic activity.
Proteolytic or starch-degrading activity on solid medium was assessed by spotting conidia on SM or starch-agar plates, respectively. The colonies were grown for 48 h at 37°C and then transferred to room temperature for another 48 h. To highlight the zone of proteolysis, SM plates were then stained with Coomassie blue dye (stain: 50% [vol/vol] methanol, 10% [vol/vol] acetic acid, 0.1% [wt/vol]) Coomassie brilliant blue) and destained in 30% methanol and 10% acetic acid. Starch plates were stained with 0.5% (wt/vol) iodine dissolved in 70% ethanol.
Azocasein assay.
Azocasein (Sigma) was dissolved at a concentration of 5 mg/ml in assay buffer containing 50 mM Tris (pH 7.5), 0.2 M NaCl, 5 mM CaCl2, and 0.05% Triton X-100 as previously described (15). Supernatants were removed from Aspergillus cultures at various times and centrifuged to pellet the cells. The azocasein solution (400 μl) was mixed with 100-μl portions of supernatants from Aspergillus cultures and incubated by shaking for 90 min at 37°C. The reactions were stopped by adding 150 μl of 12% (vol/vol) trichloroacetic acid, and the reaction mixtures were allowed to stand at room temperature for 30 min. Tubes were then centrifuged for 3 min at 8,000 × g, and 100 μl of each supernatant was added to 100 μl of 1 M NaOH. The absorbance at 436 nm of released azo dye was determined with a spectrophotometer.
Azocollagen assay.
Azocoll (Calbiochem) was suspended at a concentration of 5 mg/ml in a buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM CaCl2, and 0.01% (wt/vol) sodium azide as previously described (7). The Azocoll suspension (800 μl) was incubated with 5-μl aliquots of supernatants from Aspergillus cultures for 3 h at 37°C on a rotator. The tubes were centrifuged at 8,000 × g for 3 min, and the release of azo dye was determined by measuring the absorbance at 520 nm of each supernatant.
Preparation of fungal CF for use in cell-culture experiments.
Conidia were collected in a 0.01% (vol/vol) Tween 80 solution, washed twice in phosphate-buffered saline (PBS), and resuspended at a concentration of 1 × 106 conidia/ml in 100 ml of Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS) (Biological Industries, Beit Haemek, Israel). The FCS underwent another heat inactivation step at 70°C for 30 min to neutralize endogenous protease activity (6; Margo Moore, personal communication). Fungal cultures were grown in an orbital incubator at 200 rpm for 48 h at 37°C. Growth medium containing culture filtrate (CF) was decanted after centrifugation at 2,300 × g and filter sterilized.
Cell culture.
Human cancer cell line A549 (ATCC CLL 185), derived from a human lung carcinoma, was grown in DMEM containing 10% FCS and 1% (wt/vol) penicillin-streptomycin (Biological Industries) in 10-cm culture plates (Corning, Sigma-Aldrich). Cells were incubated at 37°C in 5.5% CO2 and a humidified atmosphere and were routinely subcultured by trypsinization every 3 to 4 days.
Hemacolor staining.
A549 cells (5 × 10−4/well) in DMEM containing 10% FCS were cultured in 96-well plates. Following incubation, the cell monolayers were fixed with methanol and stained with Hemacolor (Merck, NJ). Sodium dodecyl sulfate (0.5% [wt/vol]; 200 μl/well) was added, and the plates were read using an automated microplate reader spectrophotometer at 630 nm.
XTT cell viability assay.
A549 cells were grown in 96-well cell culture plates (Corning) to 80% confluence. CF from AF293, prtTΔ, and prtT-KI strains was added to the cells. XTT reagent [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide, disodium salt] (Biological Industries) was added after 8 to 24 h of incubation, and the colorimetric assay was performed as specified by the manufacturer. Absorbance (optical density at 490 nm) was read on an enzyme-linked immunosorbent assay reader (Spectra MAX 340; Molecular Devices).
Hemolysis assay.
Conidia from AF293, prtTΔ, and prtT-KI strains were suspended in 8 ml of 0.1% SM medium at a concentration of 10−6 conidia/ml. Fungal cultures were grown in 50-mm petri dishes for 48 h at 37°C. CF was collected from the growth medium and filter sterilized. To a microcentrifuge tube containing 950 μl CF, 50 μl (5%) of sheep red blood cells (SRBCs) (Hylabs, Rehovot, Israel) was added. The tubes were incubated in a rotor for 24 h at 37°C. After incubation, the tubes were centrifuged for 10 min at 1,200 rpm. Supernatant was collected and diluted 1:10 (vol/vol) in PBS. Calculation of the percentage of hemolysis was performed using the following formula: (absorbance of sample425 nm − absorbance of blank425 nm)/absorbance of positive control425 nm × 100. As blank and positive controls, respectively, 950 μl of 0.1% SM or 0.1% SM with 1% sodium dodecyl sulfate was mixed with 5% SRBCs and treated as described above.
Murine models for invasive pulmonary aspergillosis.
For the cyclophosphamide/cortisone acetate neutropenic model (6), 6-week-old female ICR mice were injected intraperitoneally with cyclophosphamide (150 mg/kg in PBS) at 3 days prior to and at 2 days after conidial infection. Cortisone acetate (150 mg/kg PBS with 0.1% Tween 80) was injected subcutaneously at 3 days prior to conidial infection. For the cortisone acetate immunocompromised model (6), mice were injected subcutaneously with cortisone acetate (300 mg/kg PBS with 0.1% Tween 80) at 3 days prior to conidial infection, on the day of infection, and 2 and 4 days after infection.
