Abstract
Several well-studied proteins with defined roles in Staphylococcus epidermidis biofilm formation are LPXTG motif-containing proteins. Here, we investigate the possible use of the LPXTG motif-containing protein SesC (S. epidermidis surface protein C; accession no. NP_765787) as a target for antibodies to prevent biofilm formation. In vitro and in a in vivo rat model of catheter infection, gene and protein expression analysis showed that SesC is expressed more strongly in biofilm-associated cells than in planktonic cells and is expressed particularly during the late phase of in vivo biofilm formation. Polyclonal rabbit antibodies raised against SesC reduced the fibrinogen-binding ability of S. epidermidis RP62A and Staphylococcus aureus RN4220 transformants expressing SesC, inhibited in vitro biofilm formation by S. epidermidis strains 10b and 1457, and significantly reduced the numbers of bacteria in a 1-day-old in vivo biofilm (P < 0.001, one-way analysis of variance). Our findings revealed that SesC is a promising target for prevention and treatment of S. epidermidis biofilms because it affects both the primary attachment and biofilm accumulation phases. The precise role of SesC in biofilm formation remains to be identified.
There has been substantial interest in Staphylococcus epidermidis in recent years because it is the most important cause of foreign-body infections (27, 34). Biofilm formation is a key factor in this process and is considered the most important virulence factor of S. epidermidis (6).
S. epidermidis biofilm formation is a complex, multifactorial process, involving different factors that play roles at different stages in biofilm formation. Several of the genes that have been found to play important roles in biofilm formation by S. epidermidis (for a review, see reference 21) encode LPXTG motif-containing proteins (Aap, Bhp, SdrF, and SdrG) (1, 8, 9, 15). Recently, Söderquist reported that SesI, another LPXTG protein, was present in “about one-half” of the S. epidermidis isolates causing postoperative infection following cardiac surgery and might be a bacterial adherence factor (25).
In publicly available genomes of S. epidermidis strains RP62A (11) and ATCC 12228 (37), 11 and 10 genes encoding LPXTG proteins, respectively, have been identified (2), including genes encoding the proteins mentioned above. Except for the five LPXTG proteins mentioned above, the roles of these LPXTG proteins have not been studied yet. In the present study we examined the S. epidermidis LPXTG protein SesC as a potential target for vaccination against S. epidermidis biofilms.
Bowden et al. (2) reported that the sesC gene was present in all of the 116 clinical isolates of S. epidermidis that they investigated, indicating that it might be an essential gene. Yao et al. (36), however, reported that sesC was absent in some S. epidermidis isolates, particularly isolates from the skin of healthy individuals (9 of 20 isolates).
SesC is predicted to encode a 676-amino-acid (aa) protein with a predicted molecular mass of 75 kDa. The cytoplasmic precursor of SesC contains a 35-aa N-terminal signal peptide (predicted using the SignalP server at http://www.cbs.dtu.dk/services/SignalP/), a 37-aa C-terminal LPXTG sorting signal, and a large extracellular domain. The N-terminal signal is required for sec-dependent secretion and is cleaved by signal peptidase. The C-terminal signal is needed for cleavage between the threonine and the glycine of the LPXTG motif and for attachment to peptidoglycan by sortase.
The presence of mature SesC (∼68 kDa) in the cell wall fraction of S. epidermidis RP62A in the exponential and stationary phases of growth was shown using a Western immunoblotting technique (2). All of the homologues of SesC in publicly available protein data banks had less than 70% sequence identity to SesC, and all of the homologues with identities higher than 26% were hypothetical proteins with unknown structures and functions. The closest homologue of SesC with a known function is a 341-aa fragment of clumping factor A (ClfA) (26.6% identity and 65.1% similarity in a 335-aa overlap). ClfA is a fibrinogen (Fg)-binding microbial surface component recognizing adhesive matrix molecules (MSCRAMM) of Staphylococcus aureus. However, the putative Fg-binding site of ClfA is located outside the similarity region.
Targeting specific staphylococcal biofilm-associated factors is an alternative to treatment of staphylococcal infections with antibiotics (34). Antibodies against extracellular macromolecules and surface binding proteins essential for cell-surface and cell-cell interaction and adhesion, such as polysaccharide intracellular adhesin (PIA), teichoic acids, Fbe, and Aap, have been shown to prevent biofilm formation without killing the bacteria (19, 22, 26, 35).
In this study we demonstrate that SesC is highly expressed in biofilm-associated cells and we present data showing that it is a potential target for preventing S. epidermidis biofilm formation and for treating established mature biofilms with anti-SesC antibodies.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
For DNA manipulation and recombinant protein production, Escherichia coli strains DH5α and BL21(DE3), respectively, were used. Staphylococcus spp. were grown in brain heart infusion medium (Oxoid) or tryptone soya broth (TSB) (Oxoid), except where otherwise stated. E. coli was grown in Luria-Bertani medium. Solid media consisted of the liquid media supplemented with 1 to 2% agar. When required, antibiotics were added to the media as follows: chloramphenicol, 10 μg/ml for Staphylococcus spp.; erythromycin, 10 μg/ml for Staphylococcus spp. and 500 μg/ml for E. coli; tetracycline, 5 μg/ml for Staphylococcus spp. and E. coli; and ampicillin, 100 μg/ml for E. coli.
The sesC sequence (SE2232; accession no. NP_765787) was retrieved from the National Center for Biotechnology Information from the complete genome of the non-biofilm-forming strain S. epidermidis ATCC 12228. Using the sesC sequence, primers and probes were designed with Primer Express 2.0 software (Applied Biosystems Division of Perkin-Elmer) and were purchased from Eurogentec (Seraing, Belgium). All fragments were PCR amplified using genomic DNA (gDNA) isolated from biofilm-forming S. epidermidis strain 10b (32). The primers and probe used are listed in Table 1.
TABLE 1.
