Abstract
Biofilm production is a major attribute of Enterococcus faecalis clinical isolates. Although some factors, such as sortases, autolysin, and extracellular DNA (eDNA), have been associated with E. faecalis biofilm production, the mechanisms underlying the contributions of these factors to this process have not been completely elucidated yet. In this study we define important roles for the major E. faecalis autolysin (Atn), eDNA, and sortase A (SrtA) during the developmental stages of biofilm formation under static and hydrodynamic conditions. Deletion of srtA affects the attachment stage and results in a deficiency in biofilm production. Atn-deficient mutants are delayed in biofilm development due to defects in primary adherence and DNA release, which we show to be particularly important during the accumulative phase for maturation and architectural stability of biofilms. Confocal laser scanning and freeze-dry electron microscopy of biofilms grown under hydrodynamic conditions revealed that E. faecalis produces a DNase I-sensitive fibrous network, which is important for biofilm stability and is absent in atn-deficient mutant biofilms. This study establishes the stage-specific requirements for SrtA and Atn and demonstrates a role for Atn in the pathway leading to DNA release during biofilm development in E. faecalis.
Biofilms are bacterial communities encased within an extracellular matrix composed of carbohydrates, proteins, and nucleic acids (10). They are an ideal environment for exchange of genetic materials, such as genes encoding virulence factors and antibiotic resistance determinants, among bacteria within a community (61, 76), while allowing the flow of water and nutrients, as well as ions and various small molecules, to bacteria within the community (8) and providing a protective shield against antibiotics, antimicrobial substances, and phagocytosis (33, 80). Development of a biofilm is a complex multistage process. It is initiated by primary adhesion of the bacteria to a substratum, which is followed by the formation of microcolonies and production of an exopolysaccharide matrix, and it finally culminates with the formation of a three-dimensional (3D) multicellular mature structure (48). Bacterial biofilms are important medically because they have been associated with the pathogenesis of chronic and device-related persistent infections, such as cystic fibrosis, urinary tract infections, and endocarditis (11).
Enterococcus faecalis is a gram-positive bacterium that is a commensal of the gastrointestinal tract of healthy individuals. However, it is also an important opportunistic pathogen that is responsible for a wide variety of nosocomial infections, including endocarditis, urinary tract infections, and bacteremia (14, 17, 23, 44). E. faecalis accounts for approximately 65 to 80% of all enterococcal nosocomial infections (16). The ability of E. faecalis to adhere to and develop biofilms on medical devices, such as intravascular and urinary catheters (25, 26, 36, 37, 72), is thought to contribute to its pathogenesis. The increasing resistance of enterococci to most antibiotics, including vancomycin and linezolid (62, 63), especially in biofilms (41, 59, 78), makes treatment of enterococcal infections very difficult.
Several putative virulence factors and cellular processes have been implicated in the development of biofilms in E. faecalis (36); however, very little is known about their regulation and molecular contribution to this process. One of these factors, the quorum-sensing two-component transduction signaling system encoded by the fsr locus, an important virulence factor in the pathogenesis of E. faecalis, was shown to control biofilm formation via positive regulation of the extracellular zinc metalloprotease GelE (gelatinase) and the serine protease SprE (19, 38, 54-56). These proteases were recently shown to contribute to enterococcal biofilm formation via regulation of cellular autolysis and fratricidal DNA release (70, 71). In these studies, Thomas et al. also provided the first evidence for a critical role of extracellular DNA (eDNA) in E. faecalis biofilms. eDNA is an important component of the extracellular matrix of bacterial biofilms, providing structural stability to the biofilm and protection against antimicrobials (1, 2, 42, 43, 47, 51, 52, 58, 60, 65, 77). However, the function of this macromolecule throughout the establishment and growth of E. faecalis biofilms is not well characterized.
Bacterial murein hydrolases, also referred to as autolysins, have been implicated in biofilm production (58, 60). They are important contributors to cell wall growth and regulation, as well as several lytic processes (75). Two autolysins of Staphylococcus epidermidis, AtlE and Aae, are adhesins that contribute to bacterial attachment to polymeric surfaces and biofilm formation via release of eDNA (20, 21, 58). E. faecalis produces several autolysins, which were recently identified and characterized (12, 24, 35). The major E. faecalis autolysin, Atn (also known as AtlA), is an N-acetylglucosaminidase important for daughter cell separation during cellular division (12). Disruption of atn in E. faecalis resulted in increased chaining, a defect in primary attachment, and decreased biofilm production (3, 24, 31, 37, 57, 70). Recently, Thomas et al. (70) provided evidence that inactivation of this autolysin results in a decrease in DNA release similar to that of gelatinase-deficient mutants. Furthermore, they showed that GelE and SprE can differentially cleave Atn in vitro, and this processing may underlie the mechanism of cell death and DNA release in E. faecalis during biofilm formation.
Due to their role in cell wall regulation, autolysins may affect the localization of cell wall-anchored proteins, which can be important for adhesion during biofilm development. In most gram-positive bacteria, membrane-anchored transpeptidase enzymes known as sortases are responsible for covalently anchoring cell surface proteins bearing an LPXTG motif to the cell wall (34). Thus far, only class A and class C sortases have been implicated in biofilm formation by and the virulence of E. faecalis (28). Deletion of srtC, also known as bps, which encodes sortase C (SrtC), resulted in a significant reduction in biofilm production and attenuation of virulence in a mouse model of urinary tract infection, unlike deletion of srtA, which had minor effects under similar conditions (28). However, given the ubiquitous nature of sortases and the limited knowledge of the activity and substrates of the sole SrtA characterized in E. faecalis, it is plausible that this enzyme may play an important role in E. faecalis physiology and/or pathogenesis under different conditions. For instance, SrtA was shown to anchor the plasmid-encoded protein Asc10, which is involved in the pheromone-induced aggregation of E. faecalis (30).
In this study, we determined the contributions of E. faecalis SrtA, Atn, and eDNA to the major developmental stages leading to bacterial attachment and the establishment of mature biofilms under static and hydrodynamic conditions. E. faecalis biofilm development occurs in two key steps, which involve primary attachment followed by an accumulative stage. We demonstrate that both SrtA and Atn are required for efficient primary adherence to the substratum, while Atn and eDNA promote high levels of biofilm buildup and architectural stability during the accumulative stage. In the presence of hydrodynamic shear forces, the eDNA in E. faecalis biofilms is associated with a thick and long DNase-sensitive fibrous network that is associated with lysed cells present in the biofilm in some instances, as visualized by freeze-dry microscopy, transmission electron microscopy, and confocal laser scanning microscopy (CLSM). In contrast, atn-deficient mutants are unable to produce visible DNase-sensitive extracellular fibers under these conditions, although the biofilm remains partially sensitive to DNase treatment. These findings argue for a role for Atn in the temporal regulation of DNA release. Collectively, our data underscore the importance of SrtA, the major E. faecalis autolysin, and Atn-mediated DNA release at different stages during the establishment of E. faecalis biofilms. As a critical component of the extracellular matrix of E. faecalis biofilms, eDNA may serve as a novel target for the dissolution of these structures.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Unless otherwise specified, all experiments were initiated using an overnight culture grown in brain heart infusion broth (BHI) (Becton Dickinson, Franklin Lakes, NJ) from a single colony of E. faecalis strain OG1RF from a BHI agar plate (45, 46). Liquid cultures were grown statically at 37°C for 17 to 18 h.
Genetic manipulations.