For intranasal infection in both models, mice were anesthetized by intraperitoneal injection of a solution of 250 μl xylazine (VMD, Arendonk, Belgium) and ketamine (Imalgene, Fort Dodge, IA) at concentrations of 1.0 mg/ml and 10 mg/ml, respectively (dissolved in PBS). Following anesthesia, the mice were inoculated intranasally with 2.5 × 105 (cyclophosphamide model) or 5 × 105 (cortisone acetate model) freshly harvested conidia of the AF293, prtTΔ, or prtT-KI strain in PBS plus 0.1% Tween 80. The inoculum was verified by quantitative culture. An additional control group was mock infected with PBS plus 0.1% Tween 80 alone. Mice in this group remained alive throughout the duration of the experiment. The animals were monitored for survival for 30 days postinfection. Statistical analysis of mouse survival was performed with GraphPad Prism 4 software (GraphPad Software, San Diego, CA).
For studies of tissue burden, groups of five mice were infected as described above and sacrificed on day 3 after infection. From each mouse, lungs were aseptically removed, homogenized separately in 1 ml of saline, and cultured for quantitative analysis in serial 10-fold dilutions. Statistical analysis of mouse lung burden was performed with Student's t test. Animal studies were carried out in accordance with Tel Aviv University institutional policies.
RESULTS
Primary sequence analysis of A. fumigatus prtT.
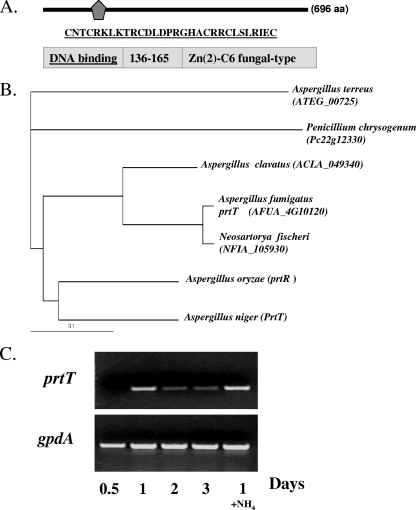
The A. fumigatus prtT gene is 2,792 nucleotides long and contains five predicted introns. PrtT (gene product of AFUA_4G10120, TIGR assembly) is 696 amino acids long and contains a predicted C6 zinc finger domain at its C terminus (Fig. 1A). Based on multiple-sequence alignment, A. fumigatus PrtT has significant sequence identity to Neosartorya fischeri hypothetical protein NFIA_105930 (92% identity in 517 amino acids), Aspergillus clavatus ACLA_049340 (79% identity in 623 amino acids), A. niger PrtT (65% identity in 632 amino acids), Aspergillus oryzae PrtR (64% identity in 628 amino acids), Aspergillus terreus ATEG_00725 (52% identity in 434 amino acids), and Penicillium chrysogenum Pc22g12330 (50% identity in 595 amino acids) (Fig. 1B). No significant similarity to other fungal species was found, suggesting that the prtT gene family is unique to the genera Aspergillus and Penicillium. Interestingly, no prtT homologs were identified in the Aspergillus nidulans genome database.
FIG. 1.
Sequence homology and expression analysis of the A. fumigatus prtT (AFUA_4G10120) gene. (A and B) A. fumigatus PrtT contains an N-terminal 29-amino-acid (aa) fungal C6 Zn finger domain (underlined) (A) and is conserved among Aspergillus and Penicillium species as show in the dendrogram (B). (C) Expression of A. fumigatus prtT. Expression was determined by RT-PCR using primers AfPrtT inner 5′ and 3′ (Table 1). Dormant conidia were incubated for 0.5, 1, 2, and 3 days in liquid SM medium at 37°C, after which total RNA was isolated and used in RT-PCR. RT-PCR with primers for gpdA, a housekeeping gene, was used as a control for the loading (lower bands). A control PCR, performed on RNA following DNase treatment, gave no visible band (data not shown). In lane 1+ NH4, the conidia were incubated in SM in the presence of 5 mM ammonia. This experiment was independently performed twice with similar results.
We showed by RT-PCR, using primers AfPrtT inner 5′ and 3′ (Table 1), that A. fumigatus prtT is not significantly expressed during germination and early growth (0 to 12 h) but is expressed throughout hyphal growth and conidiation (1 to 3 days) (Fig. 1C). Addition of ammonia, which represses secreted protease activity (4), did not alter prtT transcript levels. This suggests that modulation of prtT activity is posttranscriptional or dependent on additional factors.
Deletion of A. fumigatus prtT results in the loss of secreted protease activity.
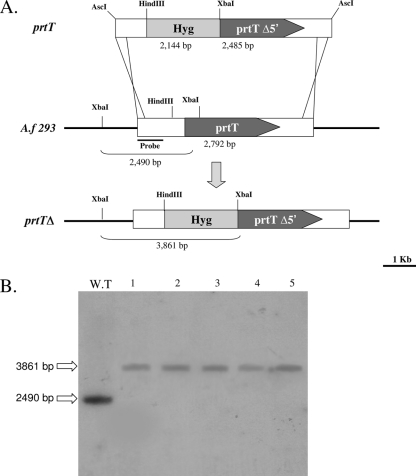
To investigate the effect of loss of function of the prtT gene in A. fumigatus, a disruption plasmid (pAfPrtT) was constructed by deletion of a 773-bp fragment, which includes 307 bp of the N-terminal prtT open reading frame, as described in Materials and Methods (Fig. 2A). After transformation of pAfPrtT into WT AF293, 15 hygromycin-resistant transformants were purified and screened by PCR for putative insertion mutants. Five putative mutants were identified and further characterized by Southern blotting. Based on this analysis, all five transformants were disrupted in the prtT gene alone (prtTΔ-D1 to -5) (Fig. 2B). Preliminary experiments conducted with the five independent disrupted strains confirmed that all exhibit the mutant phenotype, characterized by loss of halo production on SM plates. This suggests that the mutant phenotype is associated with disruption of the gene and is not due to an unlinked mutation resulting from transformation. A representative strain, the prtTΔ-D1 mutant, hereafter designated the prtTΔ mutant, was chosen for further characterization. A prtT-reconstituted strain (KI) was prepared as described in Materials and Methods and used as a control throughout this study.