Primers and probes used in this study
| Primer or probe | DNA sequence (5′-3′)a |
|---|---|
| sesC-SF | GTTGATAACCGTCAACAAGG |
| sesC-SR | CATGTTGATCTTTTGAATCCC |
| sesC-TF | AGCATCACCATCTAATAAAAACGAAA |
| sesC-TR | CCATCATTACTTTTATCGTCTTTACTATCAC |
| sesC-P | TAACAAAGAAGAATCTAGTACGACAACAAATCAATCCGA |
| sesC-RF | ACGTGCTAGCGCAGATTCAGAAAGTACATC |
| sesC-RR | GAACAGCTACAGCTGATCATCACCATCACCATCACTAGGATCCGCAT |
| 16S rRNA-F | TACACACCGCCCGTCACA |
| 16S rRNA-R | CTTCGACGGGCTAGCTCCAAAT |
| gmk-F | AAGGTGCTAAGCAAGTAAGAAAGAAATT |
| gmk-P | ATGCGTTGTTCATATTTTTAGCGCCTCCA |
| gmk-R | CAACAAGACGTTCTTTCAAGTCATCT |
| sesC-EF | TACGGGATCCCAGGTAACTTTATTAAAGGAGTATGTGTAA |
| sesC-ER | ACGTGGTACCACTAGAAGTTAATGCAAGACCATCAATTT |
Incorporated restriction sites are underlined.
A 388-bp fragment of the sesC gene in S. epidermidis 10b was PCR amplified using primers sesC-SF and sesC-SR. The amplicon was ligated into the pGEM-T Easy vector (Promega, Madison, WI), yielding pGEMsesC. Pure plasmid pGEMsesC was prepared and quantified as described previously (18, 23). Standard dilutions of a known quantity of pGEMsesC were used in real-time PCR.
A 1,359-bp fragment of sesC in S. epidermidis 10b was amplified using primers sesC-RF and sesC-RR, which incorporate flanking NheI and BamHI restriction sites and a sequence coding for a C-terminal six-His tag, respectively. This fragment was cloned into a pET11c expression vector (Stratagene, La Jolla, CA), yielding pET11csesC, which was electrotransformed into E. coli BL21(DE3). The 1,359-bp fragment of sesC encodes a 459-aa extracellular part of SesC and contains a six-His tag at the C terminus. The truncated recombinant protein was used for immunization of rabbits.
The entire coding region of the sesC gene of strain 10b was amplified using primers sesC-EF and sesC-ER, which incorporate flanking BamHI and KpnI restriction sites, respectively. The amplicon was ligated into the vectors pCN68 and pCN50 (5), yielding pCN68sesC and pCN50sesC. Plasmids pCN68, pCN50, pCN68sesC, and pCN50sesC were electroporated into S. aureus RN4220 (16), yielding strains RN-pCN68, RN-pCN50, RN-pCN68sesC, and RN-pCN50sesC, respectively. pCN50 and pCN50sesC were purified from RN4220 transformants and electroporated into S. epidermidis RP62A, yielding strains RP-pCN50 and RP-pCN50sesC, respectively. sesC gene expression in wild-type and transformed strains was quantified by TaqMan quantitative PCR.
Species identification and PCR screening for the sesC gene in clinical and commensal isolates.
We collected 239 coagulase-negative Staphylococcus sp. (CoNS) isolates from hospitalized patients (n = 215) or from the skin of healthy individuals (n = 24). Species were identified with Vitek 2 (bioMérieux). gDNA was extracted from each isolate using a Wizard gDNA purification kit (Promega) with addition of 30 μg/ml lysostaphin at the lysis step. A duplex PCR amplifying both sesC and the 16S rRNA genes was performed with all isolates. Primers for the 16S rRNA gene have been described previously (29).
Construction and purification of histidine-tagged fusion protein.
A truncated recombinant SesC (rSesC) protein was expressed in E. coli BL21(DE3) using the expression vector pET11csesC as described previously (14). Briefly, after transformation, E. coli BL2(DE3) was grown with shaking (250 rpm) at 37°C in Luria-Bertani broth with 100 μg/ml ampicillin to an optical density at 600 nm (OD600) of 0.6 to 1.0. Expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After cooling on ice, cells were harvested by centrifugation (4,000 rpm, 10 min, 4°C), resuspended in 5 ml imidazole buffer (20 mM phosphate, 0.5 M NaCl, 10 mM imidazole), and frozen at −20°C. Then the preparation was sonicated three times for 30 s. After centrifugation (30 min, 15,000 rpm, 4°C) the supernatant was used for Ni+ affinity chromatography purification of the recombinant proteins with a HisTrap kit (Amersham Pharmacia, Uppsala, Sweden). The columns were washed with 40 mM imidazole buffer, and proteins were eluted with 300 mM imidazole buffer. The purified recombinant protein was dialyzed against 10 mM HEPES buffer (pH 7.5), freeze-dried, and stored at −20°C. The purity of the recombinant protein was determined by Coomassie blue staining of a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. The sequence of the purified peptide was confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry.
Preparation and purification of polyclonal anti-SesC IgG antibodies.
Polyclonal antibodies were produced by Eurogentec (Seraing, Belgium) by immunization of rabbits with purified rSesC protein. Total immunoglobulin Gs (IgGs) from preimmune serum and antisera directed against rSesC (anti-rSesC) were purified by absorption to a protein G column (GE Healthcare) according to the manufacturer's instructions.
In order to enrich for SesC-specific IgGs, (polyclonal anti-SesC IgGs), the rSecC antigen was covalently coupled to MiniLeak (medium) affinity resin (Kem-En-Tec, Copenhagen, Denmark) as recommended by the manufacturer. A total of 1 mg of rSecC was coupled to 1 ml of resin, and the coupling efficiency was measured as described by the manufacturer. Subsequently, 5 ml of purified IgG was incubated for 3 h at room temperature with 1 ml of immunoaffinity resin and was subsequently packed into a column. After the resin was washed with 100 ml of phosphate-buffered saline (PBS) (pH 6.8) containing 0.5 M NaCl and 10 mM EDTA, immunoabsorbed material was eluted with 0.1 M glycine-HCl buffer (pH 2.7) and immediately dialyzed against PBS. After dialysis the concentration was determined spectrophotometrically at 280 nm (an optical density of 1.4 was equivalent to 1 mg/ml). The purity of the IgGs was determined by Coomassie blue staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels.
The affinity of IgGs purified from preimmune serum and of polyclonal anti-SesC IgGs isolated from antiserum against rSesC was quantified with an alkaline phosphatase-conjugated anti-rabbit Ig by performing an indirect protein enzyme-linked immunosorbent assay using standard protocols.
In vitro and in vivo gene and protein expression studies.
The in vitro model and the in vivo rat model have been described previously (12, 28, 30, 31). Slight modifications were incorporated into the anesthesia and euthanasia protocols for rats (18, 23) in accordance with guidelines of the animal ethics committee.