Genes targeted for mutation were identified based on the complete genome of E. faecalis V583 (49); all references to genomic loci below are based on this annotation (GenBank accession number AE016830). Table 1 lists the strains and plasmids used in this study. In-frame deletion constructs were made using temperature-sensitive plasmid pJRS233 (50) for genes EF3056 (pJRS333-ΔEF3056) and EF0799 (pJRS233-Δatn) as previously described (29) with primers listed in Table 2. OG1RFΔsrtA was transformed with pAL1::srtA, a pABG5-derived pAL1 plasmid expressing srtA under the rofA promoter (18, 29, 50) for complementation of srtA deletion.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant antibiotic resistancea | Characteristics | Reference(s) |
|---|---|---|---|
| E. faecalis strains | |||
| OG1RF | Rif100, Fus25 | srtA positive, srtC positive, atn positive | 45, 46 |
| OG1RFΔatn (SJH-952) | Rif100, Fus25 | srtA positive, srtC positive, atn negative | This study |
| OG1RFΔsrtA (SJH-953) | Rif100, Fus25 | srtA negative, atn positive | This study |
| OG1RFΔsrtA srtA (SJH-954) | Rif100, Fus25, Kan500 | srtA positive, atn positive | This study |
| Plasmids | |||
| pABG5 | Kan50 and Cm20 (Escherichia coli); Kan500 and Cm20 (E. faecalis) | Shuttle vector containing rofA promoter for expression in gram-positive bacteria | 18 |
| pJRS233 | Erm750 (E. coli); Erm25 (E. faecalis) | Temperature-sensitive plasmid for generation of in-frame deletions | 50 |
| pJRS233-ΔEF3056 | Erm750 (E. coli); Erm25 (E. faecalis) | Temperature-sensitive plasmid for generation of in-frame deletion of EF3056 (srtA) | 29 |
| pJRS233-Δatn | Erm750 (E. coli); Erm25 (E. faecalis) | Temperature-sensitive plasmid for generation of in-frame deletion of EF0799 (atn) | This study |
| pAL1 | Kan50 (E. coli); Kan500 (E. faecalis) | Derivative of pABG5, Cmr cassette, contains rofA promoter | 29 |
| pAL1::SrtA | Kan50 (E. coli); Kan500 (E. faecalis) | Used for complementation of srtA deletion | 29 |
The superscripts indicate the concentrations of antibiotics (in μg/ml) to which the strains and plasmids are resistant. Rif, rifampin; Fus, fusidic acid; Kan, kanamycin; Cm, chloramphenicol; Erm, erythromycin.
TABLE 2.
Primers
| Primer | Sequence (5′-3′) | Use | Reference |
|---|---|---|---|
| EF0799f1 | GCTCTAGAAAGTTGCAGCTCCGGCAATGCCACAACCGACG | Amplification of upstream region of atn | This study |
| EF0799sewr | GTTACCAAAGATAGCTATTTTTTTCAATCTTAAATTAACCAACTTTCGTCCCCAACTTTCCTTTTTTAACTTATATGT | Amplification of upstream region of atn | This study |
| EF0799sewf | GTTACCAAAGATAGCTATTTTTTTCAATCTTAAATTAACCAACTTTCGTCCCCAACTTTCCTTTTTTAACTTATATGT | Amplification of downstream region of atn | This study |
| EF0799r1 | GCTCTAGAACCCAGTTAACCGCAACTTCTGCTTCATAAGC | Amplification of downstream region of atn | This study |
| EF0799f2 | ATTTGCCACCTGCAGCCATT | Screening of atn deletion | This study |
| EF0799r2 | CCGCTCCCTCTTTCTACACG | Screening of atn deletion | This study |
| EF3056e-f1 | GGAATTCCCCCTCTACTAGCCTCCTTACCATTTTAC | Screening of srtA deletion | 29 |
| EF3056e-r1 | GGAATTCCGTTGATAATGAGTCTGCCGCTAGTGTATG | Screening of srtA deletion | 29 |
Cultivation of biofilms.
Biofilms were grown as described by Gilmore et al. (15) and modified as follows. Stationary-phase E. faecalis from overnight cultures was diluted to an optical density at 600 nm (OD600) of 0.2 in BHI broth, which was followed immediately by 1:100 dilution (106 to 107 CFU/ml) in tryptic soy broth (Becton Dickinson, Sparks, MD) supplemented with 0.25% glucose (Sigma-Aldrich, Inc., St. Louis, MO) (TSBG). A 5-ml aliquot was then placed in each well of sterile six-well flat-bottom tissue culture plates (Techno Plastic Products, St. Louis, MO), which contained a UV-sterilized polyvinyl chloride coverslip (22 by 22 mm; Fisher Scientific, Pittsburgh, PA). The plates were incubated at 37°C statically or under dynamic conditions (100 rpm) on an orbital shaker (model 3520; Lab-Line Instruments, Inc., Melrose Park, IL) for the duration of the experiments. At 24-h intervals, the media were removed by aspiration, the coverslips were washed twice with 5 ml of autoclaved water for 3 min at 100 rpm on the orbital shaker at room temperature (22 to 25°C), and fresh media were added. At selected time points, biofilms were washed as described above and air dried. Biofilms were then stained with 0.5% crystal violet (Sigma-Aldrich, Inc., St. Louis, MO) for 10 min at room temperature. Excess dye was removed with autoclaved water, and the preparations were air dried at room temperature. Biofilms were then dissolved with 500 μl of 33% acetic acid (Fisher Scientific, Fair Lawn, NJ). Solubilized crystal violet was then diluted so that the concentration fell within the linear range of a microplate reader (Molecular Devices, Sunnyvale, CA), and the absorbance (OD595) was determined. The corresponding biomass values were derived by multiplying the absorbance values by the appropriate dilution factors. Experiments were performed independently at least three times with three coverslips per condition per experiment.
Primary adherence assay.
Biofilms were grown as described above. Cells were allowed to attach for 2 h, 4 h, and 6 h at 37°C under static or shaking conditions. After the desired incubation times, the coverslips were washed once with sterile water and processed as described above. Adherent cells were quantified by crystal violet staining as described above. Experiments were performed independently in triplicate with three coverslips per condition per experiment.
DNase I assays.
DNase I (Sigma Aldrich, Inc., St. Louis, MO) was added at a final concentration of 5 μg/ml to biofilm cultures, unless otherwise stated. DNase I was either added continuously every 24 h from the beginning of the experiment or added to preformed biofilms for the times indicated below before media were removed. Experiments were performed independently in triplicate with three coverslips per condition per experiment. As a control, heat-inactivated DNase I (iDNase I) (30 min at 100°C) was used in place of active DNase I. Only DNase I was able to totally degrade 100 μg/ml salmon sperm DNA and enterococcal genomic DNA at 37°C after 1 h (see Fig. S1A and B in the supplemental material) and 24 h (data not shown), although some residual activity could be detected with iDNase I after 24 h of incubation.
CLSM and image analysis.
All confocal microscopy was performed with an LSM 510 Meta laser scanning confocal microscope (Carl Zeiss, Thornwood, NY), using a ×63 oil immersion objective. Images were acquired using the LSM Image Examiner software (Carl Zeiss, Thornwood, NY). 3D reconstruction of biofilm images and biofilm quantification were performed using Volocity software (Improvision, Inc., Waltham, MA). For microscopy, washed plastic coverslips were stained for 15 min in the dark at room temperature with 1 ml of SYTO9 mixed with propidium iodide (PI) (Molecular Probes, Eugene, OR) (3 μM each). When indicated, 1 μg/ml of wheat germ agglutinin (WGA) was added to stain carbohydrates present on the bacterial surfaces (64). Excess dye was removed by aspiration, and biofilms were washed twice with 1× phosphate-buffered saline or water. Stained coverslips were mounted on glass slides using Prolong Gold Antifade (Molecular Probes), which was followed by image acquisition. Microscopy was performed with two coverslips per condition per experiment in at least two independent experiments.
Freeze-dry electron microscopy.
Dynamic biofilms grown for 48 h in TSBG on glass coverslips were fixed with 2% glutaraldehyde in 100 mM NaCl, 30 mM HEPES, 2 mM CaCl2, 75 mM sucrose (pH 7.2) for 1 h. Samples were rinsed in deionized water and quick-frozen by forceful impact against a copper block cooled to 4°K with liquid helium. Frozen cultures were stored in liquid nitrogen until they were mounted in a Balzers 301 vacuum evaporator, in which they were freeze-dried for 20 min at −80°C and then rotary replicated with 2 nm of platinum deposited from an electron beam gun mounted at an angle of 15° above the horizontal. The Pt replica was then stabilized with a carbon film deposited from a 70° angle. Replicas were floated off the glass onto concentrated hydrofluoric acid and then transferred through several rinses of deionized water and picked up on Formvar-coated grids. Replicas were examined with a JEOL 100CX microscope and photographed with an AMT digital camera.