FIG. 2.
Disruption of the prtT gene in A. fumigatus. (A) Schematic representation of the prtT WT locus and the AscI-cut insert of plasmid pAfPrtT used for disruption. (B) Southern blot verification of the prtTΔ-D1 to -5 isolates and control AF293 WT strain. For the Southern blot analysis, genomic DNA (10 μg per well) was digested with XbaI, blotted, and hybridized with a 32P-labeled AfPrtT 5′-flanking DNA probe, resulting in fragments of ∼3.8 kb for the prtT-disrupted strains and ∼2.5 kb for the WT control strain.
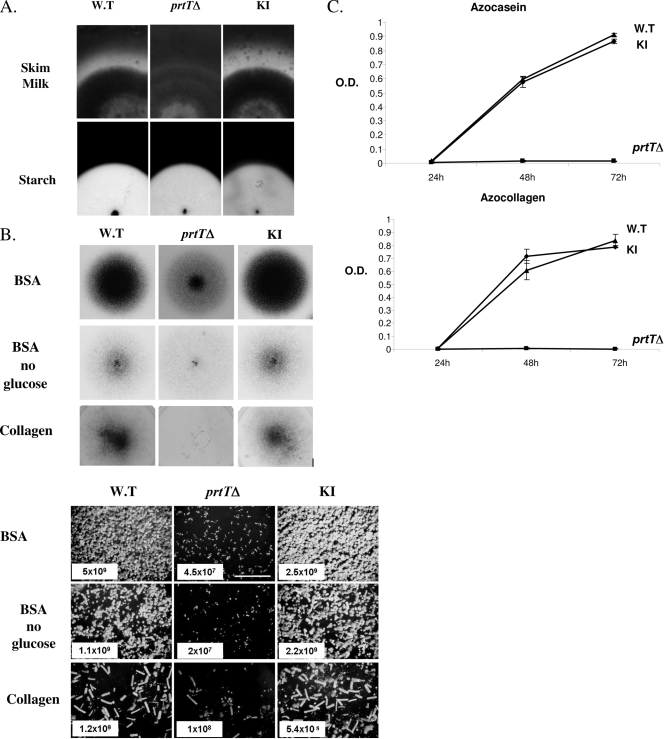
Unlike the WT and KI strains, the prtTΔ mutant did not produce a proteolytic halo when grown on SM plates (Fig. 3A, upper panels). Starch degradation was not affected in the prtTΔ strain, suggesting that it is not impaired in the general secretion of proteins such as amylases (Fig. 3A, lower panels). The prtTΔ mutant conidiated poorly on minimal medium agar plates with albumin (BSA) as the sole nitrogen or nitrogen/carbon source or with collagen as the sole nitrogen source, suggesting that under limiting conditions the inability to degrade protein affects mutant growth (Fig. 3B). The protease activities of CFs derived from the WT and KI strains showed a linear increase during the first 48 h of growth as measured by azocasein and azocollagen degradation. In contrast, the protease activity of the prtTΔ mutant remained very low (Fig. 3C). Whereas the protease activity of the WT and KI strains increased in the absence of phosphate and at acidic pH, the protease activity of the prtTΔ mutant remained almost undetectable under all conditions tested (Table 2).
FIG. 3.
Characterization of PrtT-dependent proteolytic activity and growth. (A) The prtTΔ mutant does not form a proteolytic halo when grown on SM agar plates (top panels). Conidia from the AF293 WT, prtTΔ, and reconstituted (KI) strains were point inoculated on SM plates and grown for 48 h at 37°C and then for another 48 h at room temperature. The formation of a proteolytic halo was highlighted by Coomassie blue staining. Lower panels, all three strains form normal clearance zones on starch-containing agar plates, suggesting that amylase secretion is not impaired in the prtTΔ mutant. (B) Reduced conidiation and growth of the prtTΔ mutant on agar plates containing BSA or collagen as sole nitrogen and carbon sources. The top panels illustrate the gross colonial morphology of the strains point inoculated and grown for 48 h at 37°C. The lower panels show conidiophore density (magnification, ×40) and conidial counts (total number of conidia/plate) of the strains in spread culture. (C) Time course of azocasein and azocollagen degradation by the WT, prtTΔ, and KI strains. Strains were grown for 24, 48, and 72 h at 37°C in shaking SM liquid culture. Supernatants were incubated with azocasein or azocollagen, and the proteolytic release of azo dye was determined by measuring the absorbance at 436 or 520 nm, respectively. O.D., optical density. The results are representative of three independent experiments performed in triplicate and are expressed as the means ± standard deviations for three replicates.
TABLE 2.