In vivo and in vitro extraction of DNA and RNA from sessile and planktonic bacteria and reverse transcription were performed as described previously (18, 23, 29, 31).
For in vitro studies, 20 μl of a frozen culture of S. epidermidis 10b was grown to the late exponential phase, pelleted, and resuspended in 0.9% NaCl. Seven-millimeter fragments of a commercial polyurethane intravenous catheter (Arrow International, Reading, PA) were added, and the mixture was incubated at 37°C. After various incubation periods, RNA and DNA were extracted as described previously (12, 28, 29, 31). Nucleic acid was isolated from bacteria adhering to the catheter fragments and from planktonic bacteria at zero time (n = 8) (just before the bacteria were suspended in 0.9% NaCl) and after 10 min (n = 16), 35 min (n = 16), 60 min (n = 16), 120 min (n = 16), and 180 min (n = 16). For all time points eight independent measurements from two experiments were included.
For the in vivo rat model, first-generation descendants of inbred germfree Fisher rats were used. These rats were exposed to normal rat flora from birth and were designated ex-germfree Fisher rats. Seven-millimeter catheter fragments were inoculated with a small amount of S. epidermidis 10b (1.09 × 104 cells/catheter) prior to implantation by incubation for 20 min at 37°C in a 0.9% NaCl suspension of S. epidermidis. This resulted in a 100% infection rate. Rats were anesthetized by inhalation of enflurane gas (Alyrane; Pharmacia). Rats were kept asleep during the implantation procedure by using a mixture of enflurane (20%) and oxygen (80%). A large area on the back of each rat was shaved, and the skin was disinfected with 0.5% chlorhexidine in 70% alcohol and allowed to dry. A 10-mm incision was made at the base of the tail, and the subcutis was dissected to create three subcutaneous tunnels. For each rat, eight catheter fragments were inserted at least 2 cm from the incision; the distance between any two fragments was at least 1 cm.
For catheter explantation, rats were euthanized by CO2 inhalation. The skin was disinfected before removal of the catheter fragments. All catheter fragments from the same animal were used for a single time point. In each experiment, baseline levels of gene expression in sessile bacteria before implantation were determined (time zero; n = 16). A total of 176 polyurethane catheter segments were implanted and explanted at 11 different time points. The time points used were 15 min (n = 16), 1 h (n = 16), 2 h (n = 16), 4 h (n = 16), 6 h (n = 16), 12 h (n = 16), 24 h (n = 16), 2 days (n = 16), 4 days (n = 16), 7 days (n = 16), and 14 days (n = 16). Data for each in vivo time point were obtained from 16 independent measurements obtained in two independent experiments. Nucleic acid isolation and cDNA synthesis were performed immediately after explantation as described previously (29, 31).
Using the TaqMan primers sesC-TF and sesC-TR and a dual-label probe (5′ 6-carboxyfluorescein and 3′ 6-carboxytetramethylrhodamine), sesC-P quantification of both cDNA and gDNA was performed as described previously (18, 23, 29, 31). In each run, a standard dilution of the plasmid (pGEMsesC) with a known quantity that allowed gene quantification and a negative control (distilled water) were included.
Conventional fluorescence microscopy was used to confirm SesC protein expression in sessile and planktonic cells in vitro. The method used for preparation of samples for fluorescence microscopy was similar to the method used for preparation of samples for gene expression analysis in vitro, except that samples were taken at different time points (0, 30, 60, 90, and 120 min) after inoculation and sessile cells were separated from catheter fragments as described previously (23). The bacteria were fixed using PBS containing 1.5% formaldehyde and 0.5% glutaraldehyde for 30 min and were washed with PBS. Next, the bacteria were preincubated for 20 min on ice with 2.4G2 (Fc-blocking antibody; BD Pharmingen), after which anti-SesC IgGs were added at a concentration of 5 μg per 100 μl and the preparations were incubated for another 30 min. Subsequently, the bacteria were washed twice with ice-cold PBS, and fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit antibody (BD-Pharmingen) was added for 30 min. Finally, the cells were washed twice with PBS and viewed with a fluorescence microscope (Leica, Germany) equipped with an oil immersion Plan Neofluar objective (×100; numeric aperture, 1.25).
The following preparations were used as negative controls: (i) cells incubated with preimmune IgGs and FITC-labeled goat anti-rabbit, (ii) cells incubated with polyclonal rabbit antibody against mouse Igs and FITC-labeled goat anti-rabbit, and (iii) cells incubated with only FITC-labeled goat anti-rabbit.
In vitro and in vivo biofilm inhibition assays.
The effect of IgGs (preimmune or anti-SesC) on in vitro biofilm formation during the first hour (primary attachment) and overnight (14 h) and on 1-day-old biofilms was studied as described previously (7, 26). For quantification of biofilms, 20-μl portions of frozen cultures of S. epidermidis strains 10b and 1457 (17) and a biofilm-forming sesC-negative clinical isolate of S. warneri (this study) were inoculated into 5 ml TSB and grown to the late exponential phase in a shaking incubator at 37°C. Cultures were subsequently diluted to an OD600 of 0.005 (5 × 106 CFU/ml) in fresh TSB.
To evaluate the effect of IgGs on primary attachment of S. epidermidis strains 10b and 1457, starting cultures were diluted to an OD600 of 0.005 and subsequently grown at 37°C to an OD600 of 1. Cultures then were mixed with either preimmune IgGs or anti-SesC IgGs (10 μg/ml), and after 2 h of incubation at 4°C, 200-μl portions of the mixtures were pipetted into 96-well polystyrene microtiter plates (BD Biosciences, Heidelberg, Germany) and incubated for 1 h at 37°C without shaking.
To study the effect of IgGs on biofilm formation for 14 h, cultures diluted to an OD600 of 0.005 were mixed with either preimmune IgGs or anti-SesC IgGs at concentrations of 1 to 4 μg/ml. The mixtures were incubated for 2 h at 4°C. Two hundred microliters of a mixture (106 cells per well) was added to each well of 96-well polystyrene microtiter plates and incubated overnight at 37°C without shaking.
To evaluate the effect of IgGs on 1-day-old biofilms of strains 10b and 1457, diluted overnight cultures of bacteria with an OD600 of 0.005 were pipetted into sterile 96-well polystyrene microtiter plates. Twenty-four hours later, the growth medium was replaced with fresh medium or fresh medium containing 5 μg/ml preimmune IgGs or anti-SesC IgGs and incubated at 37°C for 24 h without shaking.