Transmission electron microscopy.
For immunolocalization of DNA at the ultrastructural level, 48-h dynamic wild-type and OG1RFΔatn biofilms, with or without 1 h of treatment with 5 μg/ml of DNase I, and adherent cells from a primary attachment assay performed as described above were fixed in 4% paraformaldehyde-0.05% glutaraldehyde (Polysciences Inc., Warrington, PA) in 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-0.5 mM MgCl2 (pH 7.2) for 1 h at 4°C. Samples were then embedded in 10% gelatin and infiltrated overnight with 2.3 M sucrose-20% polyvinylpyrrolidone in PIPES-MgCl2 at 4°C. Samples were trimmed, frozen in liquid nitrogen, and sectioned with a Leica Ultracut UCT cryo-ultramicrotome (Leica Microsystems Inc., Bannockburn, IL). Sections (50 nm) were blocked with 5% fetal bovine serum-5% normal goat serum for 30 min and subsequently incubated with mouse anti-double-stranded DNA (anti-dsDNA) monoclonal antibody (Abcam, Cambridge, MA) for 1 h at room temperature. Sections were then washed in blocking buffer (5% fetal calf serum and 5% goat serum in 100 mM PIPES) and probed with 18-nm colloidal gold-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) for 1 h at room temperature. Sections were washed in PIPES buffer (100 mM PIPES, 0.5 mM MgCl2; pH 7.2), which was followed by a water rinse, and then stained with 0.3% uranyl acetate-2% polyvinyl alcohol. Samples were viewed with a JEOL 1200EX transmission electron microscope (JEOL USA Inc., Peabody, MA). All labeling experiments were conducted in parallel with controls from which the primary antibody was omitted. These controls were consistently negative with the concentration of colloidal gold-conjugated secondary antibodies used in these studies. Quantification was based on gold particle labeling in 10 to 13 independent fields at a magnification of ×30,000. Experiments were performed in duplicate.
Cellular autolysis assay.
The autolysis assay was performed as previously described using sodium phosphate buffer (57). Briefly, overnight cultures were inoculated into fresh TSBG at a final OD600 of 0.002 and grown at 37°C on an orbital shaker at 100 rpm to an OD600 of 0.6 or for 24 h. The cultures were then placed on ice for 10 min, and a 1.5-ml aliquot of each culture was removed and centrifuged at 14,000 × g for 5 min at 4°C. The supernatants were discarded, and the pellets were resuspended in 1.5 ml of ice-cold water and washed three times with centrifugation at 14,000 × g for 5 min at 4°C. After the third wash, the pellets were resuspended in 10 mM sodium phosphate buffer (pH 6.8) supplemented with 0.5 μg/ml of trypsin (Sigma Aldrich, Inc., St. Louis, MO). Portions (200 μl) of the suspensions were dispensed into 96-well flat-bottom microtiter plates in quintuplicate, and the remaining wells were filled with deionized water. Samples were incubated at 37°C, and OD600 were determined every 30 min for up to 9 h with a spectrophotometer. Autolysis was expressed as percentage of the initial OD600.
Statistical methods.
Data are expressed below as means ± standard errors of the means of biofilm values derived from the absorbance of solubilized crystal violet at 595. Comparisons of biofilm biomasses among groups were carried out using the Mann-Whitney test.
RESULTS
E. faecalis biofilm development occurs in two major steps.
In order to delineate the major stages of E. faecalis biofilm development, we monitored OG1RF biofilm formation under static conditions using crystal violet staining and CLSM for 72 h. Crystal violet staining showed a biphasic growth curve (Fig. 1A). The accumulation was slow between 0 and 24 h; this was followed by significantly faster growth during the next 48 h, and there was an approximately sixfold increase in biomass from 24 h to 72 h (Fig. 1A). CLSM images of 24-h and 72-h OG1RF biofilms stained with the DNA dyes SYTO9 and PI confirmed that there was slow accumulation of the biofilm during the first 24 h, which was followed by a rapid buildup and increases in density and thickness (see Fig. S2A in the supplemental material). Both SYTO9 (green) and PI (red) bind DNA; however, SYTO9 is membrane permeant, while PI stains only eDNA and DNA of membrane-compromised cells. The CLSM images show the presence of yellow patches throughout the biofilms, which were representative of SYTO9 and PI colocalization due to the presence of dead cells or eDNA in these structures (Fig. 1B).
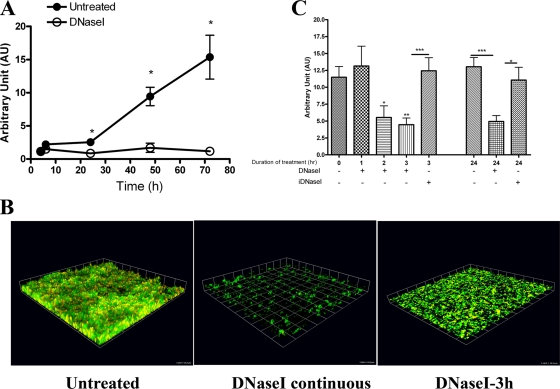
FIG. 1.
DNA is critical for OG1RF biofilms under static conditions. (A) Crystal violet staining of OG1RF biofilms grown statically in TSBG at 37°C in the presence or absence of 5 μg/ml DNase I and monitored for 72 h. The error bars indicate standard errors of the means for three to seven different experiments. *, P < 0.05, as determined by the Mann-Whitney test. (B) Seventy-two-hour static OG1RF biofilms without DNase I treatment (left panel) or treated with 5 μg/ml of DNase continuously (middle panel) or for 3 h postformation (right panel). Biofilms were then stained with SYTO9 (green) and PI (red) and visualized with CLSM. 3D reconstructions of z stacks were generated with the Volocity software. One unit on each side of each grid equals 14.3 μm. (C) Relative biomasses of 72-h OG1RF biofilms treated for 1 h, 2 h, 3 h, or 24 h with 5 μg/ml of DNase I or iDNase I at 37°C. The error bars indicate standard errors of the means for three different experiments. *, P < 0.05, as determined by the Mann-Whitney test; **, P < 0.01, as determined by the Mann-Whitney test; ***, P < 0.0005, as determined by the Mann-Whitney test.
eDNA is required for biofilm development.
To determine whether eDNA contributes to biofilm development, OG1RF biofilms were grown statically in the presence of various DNase I concentrations ranging from 0.5 μg/ml to 50 μg/ml. DNase I severely impaired the ability of OG1RF to form mature biofilms at concentrations as low as 0.5 μg/ml (data not shown). At 72 h, the biomass of OG1RF grown in the presence of 5 μg/ml DNase I was 92.3% less (P = 0.0121, Mann-Whitney test) than the biomass of an untreated control (Fig. 1A). The detrimental effect of DNase I on biofilm development was not due to bactericidal or bacteriostatic activities of the enzyme, as DNase I did not inhibit OG1RF planktonic growth (data not shown). CLSM analysis revealed that in the presence of DNase I, bacteria were able to attach to the substratum; however, they failed to form the large compact 3D structures observed with untreated cells after 72 h of incubation (Fig. 1B). In addition, approximately 10 to 15% of the cells that were present in the DNase I-treated samples were stained with PI, compared to the 30 to 50% of the cells in the untreated samples that were stained with PI (see Fig. S2B and C in the supplemental material). The lower level of PI staining in biofilms treated with DNase I suggests that there was a reduction in the number of dead cells and/or eDNA. Together, these findings indicated that the presence of eDNA in OG1RF biofilms contributes to the establishment of mature enterococcal biofilm structures.
eDNA is required for maintenance of biofilm architectural integrity.