Protease activities of the WT, mutant (prtTΔ), and reconstituted (KI) strainsa
| Substrateb | Total absorbance units/g (dry wt) myceliumc
|
||
|---|---|---|---|
| WT | prtTstrain | KI strain | |
| SM | 9.6 ± 1.4 | 0.6 ± 0.1 | 10.7 ± 0.9 |
| SM without glucose | 8.7 ± 0.1 | 1.7 ± 0.02 | 10.0 ± 0.5 |
| SM without nitrogend | 10.8 ± 0.7 | 1.3 ± 0.3 | 12.9 ± 0.7 |
| SM without phosphatee | 22.4 ± 1.2 | 1.1 ± 1 | 19.9 ± 1.6 |
| SM without sulfurf | 4.5 ± 2.5 | 0.3 ± 0.2 | 7.4 ± 1.1 |
| SM without nitrogen and phosphate | 22 ± 1 | 0.6 ± 0.4 | 21.6 ± 0.4 |
| SM without phosphate and sulfur | 18 ± 1.2 | 0.7 ± 0.4 | 21.8 ± 1.1 |
| SM without nitrogen, phosphate, and sulfur | 20.7 ± 0.5 | 0.5 ± 0.1 | 21 ± 0.5 |
| SM + NH4 | 0.6 ± 0.03 | 0.6 ± 0.05 | 0.3 ± 0.02 |
| SM, pH 5.3 | 11.8 ± 0.1 | 0.8 ± 0.8 | 13.3 ± 0.9 |
| SM, pH 7 | 7.5 ± 0.1 | 0.3 ± 0.3 | 9.1 ± 1.1 |
| SM, pH 8 | 4.6 ± 0.4 | 0.2 ± 0.2 | 8.3 ± 0.8 |
The protease activity of CFs collected after 48 h of growth on various liquid media was measured by azocasein release (see Materials and Methods).
SM was at pH 6.2 unless otherwise indicated.
Results are expressed in arbitrary units and results are the averages and standard deviations for three to six cultures.
Without Casamino Acids as a nitrogen source.
Without Na2HPO4/NaH2PO4 as phosphate sources.
Without MgSO4 as a source of sulfur.
Deletion of prtT results in reduced transcription of six A. fumigatus protease genes.
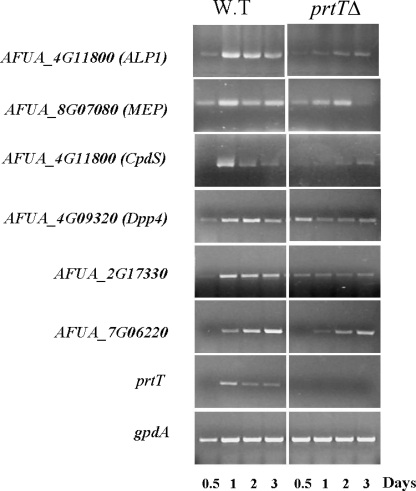
To identify proteases whose expression is altered by deletion of prtT, we assessed the expression levels of all 47 putative signal peptide-containing A. fumigatus proteases in the WT and prtTΔ mutant strains. Five other proteases lacking a signal peptide were tested as controls (see Table S1 in the supplemental material for a full gene and primer list). Total RNA was extracted after growth in liquid SM medium and analyzed by RT-PCR. Reduced expression of ALP, MEP, CpdS, Dpp4, and two uncharacterized proteases (AFUA_2G17330 and AFUA_7G06220) at most of the time points was apparent in the prtTΔ mutant strain (Fig. 4). All six proteases exhibiting reduced expression in the prtTΔ mutant contained a signal peptide.
FIG. 4.
The expression of six secreted A. fumigatus proteases is markedly reduced by deletion of prtT. Dormant conidia were incubated for 12, 24, 48, and 72 h in liquid SM medium at 37°C, after which total RNA was isolated and used in RT-PCR. Expression was determined by RT-PCR using primers shown in Table 1. Expression of prtT in the prtTΔ mutant strain was undetectable (prtT panel). RT-PCR with primers for gpdA, a housekeeping gene, was used as a control for the loading (gpdA panel). Three biological repeats of this experiment gave similar results.
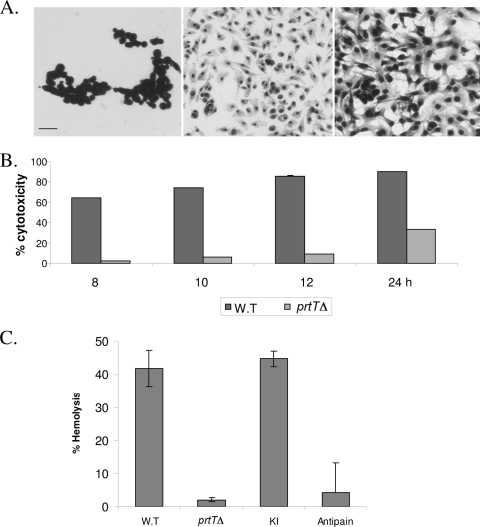
CF derived from the prtTΔ mutant show reduced killing and peeling of lung epithelial cells in culture and reduced hemolysis of SRBCs.