After this incubation, the plates were washed three times with PBS, and adherent biofilms were stained with 200 μl of 1% (wt/vol) crystal violet (Sigma) for 10 min, after which the plates were washed three times with water and dried. For quantification, 160 μl of 30% (vol/vol) acetic acid was added to each well to dissolve the stain. The OD595 of the dissolved stain was measured with a multipurpose UV/visible plate reader. The average adherence with each concentration of IgGs was determined using at least eight independent measurements obtained in at least two independent experiments. S. epidermidis strains 10b and 1457 and the clinical isolate of S. warneri in TSB without any added IgGs were used as positive controls, and TSB without bacteria was used as a negative control.
For the in vivo inhibition assay, our rat model for in vivo catheter infection was used. Seven-millimeter catheter fragments, preincubated for 20 min at 37°C with S. epidermidis 10b before implantation, were placed on ice, and eight fragments per rat were implanted immediately in ex-germfree Fisher rats. After 24 h, the rats (nine rats divided into three groups of three rats) were treated with 50 μg of anti-SesC IgG diluted in PBS (total volume, 330 μl), 50 μg of preimmune IgG diluted in PBS (total volume, 330 μl), or 330 μl of PBS via a subcutaneous injection at the place of catheter insertion.
Twenty-four hours after injection, all eight catheter fragments from each rat were explanted and used for nucleic acid extraction as described previously (29, 31). Real-time quantitative PCR of the guanylate monokinase housekeeping gene (gmk; SE0885; accession no. NP_764440) was used to determine the number of bacteria attached to the catheter fragments. As previously demonstrated (31), the number of gmk copies per catheter correlates very well with the number of CFU per catheter.
Adherence of transformants to immobilized Fg, Fn, Cn, and VWF in the presence and absence of anti-SesC IgGs.
Overnight cultures in brain heart infusion medium of transformed strains and their parental strains were precipitated and washed once with PBS. The OD600 was adjusted to 1.0, and the adherence was measured as follows. Wells of polystyrene microtiter plates were coated with human Fg (Sigma), fibronectin (Fn) (Sigma), collagen (Cn) (Sigma), and Von Willebrand factor (VWF) (Sigma) in PBS overnight at concentrations ranging from 0.1 to 100 μg/ml. Blocking was done with 2% bovine serum albumin in PBS for 1 h at 37°C. After washing, either the pure cultures (100 μl per well) were pipetted into the plates or the cultures were mixed with 5 μg/ml (final concentration) preimmune IgGs or anti-SesC IgGs and after 2 h of incubation at 4°C were added to the plates and allowed to adhere to the coated surfaces for 2 h at 37°C. After the incubation period, culture supernatants were washed, and the remaining adherent cells were stained and quantified as described above.
Statistical analysis.
All statistical analyses of the in vitro and in vivo gene expression data were performed with GraphPad Prism (GraphPad software, version 4.2; GraphPad, San Diego, CA) as described previously (18, 23, 29, 31). Since the in vitro and in vivo cDNA/gDNA ratios were not normally distributed at any time point, all data were log10 transformed in order to fulfill the requirements of normality.
For the in vitro gene expression data, two hypotheses were tested. A significant change in gene expression levels over time within one group (sessile or planktonic) was tested using a one-way analysis of variance (ANOVA). A significant difference in the evolution over time of the gene expression levels between the sessile group and the planktonic group was tested using a two-way ANOVA. When the one-way ANOVA result was significant, two-sided univariate tests with a correction for multiple comparisons were performed (Bonferroni test) to locate the significant differences.
For the in vivo gene expression data, a one-way ANOVA was used to test if there was significant change in the expression levels over time. When the one-way ANOVA result was significant, the two-sided Bonferroni multiple-comparison method was used to determine which time points differed at α = 0.05, with a correction for multiple comparisons.
For all data from bacterial adherence assays, two hypotheses were tested. A significant change in the adherence levels at different concentrations of IgGs within one group (preimmune or anti-SesC IgGs) was tested using a one-way ANOVA. A significant difference in the adherence at different concentrations of IgGs between the preimmune IgGs and anti-SesC IgG groups was tested with a two-way ANOVA. When the one-way ANOVA result was significant, two-sided univariate tests with a correction for multiple comparisons were performed (Bonferroni test) to locate the significant differences.
RESULTS
Presence of sesC in S. epidermidis and non-S. epidermidis coagulase-negative Staphylococcus spp.
Of 239 isolates from patients and healthy persons, 105 were identified as S. epidermidis and 134 isolates were identified as other CoNS, including S. hominis (n = 17), S. haemolyticus (n = 58), S. warneri (n = 43), S. capitis (n = 15), and S. saprophyticus (n = 1). All 105 S. epidermidis isolates were sesC positive, whereas the non-S. epidermidis isolates were either sesC negative (80%) or sesC positive (20%).
SesC gene and protein expression in sessile and planktonic bacteria in vitro and in sessile bacteria in vivo.
Biofilm-associated bacteria have been shown to have low metabolic activity and altered gene expression patterns (28, 30). To confirm expression of sesC in sessile and planktonic bacteria, we analyzed gene and protein expression levels.
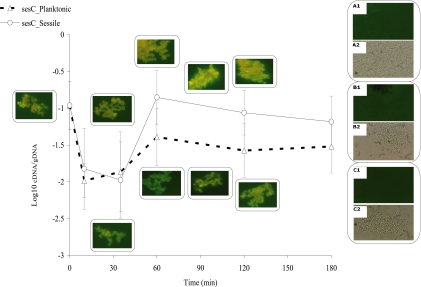
In our in vitro model, in sessile as well as planktonic bacteria, expression of sesC decreased 10-fold during the first 35 min after inoculation (P < 0.01, one-way ANOVA) and then remained at an intermediate level until the end of the experiment (at 180 min) in planktonic bacteria. In sessile bacteria, expression of this gene increased 13-fold from 35 min to 60 min (P < 0.001, one-way ANOVA) and then remained at this increased level for the remainder of the 2-h observation period (Fig. 1). However, the difference in sesC gene expression between sessile and planktonic bacteria was not statistically significant (P > 0.05, two-way ANOVA).
FIG. 1.