To further characterize the function of eDNA in enterococcal biofilms, we investigated the effects of DNase I on mature biofilms. Seventy-two-hour static OG1RF biofilms were treated with 5 μg/ml of DNase I for 1 h, 2 h, 3 h, and 24 h at 37°C, followed by crystal violet staining or CLSM analysis. The biofilm biomass was significantly reduced (∼30% less than an untreated control) after 2 h of treatment (P = 0.0428) (Fig. 1C). By 3 h, the biomass was reduced by 60% in the presence of 5 μg/ml of DNase I (P = 0.008), while iDNase I did not have a significant effect (P = 0.7602) (Fig. 1C). After 24 h, DNase I treatment reduced the biomass to 40% of the original level (P = 0.0003); iDNase I had no effect (P = 0.5079) (Fig. 1C). Disintegration of the biofilm structures was visualized at 3 h following treatment with 5 μg/ml of DNase I (Fig. 1B), in contrast to 1 h after treatment with higher DNase I concentrations (see Fig. S2D in the supplemental material), suggesting that the effects of DNase I on biofilms were dose and time dependent. Overall, the data described above indicated that eDNA is an important structural component of E. faecalis biofilms and is necessary for stability of mature structures.
E. faecalis forms a DNase I-sensitive fibrous network under hydrodynamic conditions.
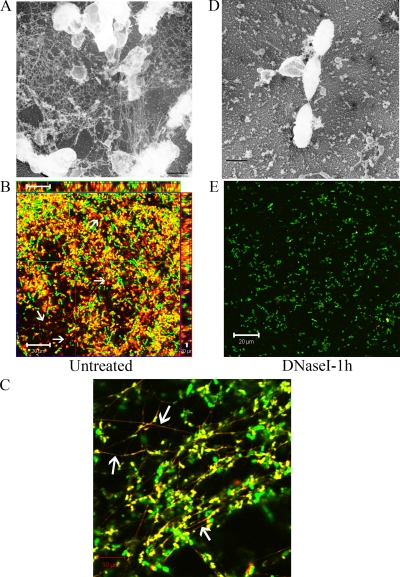
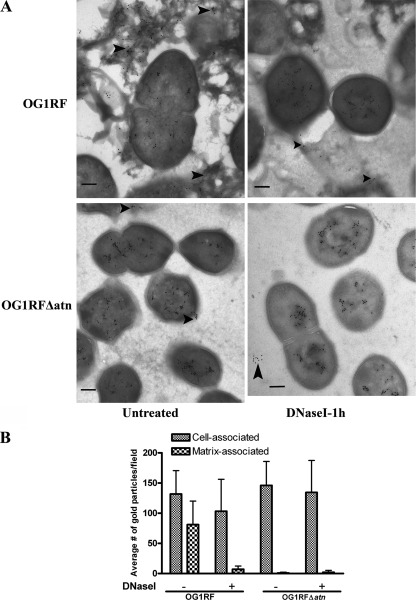
E. faecalis withstands a variety of environmental conditions involving hydrodynamics in order to efficiently colonize medical devices, which contributes to its role as an important nosocomial pathogen. To investigate the effects of shear forces on the E. faecalis biofilm development process, we cultivated OG1RF biofilms in the presence of a fluid flow generated with an orbital shaker for the duration of the experiment. Biofilms grown for 48 h were stained with SYTO9, PI, and WGA (when indicated) for CLSM analysis. Continuous shear forces during biofilm growth resulted in clustering of cells in long fibrous structures that were observed in freeze-dry electron micrographs and CLSM images (Fig. 2A to C). These extracellular fibers were stained only with the DNA dyes and not with WGA (see Fig. S3 in the supplemental material) and disappeared within 1 h after DNase I treatment (Fig. 2D and E; see Fig. S3 in the supplemental material), which correlated with the degradation of the biofilm structure. Additionally, immunogold transmission electron microscopy analyses of OG1RF hydrodynamic biofilms labeled with mouse anti-dsDNA monoclonal antibody revealed foci inside and at the surface of cells, as well as in the extracellular matrix surrounding the cells (Fig. 3A, top left panel), while examination of biofilms after 1 h of treatment with DNase I showed a reduced presence of eDNA (Fig. 3A, top right panel). Approximately 38% of the gold particles labeled the extracellular matrix, compared to ∼6% following DNase I treatment (Fig. 3B). Similar to the results for static conditions, OG1RF grown in the presence of 5 μg/ml of DNase I for 48 h failed to produce biofilms under hydrodynamic conditions, and only 2% of the gold particles were found in the extracellular milieu under such conditions (data not shown). Notably, treatment with proteinase K did not prevent OG1RF biofilm formation and did not affect the formation of the DNase I-sensitive extracellular fibers (data not shown). These results indicate that in the presence of hydrodynamic forces E. faecalis produces DNA-dependent biofilms with an architecture distinct from that of static biofilms and that eDNA is a major component of the filamentous extracellular network in biofilms formed under hydrodynamic conditions that appears to provide cell-to-cell adhesion and stability in the biofilms.
FIG. 2.
Hydrodynamic conditions induce the production of a DNase I-sensitive extracellular fibrous network. The images are freeze-dry electron micrographs (magnification, ×20,000) (A and D) and CLSM images (B, C, and E) of 48-h biofilms of OG1RF stained with SYTO9 (green) and PI (red) that were not treated (A to C) or were treated for 1 h with 5 μg/ml of DNase I (D and E). The arrows indicate SYTO9-PI-stained fibers visible in untreated OG1RF biofilms but not in DNase I-treated biofilms by CLSM. (B and E) Scale bars = 20 μm. (C) Scale bar = 10 μm. (A and D) Scale bars = 500 nm.
FIG. 3.
Atn contributes to DNA release during biofilm formation. (A) Negative staining and immunoelectron microscopy images of 48-h OG1RF and OG1RFΔatn biofilms with or without 1 h of DNase I treatment and labeled with mouse anti-dsDNA monoclonal antibody and secondary immunogold. The images are representative images of 10 random fields. Scale bars = 200 nm. (B) Cell-associated or extracellular gold particles (indicated by arrowheads in panel A) were quantified for 10 independent fields of 48-h shaken biofilms at a magnification of ×30,000.
Deletion of atn results in delayed biofilm formation.
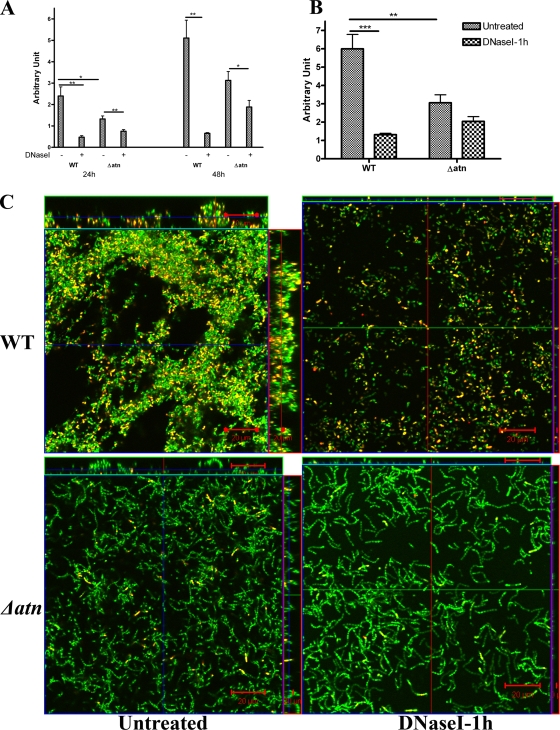
Freeze-dry electron microscope analysis revealed that lysed cells are constituents of enterococcal biofilms (see Fig. S4A in the supplemental material). We hypothesized that Atn, the major autolysin of E. faecalis, facilitates DNA release and the formation of the DNase I-sensitive extracellular fibrous network seen in the experiments described above. Thus, we generated an in-frame deletion mutation in the autolysin gene (atn) in OG1RF and studied the ability of the OG1RFΔatn (SJH-952) strain to form biofilms in the presence or absence of DNase I. Deletion of atn did not affect the planktonic growth of the mutant (data not shown). As expected, based on the role of autolysins in cell division, OG1RFΔatn formed longer chains than the parent strain (see Fig. S4B in the supplemental material) and had a reduced rate of cellular lysis, as determined by a cellular autolysis assay of cells from hydrodynamic cultures (see Fig. S4C in the supplemental material) or cells originating from static cultures (data not shown). Compared to wild-type strain OG1RF, the ability of OG1RFΔatn to produce biofilms under hydrodynamic conditions was delayed, and OG1RFΔatn produced approximately 40% less biomass than the parental strain by 48 h (Fig. 4A). Continuous DNase I treatment resulted in a ∼40% reduction in the OG1RFΔatn biomass accumulated by 48 h (P = 0.038), compared to a 87% reduction for the wild type (P = 0.0004) (Fig. 4A). Similar levels of inhibition were observed following 1 h of DNase I treatment of 48-h OG1RFΔatn biofilms. For OG1RFΔatn biofilms there was a 33% decrease in biomass (P > 0.05) following DNase I treatment, compared to a 78% reduction for the wild-type structures (P < 0.0001) (Fig. 4B and C). Similar to the results for wild-type cells, DNase I treatment was not bactericidal to OG1RFΔatn cells. Analogous results were obtained when biofilms were grown under static conditions (data not shown).