We hypothesized that the reduced secreted proteolytic activity of the prtTΔ mutant strain would impair its ability to damage cells in culture. To test this hypothesis, CF was prepared from the WT, prtTΔ, and KI strains as described in Materials and Methods. A549 lung epithelial cells were incubated in the presence of CF for 8 to 24 h. Cell adhesion was assessed microscopically following Hemacolor staining. Cell viability was measured in an XTT-based colorimetric assay. Our results indicated that when A549 cells are incubated with CF prepared from the prtTΔ strain, cytotoxicity is significantly reduced, by ∼ 90% after 12 h of incubation (Fig. 5A and B). Cytotoxicity increased after 24 h of incubation, suggesting that other fungus-secreted proteases and factors are involved in these processes. To identify additional differences between the WT and prtTΔ strain CFs, we tested their hemolytic activity on SRBCs. Interestingly, CF from the prtTΔ strain showed a ∼10-fold reduction in hemolysis compared to those from the WT and KI strains after 24 h of incubation. Hemolysis was inhibited in the presence of the serine protease inhibitor antipain, suggesting that it is dependent on proteolytic activity in the CF (Fig. 5C).
FIG. 5.
CF collected from the prtTΔ mutant shows decreased killing of A549 alveolar cells in culture and reduced hemolytic activity. (A) Confluent A549 cells were incubated in the presence of WT (left) and prtTΔ (center) CF for 12 h at 37°C with 5% CO2. Cells were subsequently stained with Hemacolor, washed, and visualized by microscopy. Control, untreated cells are shown in the right panel. Bar, 100 μm. (B) CF-induced loss of A549 cell viability (cytotoxicity), as measured by XTT assay. (C) CF-induced hemolysis of SRBCs, as measured by hemoglobin release. The results are representative of three independent experiments performed in triplicate and are expressed as the means ± standard deviations for three replicates.
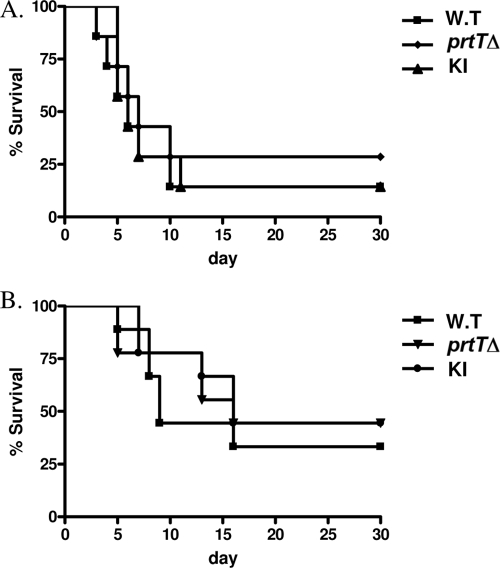
The prtTΔ mutant is not altered in virulence in infected mice.
We hypothesized that the large reduction in secreted protease activity in the prtTΔ strain would result in reduced virulence. To test this hypothesis, the virulence of the WT, KI, and prtTΔ strains was tested in two mouse models of invasive aspergillosis. In the first model, reproducing long-term immunosuppression without neutropenia (as found, for example following organ transplantation), mice were immunosuppressed with repeated doses of cortisone acetate. In the second model, reproducing profound neutropenia (as found, for example, following chemotherapy in leukemic patients), mice were immunosuppressed with a combination of cortisone acetate and cyclophosphamide. Freshly harvested conidia from the WT, prtTΔ, and KI strains were carefully counted and adjusted to the same density before intranasal administration. The number of live mice in each group was recorded daily throughout the 30-day study period. Figure 6 shows survival curves obtained during the course of the experiment for the cortisone acetate (Fig. 6A) and cyclophosphamide (Fig. 6B) models. No significant differences in virulence or fungal load (data not shown) were found for the prtTΔ mutant compared to the WT and KI strains (P > 0.3).
FIG. 6.
Deletion of prtT does not affect virulence in two murine models of pulmonary aspergillosis. Two regimens were used to immunocompromise mice: cortisone acetate alone (A) and cortisone acetate plus cyclophosphamide (neutropenia model) (B). Survival curves are shown for mice infected intranasally with an inoculum of 5 × 105 (model A) or 2.5 × 105 (model B) conidia per mouse of the AF293 WT strain (n = 10 mice) and the prtTΔ (n = 10) and reconstituted (KI) (n = 10) strains. Percent survival was monitored throughout the 30-day study period. This experiment was repeated twice with similar results.
DISCUSSION
In this report, we describe the disruption of the A. fumigatus prtT homolog and the phenotypic analysis of the mutant. prtT was originally identified in a proteolysis-deficient mutant strain (AB1.13) of A. niger generated in an effort to develop protease-deficient strains for more efficient industrial production of enzymes (20, 27). The prtT gene family encodes a C6 zinc finger-containing family of putative transcription factors involved in the regulation of secreted protease expression (27). We chose to study prtT in A. fumigatus because (i) regulation of protease expression in pathogenic filamentous fungi is poorly understood and (ii) we reasoned that deletion of a single putative transcription factor controlling the expression of multiple proteases may provide novel insights into the involvement of secreted proteases in the pathogenesis of A. fumigatus.
Deletion of prtT results in significantly reduced secreted protease activity in A. fumigatus.
The protease activity of the prtTΔ mutant on SM agar plates (halo formation) and in liquid SM using azocasein or azocollagen as a substrate was reduced to almost undetectable levels (Fig. 3). This indicated that like A. niger and A. oryzae prtT, A. fumigatus prtT is a central activator of secreted protease expression. Secreted protease activity also depends on the pH of the growth medium; the availability of carbon, nitrogen, and sulfur; and the presence of extracellular proteins (23a). For example, in A. niger expression of the aspartic protease genes pepA and pepB is repressed in the presence of ammonia, glucose, or alkaline pH (12). Similarly, regulation of protease secretion in A. fumigatus is complex and highly dependent on the composition of the medium (3, 7). We therefore assayed the protease activities of the prtTΔ mutant and WT strains under various conditions. Protease activity in the WT was repressed by ammonia or elevated pH, activated in the presence of protein as the sole nitrogen source, and markedly activated when phosphate was omitted from the medium (Table 2). In contrast, the protease activity of the prtTΔ mutant remained almost undetectable under all of these conditions, suggesting that A. fumigatus prtT is a central regulator of secreted protease activity under various environmental conditions.