In vitro sesC gene expression and extracellular presence of the SesC protein in planktonic and sessile bacteria. Gene expression is expressed as the log10 cDNA/gDNA ratio. The error bars indicate standard deviations. At each time point eight samples from two independent experiments were assessed. The fluorescence microscopy images are images of sessile bacteria (top images on the right) and planktonic bacteria (bottom images on the right) at different time points. Images A1 and A2 show bacterial cells plus preimmune IgGs and FITC-labeled goat anti-rabbit antibodies. Images B1 and B2 are images of control samples containing bacterial cells plus polyclonal rabbit antibody against mouse Igs and FITC-labeled goat anti-rabbit antibodies. Images C1 and C2 are images of control samples containing bacterial cells plus only FITC-labeled goat anti-rabbit antibodies. Images A1, B1, and C1 are fluorescence images, while images A2, B2, and C2 are bright-field images.
To confirm the results obtained in the gene expression analysis, purified polyclonal anti-SesC IgGs and FITC-labeled goat anti-rabbit antibodies were used in an immunofluorescence assay to study in vitro protein expression. Comparison of the fluorescence microscopy images of sessile and planktonic bacteria obtained at different time points confirmed that SesC was present in both sessile and planktonic bacteria. Visual comparison of images taken at different time points suggested that there was a higher level of fluorescence in sessile bacteria than in planktonic bacteria (Fig. 1). However, no quantitative measurements were obtained.
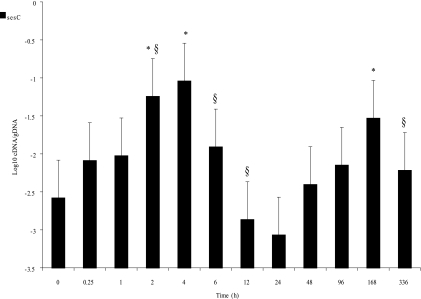
After implantation of catheters with sessile bacteria in our rat model, a 1.336-log10 or 21-fold increase in sesC expression in the sessile bacteria was observed after 2 h. The expression peaked at 4 h (1.47-log10 or 34-fold increase, a significant change over time [P < 0.001, one-way ANOVA]). This increase was followed by a 2.026-log10 or 106-fold decrease in gene expression in the next 20 h (P < 0.001, one-way ANOVA). However, after 24 h, gene expression again increased up to a maximum value at 168 h (P < 0.001, one-way ANOVA) and was again lower at 336 h after implantation (P < 0.05, one-way ANOVA) (Fig. 2).
FIG. 2.
Levels of sesC expression over time in vivo for 2 weeks after implantation. The expression level is expressed as the log10 cDNA/gDNA ratio. The error bars indicate standard deviations. At each time point 16 samples from two independent experiments were assessed. Significant differences for time points compared to time zero and previous time points are indicated by asterisks and section signs, respectively.
Biofilm inhibition by purified preimmune IgGs and polyclonal anti-SesC IgGs.
Binding of the purified preimmune IgGs and of the anti-SesC IgGs was tested by performing an indirect enzyme-linked immunosorbent assay with the 65-kDa rSesC. Tests performed with increasing concentrations of purified preimmune IgGs and anti-SesC IgGs showed that there was no binding of the preimmune IgGs and that there was dose-dependent binding of the purified anti-SesC IgGs (data not shown).
Different modes of preincubation of bacteria with IgGs (preimmune IgGs or polyclonal anti-SesC IgGs) and of incubation of bacteria and IgGs in 96-well polystyrene microtiter plates were tested in our in vitro model for biofilm formation.
Preincubation of strains 10b and 1457 for 2 h with anti-SesC IgGs or preimmune IgGs at 4°C and subsequent incubation for 1 h in polystyrene wells led to a significant reduction in initial attachment compared to controls for anti-SesC IgGs (P < 0.001, one-way-ANOVA) but not for preimmune IgGs (Fig. 3).
FIG. 3.
Effect of anti-SesC IgGs on primary attachment of S. epidermidis 10b and 1457 to polystyrene surfaces. Bacteria were mixed with IgGs, incubated for 2 h at 4°C, and then pipetted into wells. After 1 h of incubation at 37°C, the plates were washed and stained with crystal violet, and the OD595 was measured. The error bars indicate standard deviations. The data are the averages of eight measurements obtained in two independent experiments.
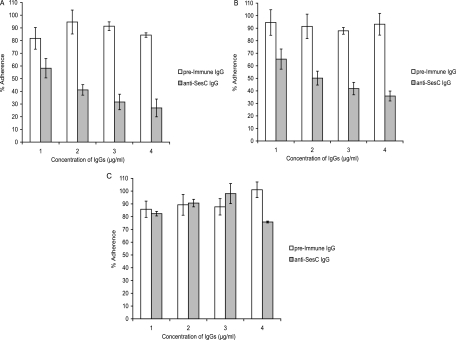
Two hours of preincubation of bacteria with IgGs followed by overnight (14-h) incubation in the 96-well polystyrene microtiter plates showed that the polyclonal anti-SesC IgGs purified from rSesC-induced rabbit antisera exhibited dose-dependent S. epidermidis biofilm inhibition activity, whereas the purified IgGs from preimmune serum showed only low activity that was dose independent (Fig. 4). Increasing the concentration of polyclonal anti-SesC IgGs from 1 to 4 μg/ml increased the inhibition of S. epidermidis strain 10b and 1457 biofilms from 40 to 70% (P < 0.001, one-way-ANOVA) (Fig. 4A and B). For both strains, the inhibition effects seen with anti-SesC IgGs were significantly different from those observed with preimmune IgGs (P < 0.001, two-way-ANOVA). A sesC-negative biofilm-positive S. warneri strain was included as a control; for this strain the inhibitory effect of anti-SesC IgGs was not different from that of the preimmune serum and was not dose dependent (Fig. 4C). For all three strains, the effect of preimmune IgGs on biofilm formation was not significant and was dose independent (P > 0.05, one-way ANOVA).
FIG. 4.
Inhibition of biofilm formation by S. epidermidis strains 10b (A) and 1457 (B) and a sesC-negative biofilm-positive clinical isolate of S. warneri (C) with increasing concentrations of preimmune and anti-SesC IgGs. Overnight cultures were diluted to an OD600 of 0.005, mixed with the indicated concentrations of IgGs, and incubated for 2 h at 4°C and then overnight at 37°C. The formation of biofilms was measured using the stain crystal violet. The error bars indicate standard errors. For each concentration of IgGs at least nine independent measurements from three independent experiments were assessed. One hundred percent adherence was defined as biofilm formation in TSB without any IgG.