FIG. 4.
Atn is required for production of DNase-sensitive biofilms under hydrodynamic conditions. (A and B) Crystal violet staining of OG1RF (WT) and OG1RFΔatn (Δatn) biofilms treated continuously for 24 h and 48 h (A) or for 1 h after 48 h of growth (B) with 5 μg/ml of DNase I under hydrodynamic conditions in TSBG. The error bars indicate standard errors of the means for three or four independent experiments. *, P < 0.05, as determined by the Mann-Whitney test; **, P < 0.005, as determined by the Mann-Whitney test; ***, P < 0.0005, as determined by the Mann-Whitney test. (C) CLSM-acquired z-stack images of 48-h biofilms of OG1RF and OG1RFΔatn that were not treated or were treated for 1 h with 5 μg/ml of DNase I and stained with SYTO9 and PI. Long chains of bacteria were evident in OG1RFΔatn mutant biofilms. DNase I treatment dramatically reduced the biomass of the OG1RF biofilm, but not the biomass of the OG1RFΔatn biofilm. Furthermore, few SYTO9-PI-positive bacterial cells (yellow) were visible in the OG1RFΔatn mutant biofilm compared to the wild-type biofilm. Scale bars = 20 μm.
CLSM analysis of 48-h dynamic OG1RFΔatn biofilms revealed the absence of DNase-sensitive extracellular fibers (see Fig. S4B in the supplemental material) and very few PI-stained cells relative to the total number of stained cells compared to the results for wild-type OG1RF biofilms (Fig. 4C; see Fig. S4D and E in the supplemental material). Additionally, reduced amounts of eDNA were detected in the extracellular matrix of the mutant biofilms using cryoimmunoelectron microscopy with murine anti-dsDNA monoclonal antibody labeling (Fig. 3). Complementation studies could not be performed with OG1RFΔatn expressing atn from a vector, perhaps due to the potential cytotoxic effects of Atn in the recipients, as previously suggested (3). Nonetheless, the findings described above provide evidence supporting the hypothesis that Atn and cellular lysis play a role in DNA release during enterococcal biofilm formation. However, the ability of OG1RFΔatn to form delayed, but partially DNase-sensitive, biofilms suggests that an Atn-independent mechanism of DNA release exists as well.
Atn promotes DNA-independent attachment of E. faecalis to plastic surfaces.
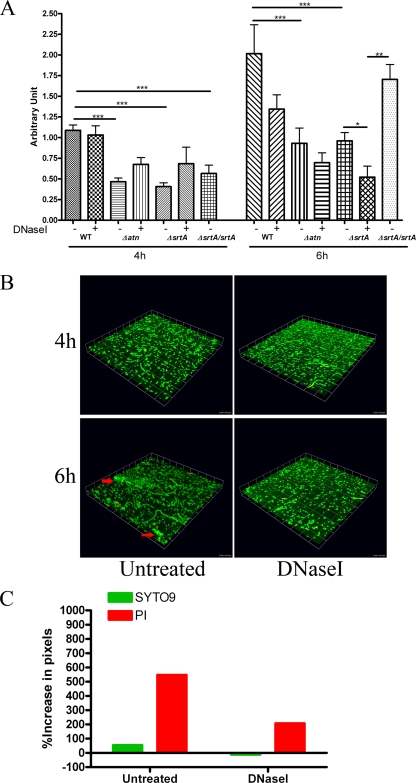
To assess the role of eDNA and Atn in initial attachment of E. faecalis to plastic coverslips, we performed an assay of primary attachment in the presence or absence of DNase I under static and shaking conditions using OG1RF and OG1RFΔatn. Very few cells were adhered to the substratum after 2 h in either condition (data not shown). By 4 h and 6 h, significantly (P < 0.0001) fewer OG1RFΔatn cells (40%) than wild-type strain cells were adhered to the surface, suggesting that Atn is required for this process. In contrast to atn deletion, DNase I treatment did not affect the primary attachment at 4 h for both wild-type strain OG1RF and OG1RFΔatn. Similar levels of wild-type cells were attached to the surfaces regardless of the presence of DNase I at 4 h (P = 0.3527) (Fig. 5A), indicating that eDNA is not critical for the initial attachment stage during biofilm development. Six hours of continuous DNase I treatment prevented microcolony formation (Fig. 5B), and this corresponded with a 33.3% reduction in the number of adherent cells. Albeit not statistically significant (P = 0.4359), this reduction suggests that DNase I affects microcolony formation, which was readily visible at 6 h only in untreated samples (Fig. 5B). Furthermore, we observed a >500% increase in PI staining from 4 h to 6 h, compared to a ∼200% increase in the presence of DNase I (Fig. 5C), arguing that cell death and eDNA contribute to microcolony formation.
FIG. 5.
Primary attachment requires srtA and atn, but not eDNA. (A) OG1RF (WT), OG1RFΔatn (Δatn), OG1RFΔsrtA (ΔsrtA), and OG1RFΔsrtA srtA (ΔsrtA/srtA) were incubated statically on plastic coverslips for 4 to 6 h at 37°C in TSBG with or without 5 μg/ml of DNase I. Biofilms were stained with crystal violet and then solubilized. The degree of primary attachment was determined based on the OD595 of solubilized crystal violet. The error bars indicate the standard errors of the means for at least three independent experiments. *, P < 0.05, as determined by the Mann-Whitney test; **, P < 0.01, as determined by the Mann-Whitney test; ***, P < 0.0001, as determined by the Mann-Whitney test. (B) Representative CLSM images of 4-h and 6-h adherent cells in the presence or absence of DNase I following staining with SYTO9 and PI. The arrows indicate microcolonies in 6-h samples. One unit on each side of each grid equals 14.3 μm. (C) Quantification of the change in fluorescence of SYTO9 and PI relative to the total fluorescence from adhering cells from 4 h to 6 h postinoculation. The graph was generated from data obtained from 12 randomly chosen fields from two independent experiments using the Volocity software.
Deletion of srtA results in a defect in primary adhesion.
Given that autolysins are involved in cell wall growth and regulation, we reasoned that deletion of atn may cause mislocalization of adhesion factors critical for primary attachment and biofilm development. Sortases are ubiquitous enzymes with a critical role in the adhesion and virulence of gram-positive bacteria. Given the role of these enzymes in protein sorting, we hypothesized that a SrtA-dependent substrate on the cell surface may contribute to the initial stages of biofilm development. To investigate the role of SrtA in biofilm formation under static and hydrodynamic conditions, we generated an in-frame srtA deletion in OG1RF, resulting in OG1RFΔsrtA. Deletion of srtA has no effect on cell viability and cell growth (data not shown). Similar to OG1RFΔatn, OG1RFΔsrtA was also significantly deficient in primary attachment under static conditions (P = 0.0001) (Fig. 5A) and shaking conditions (data not shown). Forty percent fewer OG1RFΔsrtA cells than parental strain cells adhered to the coverslips after 4 h of incubation. The complemented strain OG1RFΔsrtA srtA exhibited wild-type levels of adherence by 6 h, showing significantly more adherence than OG1RFΔsrtA (P = 0.0022) (Fig. 5A). It is not clear why the complementation effect was delayed and thus not observed at 4 h; however, this may have been the result of plasmid loss in the complemented strain population, which allowed only a small subset to efficiently adhere to the substratum, leading to the observed delay. Overall, the findings described above indicate that SrtA and likely a SrtA-dependent substrate(s) play a role in promoting efficient initiation of biofilm development in E. faecalis.