Identification of A. fumigatus prtT-regulated proteases.
We measured the RNA levels of all 47 signal peptide-containing proteases in A. fumigatus prtTΔ mutant and WT strains growing on SM medium. Of these, seven genes were not expressed in either strain under these conditions (see Table S1 in the supplemental material). Of the 40 protease genes expressed in the WT strain, we identified six gene products exhibiting reduced expression in the prtTΔ mutant (Fig. 4). Two of them, ALP1 (serine endoprotease) (17, 24, 29, 34) and MEP (a metallo-endoprotease) (13, 23), have been extensively studied. Together they are responsible for most of the proteolytic activity detected in A. fumigatus CF at neutral pH (13). The third, AFUA_6G00310, is a homolog of CpdS, a secreted serine carboxypeptidase isolated from Aspergillus saitoi (5) which has not been characterized in A. fumigatus. The fourth, Dpp4 (dipeptidyl-peptidase IV), is an antigenic secreted protein which cleaves dipeptides from proteins and peptides (2). It is able to bind to and release a dipeptide from collagen, the main component of the lung basal membrane. Deletion of this gene in A. fumigatus has not been described. The remaining two proteases, AFUA_2G17330 (an extracellular serine carboxypeptidase) and AFUA_7G06220 (a serine protease) have never been characterized. Disruption of A. niger prtT results in the absence of expression of four proteases (PepA, -B, -D, and -F) (27). Only one of these, PepD, is a homolog of A. fumigatus Alp1. Disruption of A. oryzae prtT results in the absence of expression of two proteases, Alp and NP1, homologous to A. fumigatus ALP1 and MEP, respectively (27). These results suggest that PrtT-dependent regulation of protease expression differs somewhat among the aspergilli.
CF from the prtT deletion strain exhibits reduced killing of lung epithelial cells and lysis of erythrocytes.
Our previous studies have shown that secreted A. fumigatus proteases disrupt A549 cell actin fibers and focal adhesions, leading to cell detachment and death (15). Those results suggested that A. fumigatus breaches the alveolar epithelial cell barrier by secreting proteases that act together to disorganize the actin cytoskeleton and destroy cell attachment to the substrate by disrupting focal adhesions. In this study, prtTΔ mutant-derived CF exhibited a greatly reduced capacity to cause A549 cell detachment (Fig. 5A) and subsequent cell death (Fig. 5B). We also tested the hemolytic activity of prtTΔ mutant-derived CF because we reasoned that it is an important virulence trait for iron acquisition and during dissemination and invasion of blood vessels. A. fumigatus CF contains Asp-hemolysin, a secreted 14-kDa protein with hemolytic activity. However, it is active only after purification from the CF (39). Here we demonstrate for the first time in A. fumigatus a correlation between protease activity and hemolysis: prtTΔ mutant-derived CF showed only residual hemolytic activity, and WT-derived CF lost most of its hemolytic activity following the addition of protease inhibitors. The mechanism governing this phenomenon is unknown, but hemolytic activity of secreted bacterial proteases has been reported (8, 19, 32) and has been attributed to the proteolytic cleavage of transmembrane erythrocyte proteins, leading to a perturbation of membrane stability.
The A. fumigatus prtT deletion strain exhibits normal virulence in infected mice.
Using two low-dose models of murine invasive aspergillosis, we demonstrated that the prtT deletion strain is not attenuated in virulence. These results confirm previous reports showing that deletions of single (ALP1, MEP, and PEP) and multiple (ALP1/MEP) proteases do not affect A. fumigatus virulence in cortisone acetate- or cyclophosphamide-immunocompromised mice (13, 23, 30, 33, 35).
How can these in vivo results be explained in light of our findings in vitro of greatly reduced cell desquamation and killing by prtTΔ mutant-derived CF? One possibility is that conditions in the lungs activate substantial protease expression even in the absence of prtT, although this is unlikely taking into account our results showing that the prtTΔ mutant exhibited greatly reduced protease activity under various growth conditions. Another possibility is that WT A. fumigatus proteases are secreted inside the lungs at lower levels relative to their concentration in the CF. Alternatively, these proteases may be inhibited by endogenous protease inhibitors in the lungs. For example, lung α1 protease inhibitors protect lung elastin from degradation by secreted neutrophil proteases (25). Yet another possibility is that the ability of epithelial cells to withstand proteolytic damage in the lungs may be greater than that in culture.
Other secreted proteases, whose expression is affected by additional, as-yet-uncharacterized transcription factors, may also be generated during A. fumigatus lung infection. A recent transcriptional analysis of A. fumigatus genes expressed during early lung infection in mice identified increased an abundance of transcripts for the secreted proteases MEP, Dpp4, Dpp5, AFUA_6G10250, and AFUA_3G07850 (21). Of these, the prtTΔ mutant exhibits reduced transcription of MEP and Dpp4. It is conceivable that the other three upregulated proteases also contribute to virulence.