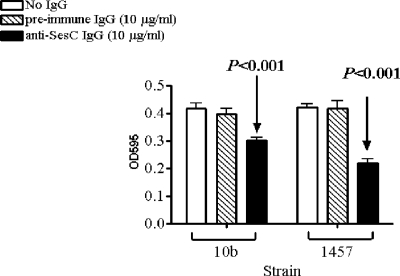
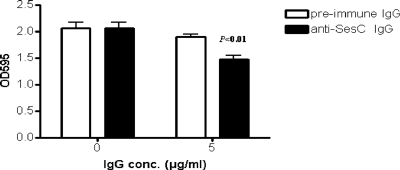
The effect of anti-SesC IgGs on 1-day-old biofilms of strain 10b was significant (P < 0.001, one-way ANOVA) compared to the effect on nontreated biofilms or biofilms treated with preimmune IgGs (Fig. 5). However, anti-SesC IgGs had no effect on 1-day-old biofilms of S. epidermidis strain 1457 (data not shown).
FIG. 5.
Effect of anti-SesC IgGs on 1-day-old biofilms of S. epidermidis 10b on polystyrene surfaces in vitro. After 24 h of incubation at 37°C the growth media were replaced with fresh TSB (control) or TSB containing 5 μg/ml preimmune or anti-SesC IgGs and then incubated for 24 h at 37°C. The remaining biofilms were stained with crystal violet and quantified by determining the OD595. The error bars indicate standard deviations. The data are the averages of eight measurements obtained in two independent experiments.
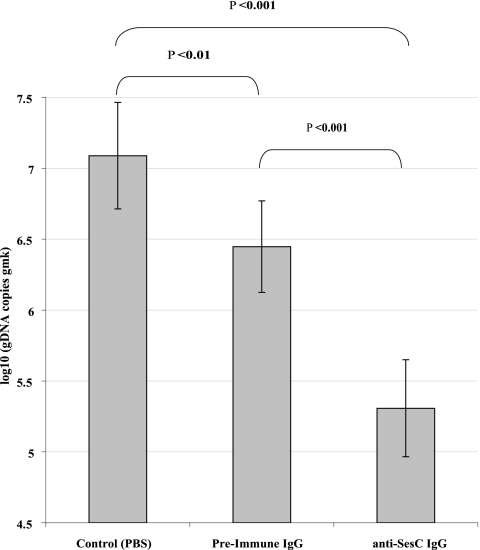
In the in vivo rat model, the number of sessile bacteria was determined after 1 day by quantification of the gDNA copies of the housekeeping gene gmk. Injection of 50 μg preimmune IgGs reduced the number of biofilm-associated S. epidermidis 10b bacteria 0.64 log10 or 4-fold (P < 0.01, one-way ANOVA), whereas injection of 50 μg anti-SesC IgGs decreased the number of biofilm-associated bacteria 1.78 log10 or 60.42-fold and 1.15 log10 or 14.35-fold (P < 0.001, one-way ANOVA) compared to the control group treated with PBS and the group treated with preimmune IgG, respectively (Fig. 6).
FIG. 6.
Effect of anti-SesC IgGs on 1-day-old biofilms of S. epidermidis 10b in vivo. One-day-old biofilms on catheter fragments implanted in three groups of rats (three rats per group) were treated with 50 μg preimmune IgGs or anti-SesC IgGs diluted in PBS (total volume, 330 μl) or with PBS via subcutaneous injection at the site of catheter insertion. The next day, catheter fragments were explanted, and the number of sessile bacteria was determined by quantification of the gDNA copies of the housekeeping gene gmk. The error bars indicate standard deviations. The data for each group were obtained using bacteria adhering to 24 catheter fragments implanted in three rats.
SesC is a potential Fg-binding MSCRAMM.
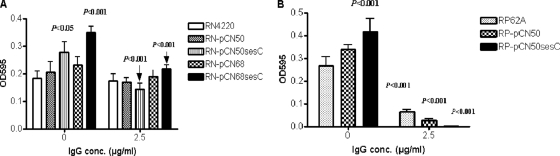
To explore the function of SesC, we expressed SesC in the sesC-negative strain S. aureus RN4220 and overexpressed SesC in the sesC-positive strain S. epidermidis RP62A by transformation of these strains with pCN68sesC and pCN50sesC. The copy number of pCN68 in S. aureus is higher than that of pCN50. RN transformants expressing SesC (RN-pCN68sesC and RN-pCN50sesC) and the RP transformant RP-pCN50sesC exhibited greater Fg-binding ability than their parental strains (Fig. 7). The levels of binding of these transformants to other host extracellular matrix (ECM) proteins (Fn, Cn, and VWF) were similar to the levels of binding of their parental strains (data not shown). RN-pCN68sesC had significantly greater Fg-binding ability than RN4220 and RN-pCN68 (P < 0.001 and P < 0.01, one-way ANOVA). Compared to their parental strains, RN-pCN50sesC and RP-pCN50sesC also exhibited significantly greater Fg-binding ability (P < 0.05 and P < 0.01, respectively, one-way ANOVA). However, the Fg-binding abilities of RN-pCN50sesC and RP-pCN50sesC were not significantly different from those of their parental strains transformed by mock plasmids (RN-pCN50 and RP-pCN50). There was no significant difference between the Fg-binding levels of RN4220 and RN-pCN50 or between the Fg-binding levels of RP62A and RP-pCN50.
FIG. 7.
Fg-binding abilities of RN4220 (A) and RP62A (B) transformants in the presence and absence of anti-SesC IgG. The abilities of strains RN4220 and RP62A and their transformants to bind immobilized Fg were measured using plates coated with Fg. The binding activities of RN and RP strains were determined on plates coated with Fg at concentrations of 10 and 1 μg/ml, respectively.
Addition of anti-SesC IgGs reduced the Fg-binding abilities of RN-pCN68sesC and RN-pCN50sesC to the wild-type strain levels and also reduced the Fg-binding abilities of wild-type and transformed RP strains (Fig. 7).
Anti-SesC IgGs had no effect on the abilities of RN and RP wild-type strains and transformed strains to bind to Fn, Cn, and VWF (data not shown).