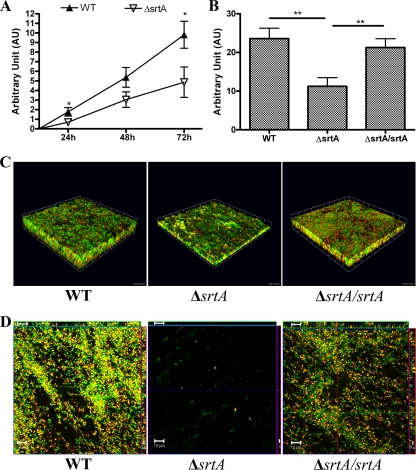
Deletion of srtA results in defects in biofilm production.
OG1RFΔsrtA was significantly defective in biofilm formation under static conditions; it produced ∼60% less biomass than the wild type at 24 h (P = 0.0142) and exhibited less accumulated biomass than the wild type at later time points (∼40% less biomass) (P = 0.0244) (Fig. 6A). The complemented strain OG1RFΔsrtA srtA produced wild-type biofilm levels under static conditions (Fig. 6B and C). DNase I treatment prevented OG1RFΔsrtA and the complemented strain from forming a 3D biofilm structure like the wild type (data not shown). Additionally, we investigated the role of srtA in biofilm formation under fluid flow conditions. The impairment of biofilm formation by the srtA-deficient mutant under hydrodynamic conditions was even more pronounced than the impairment observed under static conditions (Fig. 6D). OG1RFΔsrtA srtA exhibited a wild-type phenotype (Fig. 6D), forming mature biofilms containing SYTO9-PI-stained extracellular fibers. These findings suggest that DNA release and extracellular fibrous network formation occur independent of SrtA.
FIG. 6.
SrtA promotes high levels of DNA-dependent biofilm development. (A) Crystal violet-based quantification of OG1RF (WT) and OG1RFΔsrtA (ΔsrtA) biofilm development. (B) Crystal violet-based quantification of 72-h OG1RF, OG1RFΔsrtA, and OG1RFΔsrtA srtA (ΔsrtA/srtA) biofilms. The error bars indicate the standard errors of the means for three different experiments *, P < 0.05, as determined by the Mann-Whitney test; **, P < 0.01, as determined by the Mann-Whitney test. (C) 3D reconstruction of CLSM-acquired images of static 72-h-old biofilms of OG1RF, OG1RFΔsrtA, and OG1RFΔsrtA srtA grown statically and stained with SYTO9 (green) and PI (red). The CLSM images are representative images from the same experiment. One unit on each side of the grid equals 14.3 μm. (D) CLSM images of 48-h biofilms under hydrodynamic conditions for OG1RF, OG1RFΔsrtA, and OG1RFΔsrtA srtA stained with SYTO9 and PI. Scale bars = 10 μm.
DISCUSSION
Bacterial biofilms have been associated with the establishment of chronic and persistent infections due to their ability to withstand harsh environmental stress conditions, including low nutrient availability, temperature fluctuations, antibiotic and antimicrobial treatments, and host responses (7, 10, 11, 67). Several genetic factors and cellular processes, such as sortases and autolysis, as well as eDNA, have been implicated in E. faecalis biofilm formation (6, 13, 19, 22, 28, 31, 36, 40, 68-72); however, the temporal and spatial contributions of these factors and processes during the development process remain to be characterized. In this study, we delineated two major steps during E. faecalis biofilm formation: an initial attachment stage involving binding to abiotic surfaces, followed by an accumulative phase during which intercellular interactions generate mature multicellular 3D biofilm structures. The data for biofilm development after atn and srtA deletion under static and hydrodynamic conditions support the hypothesis that there is a temporal requirement for Atn, eDNA, and SrtA during biofilm development. Our data demonstrated that Atn and SrtA were important factors in the DNA-independent initial attachment step. Additionally, Atn played a role during the accumulative stage and in DNA release, which we demonstrated to be crucial for the growth, maturation, and structural stability of E. faecalis biofilms.
Previous reports have implicated Atn in primary attachment and biofilm production (31, 37). Unlike autolysins described for other gram-positive bacteria (21, 58), Atn contributed to initial adhesion in a DNA-independent fashion. Since Atn is important for cellular growth as well as cell lysis, the adherence defect observed after atn deletion could be the result of increased chaining of the mutants (9, 57), leading to a reduction in binding sites or an increase in cell surface charges, causing a lower cell deposition rate. Repulsive interactions between abiotic surfaces and bacteria, as well as between adhered cells and incoming cells, due to surface charges were shown to influence the initial adhesion of bacteria to substrata (73). Alternatively, defects in autolytic processes may result in modification of the cell wall structure and mislocalization of surface proteins required for attachment during biofilm formation.
In streptococcal species, disruption of srtA and SrtA-dependent proteins also resulted in decreased biofilm production and bacterial colonization of abiotic surfaces (32, 79). In contrast to previous reports for E. faecalis (28), we show that deletion of srtA severely hampered adherence to plastic substrata and subsequent biofilm growth, especially under hydrodynamic conditions. Our data corroborate findings of Kristich et al. (31), who showed that transposon-mediated disruption of srtA resulted in an approximately 30% decrease in biofilm biomass compared to the biofilm biomass of the parental strain using the microtiter biofilm assay. Thus, we argue that SrtA plays a critical role in biofilm development in E. faecalis, especially under hydrodynamic conditions.
Our data reveal that in addition to its role in primary attachment, Atn is involved in DNA release during the accumulative phase of biofilm development. We corroborated previous findings highlighting the importance of eDNA for the development of mature E. faecalis biofilms (71). Recently, Thomas et al. demonstrated that gelatinase (GelE) and serine protease (SprE) contribute to enterococcal biofilm formation by regulating cellular autolysis (71). GelE was shown to be important for DNA release during planktonic and biofilm modes of growth, in contrast to SprE, which was shown to be a negative regulator of autolysis and biofilm production. Thomas et al. showed that DNase I treatment impaired development of a biofilm by the vancomycin-resistant strain E. faecalis V583. These investigators suggested that there is a fratricidal mechanism underlying cell death and DNA release, which may be a result of the interactions between GelE and Atn (70), suggesting that this crucial event in biofilm development is under control of an unknown regulatory pathway. Similarly, our data suggest that there is a regulatory pathway leading to the activation of Atn at a specific time point for DNA release that is crucial for biofilm development. It is possible that when a critical mass is reached following attachment, the fsr quorum-sensing signaling pathway is activated, resulting in expression of autolytic factors, such as GelE, SprE, and Atn, which would cause cell death and DNA release and ultimately culminate in biofilm production and maturation. However, early reports also showed the ability of certain clinical isolates to produce biofilms regardless of the presence or production of gelatinase (39). We also found that atn mutants are able to produce DNA-dependent biofilms, albeit in a delayed fashion, consistent with slower accumulation of eDNA possibly due to the actions of other autolytic factors present in E. faecalis (53). Mesnage et al. characterized two additional peptidoglycan hydrolases, AtlB and AtlC, in the JH2-2 strain of E. faecalis, which they showed to have compensatory functions in cell separation and autolysis in the absence of the major N-acetylglucosaminidase, AtlA (Atn) (35). It is likely that DNA release is the result of the synergistic activities of these autolytic enzymes during the establishment of E. faecalis biofilms.