In summary we carried out an initial characterization of the A. fumigatus prtT gene, encoding a putative transcription factor controlling the expression of multiple secreted proteases. Future work will utilize transcriptomics and proteomics to provide a systems biology view of the function of A. fumigatus prtT to identify additional factors regulating the expression of secreted proteases and the upstream signaling and sensing pathways which modulate them.
Supplementary Material
Acknowledgments
This work was funded by Israel Academy of Sciences grant 280/06 and an Israel Ministry of Health grant to N.O.
Editor: A. Casadevall
Footnotes
Published ahead of print on 29 June 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Bainbridge, B. W. 1971. Macromolecular composition and nuclear division during spore germination in Aspergillus nidulans. J. Gen. Microbiol. 66319-325. [DOI] [PubMed] [Google Scholar]
- 2.Beauvais, A., M. Monod, J. Wyniger, J. P. Debeaupuis, E. Grouzmann, N. Brakch, J. Svab, A. G. Hovanessian, and J. P. Latge. 1997. Dipeptidyl-peptidase IV secreted by Aspergillus fumigatus, a fungus pathogenic to humans. Infect. Immun. 653042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouchara, J. P., G. Larcher, F. Joubaud, P. Penn, G. Tronchin, and D. Chabasse. 1993. Extracellular fibrinogenolytic enzyme of Aspergillus fumigatus: substrate-dependent variations in the proteinase synthesis and characterization of the enzyme. FEMS Immunol. Med. Microbiol. 781-91. [DOI] [PubMed] [Google Scholar]
- 4.Braaksma, M., and P. J. Punt. 2007. Aspergillus as a cell factory for protein production: controlling protease activity in fungal production, p.441-456. In S. A. Osmani and G. H. Goldman (ed.), The aspergilli. CRC Press, San Francisco, CA.
- 5.Chiba, Y., T. Midorikawa, and E. Ichishima. 1995. Cloning and expression of the carboxypeptidase gene from Aspergillus saitoi and determination of the catalytic residues by site-directed mutagenesis. Biochem. J. 308405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ejzykowicz, D. E., M. M. Cunha, S. Rozental, N. V. Solis, F. N. Gravelat, D. C. Sheppard, and S. G. Filler. 2009. The Aspergillus fumigatus transcription factor Ace2 governs pigment production, conidiation and virulence. Mol. Microbiol. 72155-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gifford, A. H., J. R. Klippenstein, and M. M. Moore. 2002. Serum stimulates growth of and proteinase secretion by Aspergillus fumigatus. Infect. Immun. 7019-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goguen, J. D., N. P. Hoe, and Y. V. Subrahmanyam. 1995. Proteases and bacterial virulence: a view from the trenches. Infect. Agents Dis. 447-54. [PubMed] [Google Scholar]
- 9.Hsu, J. T., H. C. Wang, G. W. Chen, and S. R. Shih. 2006. Antiviral drug discovery targeting to viral proteases. Curr. Pharm. Des. 121301-1314. [DOI] [PubMed] [Google Scholar]
- 10.Iadarola, P., G. Lungarella, P. A. Martorana, S. Viglio, M. Guglielminetti, E. Korzus, M. Gorrini, E. Cavarra, A. Rossi, J. Travis, and M. Luisetti. 1998. Lung injury and degradation of extracellular matrix components by Aspergillus fumigatus serine proteinase. Exp. Lung Res. 24233-251. [DOI] [PubMed] [Google Scholar]
- 11.Jadoun, J., Y. Shadkchan, and N. Osherov. 2004. Disruption of the Aspergillus fumigatus argB gene using a novel in vitro transposon-based mutagenesis approach. Curr. Genet. 45235-241. [DOI] [PubMed] [Google Scholar]
- 12.Jarai, G., and F. Buxton. 1994. Nitrogen, carbon, and pH regulation of extracellular acidic proteases of Aspergillus niger. Curr. Genet. 26238-244. [DOI] [PubMed] [Google Scholar]
- 13.Jaton-Ogay, K., S. Paris, M. Huerre, M. Quadroni, R. Falchetto, G. Togni, J. P. Latge, and M. Monod. 1994. Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol. Microbiol. 14917-928. [DOI] [PubMed] [Google Scholar]
- 14.Kauffman, H. F., J. F. Tomee, M. A. van de Riet, A. J. Timmerman, and P. Borger. 2000. Protease-dependent activation of epithelial cells by fungal allergens leads to morphologic changes and cytokine production. J. Allergy Clin. Immunol. 1051185-1193. [DOI] [PubMed] [Google Scholar]
- 15.Kogan, T. V., J. Jadoun, L. Mittelman, K. Hirschberg, and N. Osherov. 2004. Involvement of secreted Aspergillus fumigatus proteases in disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected A549 lung pneumocytes. J. Infect. Dis. 1891965-1973. [DOI] [PubMed] [Google Scholar]
- 16.Kolattukudy, P. E., J. D. Lee, L. M. Rogers, P. Zimmerman, S. Ceselski, B. Fox, B. Stein, and E. A. Copelan. 1993. Evidence for possible involvement of an elastolytic serine protease in aspergillosis. Infect. Immun. 612357-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunert, J. 2001. Further studies on the multiple forms of protease ALP of Aspergillus fumigatus. Mycoses 44307-310. [PubMed] [Google Scholar]
- 18.Latge, J. P. 2001. The pathobiology of Aspergillus fumigatus. Trends Microbiol. 9382-389. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C. Y., M. F. Cheng, M. S. Yu, and M. J. Pan. 2002. Purification and characterization of a putative virulence factor, serine protease, from Vibrio parahaemolyticus. FEMS Microbiol. Lett. 20931-37. [DOI] [PubMed] [Google Scholar]
- 20.Mattern, I. E., J. M. van Noort, P. van den Berg, D. B. Archer, I. N. Roberts, and C. A. van den Hondel. 1992. Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol. Gen. Genet. 234332-336. [DOI] [PubMed] [Google Scholar]
- 21.McDonagh, A., N. D. Fedorova, J. Crabtree, Y. Yu, S. Kim, D. Chen, O. Loss, T. Cairns, G. Goldman, D. Armstrong-James, K. Haynes, H. Haas, M. Schrettl, G. May, W. C. Nierman, and E. Bignell. 2008. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog 4e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monod, M., S. Capoccia, B. Lechenne, C. Zaugg, M. Holdom, and O. Jousson. 2002. Secreted proteases from pathogenic fungi. Int. J. Med. Microbiol. 292405-419. [DOI] [PubMed] [Google Scholar]
- 23.Monod, M., S. Paris, D. Sanglard, K. Jaton-Ogay, J. Bille, and J. P. Latge. 1993. Isolation and characterization of a secreted metalloprotease of Aspergillus fumigatus. Infect. Immun. 614099-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Monod, M., O. Jousson, and U. Reichard. 2009. Aspergillus fumigatus secreted proteases, p. 87-106. In J.-P. Latgé and W. J. Steinbach (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC.