DISCUSSION
Primary attachment of S. epidermidis to unmodified polymer surfaces is mediated by several protein and carbohydrate factors, such as PS/A, AtlE, and surface proteins SSP-1 and -2 (21). In vivo medical devices are quickly coated with adsorbed blood plasma proteins that can interact with S. epidermidis surface factors and promote binding. Once attached, S. epidermidis proliferates and accumulates as multilayer cell clusters. In addition to PIA, proteins are also important for biofilm formation. Therefore, antibodies directed against S. epidermidis surface targets involved in attachment or accumulation could be interesting alternatives to current antibiotic-dependent treatment of biofilms.
Here we show that SesC is an extracellular protein that is expressed more in sessile bacteria than in planktonic bacteria and that antibodies targeted against SesC reduce biofilm formation on polystyrene microtiter plates in vitro and on subcutaneously implanted polyurethane catheters in vivo.
In our gene expression analysis, we observed that in vitro and in vivo adherence to polyurethane catheters did not sharply increase sesC expression. The initial adherence takes place very rapidly after inoculation and is usually complete within the first hour. Upregulation of sesC gene expression occurred after 1 h in vitro, after 2 h in vivo, and during the late stages of in vivo foreign-body infections. This appears to indicate that SesC is associated more with the accumulation phase and with persistence of a biofilm than with the initial adhesion. This is consistent with the fluorescence data. The fact that more SesC was present on the surface of sessile cells than on planktonic cells suggests that SesC has a specific function in biofilm formation.
The precise function of SesC is not known. We found that the sesC gene was present in all S. epidermidis strains but was not universally present in non-S. epidermidis CoNS isolates. All sesC-negative non-S. epidermidis CoNS strains were biofilm negative, except a few (3 of 107) agr-negative isolates that were biofilm positive. The agr quorum-sensing system is one of the global regulators in Staphylococcus spp. that have important functions in the regulation of the biofilm phenotype. In S. epidermidis, agr represses biofilm formation (33). On the other hand, sesC-positive non-S. epidermidis CoNS can be either biofilm positive or biofilm negative. This observation suggests that sesC might code for an essential function in S. epidermidis biofilm formation.
So far, we have failed to knock out sesC in S. epidermidis, and we have not found a natural S. epidermidis sesC mutant. Expression of sesC in the sesC-negative strain S. aureus RN4220 or overexpression of sesC in the sesC-positive strain S. epidermidis RP62A did have a clear effect on Fg binding but not on binding to other host ECM proteins. This finding suggests that SesC can act as an Fg-binding MSCRAMM. However, the truncated rSesC did not show any Fg-binding activity (data not shown).
Several recent studies have shown that application of antibodies against surface components of S. epidermidis can affect the rate of biofilm formation or the adherence of bacteria to medical devices in vitro. Cerca et al. showed that antibodies against PIA readily penetrated a biofilm and bound to the sessile cells (4). Sessile bacteria were, however, more resistant to opsonic killing than their planktonic counterparts. Using polyclonal antibodies against Fbe, Pei and Flock blocked adherence of S. epidermidis to Fg-coated catheters in vitro (22). Sun et al. showed that monoclonal antibodies against AAP can significantly reduce the accumulation phase during biofilm formation by S. epidermidis in vitro (26).
Our in vitro experiments showed that rabbit polyclonal anti-SesC IgGs could significantly reduce primary attachment of S. epidermidis to the abiotic and Fg-coated surfaces of polystyrene plates and inhibit biofilm formation by S. epidermidis strains 10b and 1457 to less than 40% of control in a dose-dependent manner. In contrast, the effect of rabbit polyclonal anti-SesC IgGs on primary attachment of S. epidermidis to the surfaces of plates coated with other host ECM proteins or on biofilm formation by a sesC-negative biofilm-positive S. warneri strain was not significant, dose independent, and limited. Anti-SesC IgGs could also affect 1-day-old biofilms of strain 10b on polystyrene microtiter plates in vitro and on subcutaneously implanted polyurethane catheters in vivo.
Different explanations are possible. It is possible that in vivo opsonic activity plays a role in addition to the direct effect of antibody binding on the activity of SesC, as Rennermalm et al. have previously described for Fbe (24). Other workers have shown that antigen-antibody binding may inhibit ligand-receptor interactions or may lead to conformational changes (3, 10, 20). The inhibition of biofilm formation by preimmune IgGs could be due to cross-reactive IgGs directed against other surface-exposed antigens of Staphylococcus spp.
Gene expression data support the conclusion that SesC is involved in the accumulation phase and persistence of biofilms, whereas biofilm inhibition studies show that this protein is involved in primary attachment to naked and Fg-coated surfaces of polystyrene microtiter plates as well. An example of a protein which plays a role in attachment to abiotic surfaces and also shows host matrix protein-binding activity is AtlE (13).
Anti-SesC IgGs could restore the Fg-binding level of the RN transformants to the Fg-binding level of their parental strain and significantly reduce the Fg-binding ability of RP62A and its transformants, but they had no effect on the Cn-, Fn-, and VWF-binding abilities of the strains tested. These data indicate that anti-SesC IgGs efficiently and specifically inhibit the SesC function and have no effect on the function of other MSCRAMMs.
In conclusion, the effect of antibodies against SesC on S. epidermidis biofilm formation suggests that SesC might be a promising target for inhibition of S. epidermidis biofilm formation. The expression of SesC at the gene and protein levels in sessile S. epidermidis is in line with the biofilm inhibition data and support a role for SesC in S. epidermidis biofilm formation. The sesC gene is present in all biofilm-positive and -negative S. epidermidis isolates, suggesting that SesC may be a factor whose presence is necessary but not sufficient for biofilm formation.
Acknowledgments
This work was supported by the Glaxo-Wellcome Chair in Medical Microbiology (J. Van Eldere) at the Catholic University of Leuven (Leuven, Belgium), by the Sophia Foundation for Medical Research, and by the Erasmus MC—Sophia Children's Hospital, Rotterdam, The Netherlands.
We thank Caroline Massonet, Valerie Pintens, Marcel Sluijter, Theo Hoogeboezem, and Gholamreza Hassanzadeh Ghassabeh for their excellent assistance.
Editor: A. Camilli
Footnotes
Published ahead of print on 15 June 2009.