eDNA is a crucial structural component of the biofilms of both gram-negative and gram-positive bacteria and is important for primary attachment, cell-to-cell adhesion, mature 3D biofilm architecture, and antimicrobial activities (65, 66, 71, 74, 77). It is an important component of the extracellular matrix of E. faecalis biofilms and is involved in cellular clustering during biomass accumulation. Although eDNA has similar functions under static and hydrodynamic conditions, its spatial contribution may vary. Static biofilms tend to be more compact with higher cell densities than biofilms formed in the presence of shear forces, as visualized by CLSM. Freeze-dry electron microscopy and CLSM revealed that E. faecalis produces a DNase I-sensitive fibrous network during development of biofilms under hydrodynamic conditions. This filamentous network is reminiscent of a stable fibrous eDNA network described for the aquatic gram-negative bacterium strain F8 (4, 5). The formation of extracellular fibrous DNA networks has also been reported for biofilms of nontypeable Haemophilus influenzae formed in vivo (27). Importantly, Kristich et al. (31) reported the presence of a filamentous extracellular network in enterococcal biofilms of both wild-type and transposon-inactivated atn mutant strains grown statically on nitrocellulose membranes. However, the composition of this structure remains unknown.
Biofilm formation is a complex and highly regulated process involving a combination of several genetic and environmental factors acting at different stages. Our study uncovered a critical function for SrtA and provided strong evidence for the role of Atn in DNA release and biofilm production. Further studies are required to identify the SrtA-dependent proteins important for biofilm formation. As an opportunistic pathogen, E. faecalis may use several mechanisms for establishing biofilms. eDNA is a critical component of the extracellular matrix of DNA-dependent biofilms produced by E. faecalis, as well as E. faecium and E. gallinarum clinical isolates (data not shown). eDNA is crucial for biofilm integrity and bacterial survival. The events mediating DNA release, localization, and interactions with other components of this protective structure require further investigation. Understanding the regulation underlying the formation of the extracellular matrix of enterococcal biofilms could be of great importance in the eradication of these pathogens.
Supplementary Material
Acknowledgments
We thank Wandy Beatty of the Molecular Microbiology Imaging Facility at Washington University School of Medicine in St. Louis for her technical assistance with transmission electron microscopy.
This work was funded by NIH grants DK64540, DK51406 (S.J.H.), and AI46433 (M.G.C.), by AHA postdoctoral fellowship 0625736Z (K.A.K.), and by Medical Scientist Training Program grant T32 GM07200 (A.L.K.).
Editor: A. Camilli
Footnotes
Published ahead of print on 15 June 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 591114-1128. [DOI] [PubMed] [Google Scholar]
- 2.Barken, K. B., S. J. Pamp, L. Yang, M. Gjermansen, J. J. Bertrand, M. Klausen, M. Givskov, C. B. Whitchurch, J. N. Engel, and T. Tolker-Nielsen. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 102331-2343. [DOI] [PubMed] [Google Scholar]
- 3.Beliveau, C., C. Potvin, J. Trudel, A. Asselin, and G. Bellemare. 1991. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J. Bacteriol. 1735619-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bockelmann, U., A. Janke, R. Kuhn, T. R. Neu, J. Wecke, J. R. Lawrence, and U. Szewzyk. 2006. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol. Lett. 26231-38. [DOI] [PubMed] [Google Scholar]
- 5.Bockelmann, U., H. Lunsdorf, and U. Szewzyk. 2007. Ultrastructural and electron energy-loss spectroscopic analysis of an extracellular filamentous matrix of an environmental bacterial isolate. Environ. Microbiol. 92137-2144. [DOI] [PubMed] [Google Scholar]
- 6.Bourgogne, A., K. V. Singh, K. A. Fox, K. J. Pflughoeft, B. E. Murray, and D. A. Garsin. 2007. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J. Bacteriol. 1896490-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady, R. A., J. G. Leid, J. H. Calhoun, J. W. Costerton, and M. E. Shirtliff. 2008. Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol. Med. Microbiol. 5213-22. [DOI] [PubMed] [Google Scholar]
- 8.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 1320-26. [DOI] [PubMed] [Google Scholar]
- 9.Cornett, J. B., B. E. Redman, and G. D. Shockman. 1978. Autolytic defective mutant of Streptococcus faecalis. J. Bacteriol. 133631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton, J. W. 2001. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 950-52. [DOI] [PubMed] [Google Scholar]
- 11.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 2841318-1322. [DOI] [PubMed] [Google Scholar]
- 12.Eckert, C., M. Lecerf, L. Dubost, M. Arthur, and S. Mesnage. 2006. Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J. Bacteriol. 1888513-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabretti, F., C. Theilacker, L. Baldassarri, Z. Kaczynski, A. Kropec, O. Holst, and J. Huebner. 2006. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 744164-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felmingham, D., A. P. Wilson, A. I. Quintana, and R. N. Gruneberg. 1992. Enterococcus species in urinary tract infection. Clin. Infect. Dis. 15295-301. [DOI] [PubMed] [Google Scholar]
- 15.Gilmore, K. S., P. Srinivas, D. R. Akins, K. L. Hatter, and M. S. Gilmore. 2003. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect. Immun. 714759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore, M. S., P. S. Coburn, S. R. Nallapareddy, and B. E. Murray. 2002. Enterococcal virulence, p. 301. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, and B. E. Murray (ed.), The enterococci: pathogenesis, molecular biology, and antimicrobial resistance. ASM Press, Washington, DC.
- 17.Graninger, W., and R. Ragette. 1992. Nosocomial bacteremia due to Enterococcus faecalis without endocarditis. Clin. Infect. Dis. 1549-57. [DOI] [PubMed] [Google Scholar]
- 18.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 1821529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 1865629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 241013-1024. [DOI] [PubMed] [Google Scholar]
- 21.Heilmann, C., G. Thumm, G. S. Chhatwal, J. Hartleib, A. Uekotter, and G. Peters. 2003. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 1492769-2778. [DOI] [PubMed] [Google Scholar]
- 22.Hufnagel, M., S. Koch, R. Creti, L. Baldassarri, and J. Huebner. 2004. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J. Infect. Dis. 189420-430. [DOI] [PubMed] [Google Scholar]
- 23.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joris, B., S. Englebert, C. P. Chu, R. Kariyama, L. Daneo-Moore, G. D. Shockman, and J. M. Ghuysen. 1992. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 70257-264. [DOI] [PubMed] [Google Scholar]
- 25.Joyanes, P., A. Pascual, L. Martinez-Martinez, A. Hevia, and E. J. Perea. 1999. In vitro adherence of Enterococcus faecalis and Enterococcus faecium to plastic biomaterials. Clin. Microbiol. Infect. 5382-386. [DOI] [PubMed] [Google Scholar]
- 26.Joyanes, P., A. Pascual, L. Martinez-Martinez, A. Hevia, and E. J. Perea. 2000. In vitro adherence of Enterococcus faecalis and Enterococcus faecium to urinary catheters. Eur. J. Clin. Microbiol. Infect. Dis. 19124-127. [DOI] [PubMed] [Google Scholar]
- 27.Jurcisek, J. A., and L. O. Bakaletz. 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 1893868-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp, K. D., K. V. Singh, S. R. Nallapareddy, and B. E. Murray. 2007. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect. Immun. 755399-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kline, K. A., A. L. Kau, S. L. Chen, A. Lim, J. S. Pinkner, J. Rosch, S. R. Nallapareddy, B. E. Murray, B. Henriques-Normark, W. Beatty, M. G. Caparon, and S. J. Hultgren. 2009. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J. Bacteriol. 1913237-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristich, C. J., D. A. Manias, and G. M. Dunny. 2005. Development of a method for markerless genetic exchange in Enterococcus faecalis and its use in construction of a srtA mutant. Appl. Environ. Microbiol. 715837-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kristich, C. J., V. T. Nguyen, T. Le, A. M. Barnes, S. Grindle, and G. M. Dunny. 2008. Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl. Environ. Microbiol. 743377-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levesque, C. M., E. Voronejskaia, Y. C. Huang, R. W. Mair, R. P. Ellen, and D. G. Cvitkovitch. 2005. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect. Immun. 733773-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mesnage, S., F. Chau, L. Dubost, and M. Arthur. 2008. Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J. Biol. Chem. 28319845-19853. [DOI] [PubMed] [Google Scholar]