- 24.Moutaouakil, M., M. Monod, M. C. Prevost, J. P. Bouchara, S. Paris, and J. P. Latge. 1993. Identification of the 33-kDa alkaline protease of Aspergillus fumigatus in vitro and in vivo. J. Med. Microbiol. 39393-399. [DOI] [PubMed] [Google Scholar]
- 25.Mulgrew, A. T., C. C. Taggart, and N. G. McElvaney. 2007. Alpha-1-antitrypsin deficiency: current concepts. Lung 185191-201. [DOI] [PubMed] [Google Scholar]
- 26.Osherov, N. 2007. The virulence of Aspergillus fumigatus, p. 185-293. In K. Kavanagh (ed.), New insights in medical mycology. Springer, Dordrecht, The Netherlands.
- 27.Punt, P. J., F. H. Schuren, J. Lehmbeck, T. Christensen, C. Hjort, and C. A. van den Hondel. 2008. Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet. Biol. 451591-1599. [DOI] [PubMed] [Google Scholar]
- 28.Punt, P. J., and C. A. van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216447-457. [DOI] [PubMed] [Google Scholar]
- 29.Reichard, U., S. Buttner, H. Eiffert, F. Staib, and R. Ruchel. 1990. Purification and characterisation of an extracellular serine proteinase from Aspergillus fumigatus and its detection in tissue. J. Med. Microbiol. 33243-251. [DOI] [PubMed] [Google Scholar]
- 30.Reichard, U., M. Monod, F. Odds, and R. Ruchel. 1997. Virulence of an aspergillopepsin-deficient mutant of Aspergillus fumigatus and evidence for another aspartic proteinase linked to the fungal cell wall. J. Med. Vet. Mycol. 35189-196. [DOI] [PubMed] [Google Scholar]
- 31.Romano, J., G. Nimrod, N. Ben-Tal, Y. Shadkchan, K. Baruch, H. Sharon, and N. Osherov. 2006. Disruption of the Aspergillus fumigatus ECM33 homologue results in rapid conidial germination, antifungal resistance and hypervirulence. Microbiology 1521919-1928. [DOI] [PubMed] [Google Scholar]
- 32.Saito, T., K. Ishihara, T. Kato, and K. Okuda. 1997. Cloning, expression, and sequencing of a protease gene from Bacteroides forsythus ATCC 43037 in Escherichia coli. Infect. Immun. 654888-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, J. M., C. M. Tang, S. Van Noorden, and D. W. Holden. 1994. Virulence of Aspergillus fumigatus double mutants lacking restriction and an alkaline protease in a low-dose model of invasive pulmonary aspergillosis. Infect. Immun. 625247-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, C. M., J. Cohen, and D. W. Holden. 1992. An Aspergillus fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol. Microbiol. 61663-1671. [DOI] [PubMed] [Google Scholar]
- 35.Tang, C. M., J. Cohen, T. Krausz, S. Van Noorden, and D. W. Holden. 1993. The alkaline protease of Aspergillus fumigatus is not a virulence determinant in two murine models of invasive pulmonary aspergillosis. Infect. Immun. 611650-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomee, J. F., A. T. Wierenga, P. S. Hiemstra, and H. K. Kauffman. 1997. Proteases from Aspergillus fumigatus induce release of proinflammatory cytokines and cell detachment in airway epithelial cell lines. J. Infect. Dis. 176300-303. [DOI] [PubMed] [Google Scholar]
- 37.van den Hombergh, J. P., P. J. van de Vondervoort, N. C. van der Heijden, and J. Visser. 1995. New protease mutants in Aspergillus niger result in strongly reduced in vitro degradation of target proteins; genetical and biochemical characterization of seven complementation groups. Curr. Genet. 28299-308. [DOI] [PubMed] [Google Scholar]
- 38.Wladyka, B., and K. Pustelny. 2008. Regulation of bacterial protease activity. Cell Mol. Biol. Lett. 13212-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokota, K., H. Shimada, A. Kamaguchi, and O. Sakaguchi. 1977. Studies on the toxin of Aspergillus fumigatus. VII. Purification and some properities of hemolytic toxin (asp-hemolysin) from CFs and mycelia. Microbiol. Immunol. 2111-22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.