REFERENCES
- 1.Arrecubieta, C., M. H. Lee, A. Macey, T. J. Foster, and F. D. Lowy. 2007. SdrF, a Staphylococcus epidermidis surface protein, binds type I collagen. J. Biol. Chem. 28218767-18776. [DOI] [PubMed] [Google Scholar]
- 2.Bowden, M. G., W. Chen, J. Singvall, Y. Xu, S. J. Peacock, V. Valtulina, P. Speziale, and M. Hook. 2005. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 1511453-1464. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. C., and M. E. Koshland. 1977. Evidence for a long-range conformational change induced by antigen binding to IgM antibody. Proc. Natl. Acad. Sci. USA 745682-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerca, N., K. K. Jefferson, R. Oliveira, G. B. Pier, and J. Azeredo. 2006. Comparative antibody-mediated phagocytosis of Staphylococcus epidermidis cells grown in a biofilm or in the planktonic state. Infect. Immun. 744849-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charpentier, E., A. I. Anton, P. Barry, B. Alfonso, Y. Fang, and R. P. Novick. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 706076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, G. D., W. A. Simpson, A. L. Bisno, and E. H. Beachey. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 1832888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, S. L., S. Gurusiddappa, K. W. McCrea, S. Perkins, and M. Hook. 2001. SdrG, a fibrinogen-binding bacterial adhesin of the microbial surface components recognizing adhesive matrix molecules subfamily from Staphylococcus epidermidis, targets the thrombin cleavage site in the Bbeta chain. J. Biol. Chem. 27627799-27805. [DOI] [PubMed] [Google Scholar]
- 10.Einhauer, A., and A. Jungbauer. 2003. Complex formation of a calcium-dependent antibody: a thermodynamical consideration. J. Chromatogr. A 100981-87. [DOI] [PubMed] [Google Scholar]
- 11.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 1872426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA, and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 401439-1447. [DOI] [PubMed] [Google Scholar]
- 13.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 241013-1024. [DOI] [PubMed] [Google Scholar]
- 14.Hermans, P. W. M., P. V. Adrian, C. Albert, S. Estevao, T. Hoogenboezem, I. H. T. Luijendijk, T. Kamphausen, and S. Hammerschmidt. 2006. The streptococcal lipoprotein rotamase A (SlrA) is a functional peptidyl-prolyl isomerase involved in pneumococcal colonisation. J. Biol. Chem. 281968-976. [DOI] [PubMed] [Google Scholar]
- 15.Hussain, M., M. Herrmann, C. von Eiff, F. Perdreau-Remington, and G. Peters. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 17.Mack, D., N. Siemssen, and R. Laurs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 602048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massonet, C., V. Pintens, R. Merckx, J. Anne, E. Lammertyn, and J. Van Eldere. 2006. Effect of iron on the expression of sirR and sitABC in biofilm-associated Staphylococcus epidermidis. BMC Microbiol. 6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenney, D., K. Pouliot, Y. Wang, V. Murthy, M. Ulrich, G. Doring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 2000. Vaccine potential of poly-1-6-beta-d-N-succinylglucosamine, an immunoprotective surface polysaccharide of Staphylococcus aureus and Staphylococcus epidermidis. J. Biotechnol. 8337-44. [DOI] [PubMed] [Google Scholar]
- 20.Oda, M., H. Kozono, H. Morii, and T. Azuma. 2003. Evidence of allosteric conformational changes in the antibody constant region upon antigen binding. Int. Immunol. 15417-426. [DOI] [PubMed] [Google Scholar]
- 21.Otto, M. 2004. Virulence factors of the coagulase-negative staphylococci. Front. Biosci. 9841-863. [DOI] [PubMed] [Google Scholar]
- 22.Pei, L., and J. I. Flock. 2001. Functional study of antibodies against a fibrogenin-binding protein in Staphylococcus epidermidis adherence to polyethylene catheters. J. Infect. Dis. 18452-55. [DOI] [PubMed] [Google Scholar]
- 23.Pintens, V., C. Massonet, R. Merckx, S. Vandecasteele, W. E. Peetermans, J. K. M. Knobloch, and J. Van Eldere. 2008. The role of sigma(B) in persistence of Staphylococcus epidermidis foreign body infection. Microbiology 1542827-2836. [DOI] [PubMed] [Google Scholar]
- 24.Rennermalm, A., M. Nilsson, and J. I. Flock. 2004. The fibrinogen binding protein of Staphylococcus epidermidis is a target for opsonic antibodies. Infect. Immun. 723081-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soderquist, B. 2007. Surgical site infections in cardiac surgery: microbiology. APMIS 1151008-1011. [DOI] [PubMed] [Google Scholar]
- 26.Sun, D. Q., M. A. Accavitti, and J. D. Bryers. 2005. Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin. Diagn. Lab. Immunol. 1293-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vadyvaloo, V., and M. Otto. 2005. Molecular genetics of Staphylococcus epidermidis biofilms on indwelling medical devices. Int. J. Artif. Organs 281069-1078. [DOI] [PubMed] [Google Scholar]
- 28.Vandecasteele, S. J., W. E. Peetermans, A. Carbonez, and J. Van Eldere. 2004. Metabolic activity of Staphylococcus epidermidis is high during initial and low during late experimental foreign-body infection. J. Bacteriol. 1862236-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, and J. Van Eldere. 2001. Quantification of the expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J. Bacteriol. 1837094-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, and J. Van Eldere. 2003. Expression of biofilm-associated genes in Staphylococcus epidermidis during in vitro and in vivo foreign body infections. J. Infect. Dis. 188730-737. [DOI] [PubMed] [Google Scholar]
- 31.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, M. Van Ranst, and J. Van Eldere. 2002. Use of gDNA as internal standard for gene expression in staphylococci in vitro and in vivo. Biochem. Biophys. Res. Commun. 291528-534. [DOI] [PubMed] [Google Scholar]
- 32.Van Wijngaerden, E., W. E. Peetermans, J. Vandersmissen, S. Van Lierde, H. Bobbaers, and J. Van Eldere. 1999. Foreign body infection: a new rat model for prophylaxis and treatment. J. Antimicrob. Chemother. 44669-674. [DOI] [PubMed] [Google Scholar]
- 33.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188706-718. [DOI] [PubMed] [Google Scholar]
- 34.Vuong, C., and M. Otto. 2002. Staphylococcus epidermidis infections. Microbes Infect. 4481-489. [DOI] [PubMed] [Google Scholar]
- 35.Williams, R. J., B. Henderson, L. J. Sharp, and S. P. Nair. 2002. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 706805-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao, Y., D. E. Sturdevant, A. Villaruz, L. Xu, Q. Gao, and M. Otto. 2005. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect. Immun. 731856-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 491577-1593. [DOI] [PubMed] [Google Scholar]