- 36.Mohamed, J. A., and D. B. Huang. 2007. Biofilm formation by enterococci. J. Med. Microbiol. 561581-1588. [DOI] [PubMed] [Google Scholar]
- 37.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 723658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohamed, J. A., and B. E. Murray. 2006. Influence of the fsr locus on biofilm formation by Enterococcus faecalis lacking gelE. J. Med. Microbiol. 551747-1750. [DOI] [PubMed] [Google Scholar]
- 39.Mohamed, J. A., and B. E. Murray. 2005. Lack of correlation of gelatinase production and biofilm formation in a large collection of Enterococcus faecalis isolates. J. Clin. Microbiol. 435405-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohamed, J. A., F. Teng, S. R. Nallapareddy, and B. E. Murray. 2006. Pleiotropic effects of 2 Enterococcus faecalis sagA-like genes, salA and salB, which encode proteins that are antigenic during human infection, on biofilm formation and binding to collagen type I and fibronectin. J. Infect. Dis. 193231-240. [DOI] [PubMed] [Google Scholar]
- 41.Molinari, G., V. Pugliese, G. C. Schito, and C. A. Guzman. 1996. Bacteria involved in the blockage of biliary stents and their susceptibility to antibacterial agents. Eur. J. Clin. Microbiol. Infect. Dis. 1588-92. [DOI] [PubMed] [Google Scholar]
- 42.Moscoso, M., E. Garcia, and R. Lopez. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 1887785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulcahy, H., L. Charron-Mazenod, and S. Lewenza. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 346-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murray, B. E., F. Y. An, and D. B. Clewell. 1988. Plasmids and pheromone response of the beta-lactamase producer Streptococcus (Enterococcus) faecalis HH22. Antimicrob. Agents Chemother. 32547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 1755216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nemoto, K., K. Hirota, K. Murakami, K. Taniguti, H. Murata, D. Viducic, and Y. Miyake. 2003. Effect of Varidase (streptodornase) on biofilm formed by Pseudomonas aeruginosa. Chemotherapy 49121-125. [DOI] [PubMed] [Google Scholar]
- 48.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 5449-79. [DOI] [PubMed] [Google Scholar]
- 49.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2992071-2074. [DOI] [PubMed] [Google Scholar]
- 50.Perez-Casal, J. F., H. F. Dillon, L. K. Husmann, B. Graham, and J. R. Scott. 1993. Virulence of two Streptococcus pyogenes strains (types M1 and M3) associated with toxic-shock-like syndrome depends on an intact mry-like gene. Infect. Immun. 615426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petersen, F. C., D. Pecharki, and A. A. Scheie. 2004. Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J. Bacteriol. 1866327-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen, F. C., L. Tao, and A. A. Scheie. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 1874392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfeffer, J. M., H. Strating, J. T. Weadge, and A. J. Clarke. 2006. Peptidoglycan O acetylation and autolysin profile of Enterococcus faecalis in the viable but nonculturable state. J. Bacteriol. 188902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pillai, S. K., G. Sakoulas, G. M. Eliopoulos, R. C. Moellering, Jr., B. E. Murray, and R. T. Inouye. 2004. Effects of glucose on fsr-mediated biofilm formation in Enterococcus faecalis. J. Infect. Dis. 190967-970. [DOI] [PubMed] [Google Scholar]
- 55.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 1833372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 682579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin, X., K. V. Singh, Y. Xu, G. M. Weinstock, and B. E. Murray. 1998. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 422883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin, Z., Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin, and D. Qu. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 1532083-2092. [DOI] [PubMed] [Google Scholar]
- 59.Raad, I. I., H. A. Hanna, M. Boktour, G. Chaiban, R. Y. Hachem, T. Dvorak, R. Lewis, and B. E. Murray. 2005. Vancomycin-resistant Enterococcus faecium: catheter colonization, esp gene, and decreased susceptibility to antibiotics in biofilm. Antimicrob. Agents Chemother. 495046-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rice, K. C., E. E. Mann, J. L. Endres, E. C. Weiss, J. E. Cassat, M. S. Smeltzer, and K. W. Bayles. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1048113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts, A. P., J. Pratten, M. Wilson, and P. Mullany. 1999. Transfer of a conjugative transposon, Tn5397 in a model oral biofilm. FEMS Microbiol. Lett. 17763-66. [DOI] [PubMed] [Google Scholar]
- 62.Ruggero, K. A., L. K. Schroeder, P. C. Schreckenberger, A. S. Mankin, and J. P. Quinn. 2003. Nosocomial superinfections due to linezolid-resistant Enterococcus faecalis: evidence for a gene dosage effect on linezolid MICs. Diagn. Microbiol. Infect. Dis. 47511-513. [DOI] [PubMed] [Google Scholar]
- 63.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 331588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sizemore, R. K., J. J. Caldwell, and A. S. Kendrick. 1990. Alternate Gram staining technique using a fluorescent lectin. Appl. Environ. Microbiol. 562245-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spoering, A. L., and M. S. Gilmore. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9133-137. [DOI] [PubMed] [Google Scholar]
- 66.Steinberger, R. E., and P. A. Holden. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 715404-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358135-138. [DOI] [PubMed] [Google Scholar]
- 68.Tendolkar, P. M., A. S. Baghdayan, and N. Shankar. 2006. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J. Bacteriol. 1882063-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Theilacker, C., P. Sanchez-Carballo, I. Toma, F. Fabretti, I. Sava, A. Kropec, O. Holst, and J. Huebner. 2009. Glycolipids are involved in biofilm accumulation and prolonged bacteraemia in Enterococcus faecalis. Mol. Microbiol. 711055-1069. [DOI] [PubMed] [Google Scholar]
- 70.Thomas, V. C., Y. Hiromasa, N. Harms, L. Thurlow, J. Tomich, and L. Hancock. 2009. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 721022-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas, V. C., L. R. Thurlow, D. Boyle, and L. E. Hancock. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 1905690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 674538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Merode, A. E., H. C. van der Mei, H. J. Busscher, and B. P. Krom. 2006. Influence of culture heterogeneity in cell surface charge on adhesion and biofilm formation by Enterococcus faecalis. J. Bacteriol. 1882421-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vlassov, V. V., P. P. Laktionov, and E. Y. Rykova. 2007. Extracellular nucleic acids. Bioessays 29654-667. [DOI] [PubMed] [Google Scholar]
- 75.Vollmer, W., and U. Bertsche. 2008. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta 17781714-1734. [DOI] [PubMed] [Google Scholar]
- 76.Wang, B. Y., B. Chi, and H. K. Kuramitsu. 2002. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol. Immunol. 17108-112. [DOI] [PubMed] [Google Scholar]
- 77.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 2951487. [DOI] [PubMed] [Google Scholar]
- 78.Wiederhold, N. P., E. A. Coyle, Raad, I. I., R. A. Prince, and R. E. Lewis. 2005. Antibacterial activity of linezolid and vancomycin in an in vitro pharmacodynamic model of gram-positive catheter-related bacteraemia. J. Antimicrob. Chemother. 55792-795. [DOI] [PubMed] [Google Scholar]
- 79.Yamaguchi, M., Y. Terao, T. Ogawa, T. Takahashi, S. Hamada, and S. Kawabata. 2006. Role of Streptococcus sanguinis sortase A in bacterial colonization. Microbes Infect. 82791-2796. [DOI] [PubMed] [Google Scholar]
- 80.Yasuda, H., Y. Ajiki, J. Aoyama, and T. Yokota. 1994. Interaction between human polymorphonuclear leucocytes and bacteria released from in-vitro bacterial biofilm models. J. Med. Microbiol. 41359-367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.