Abstract
Sialic acids comprise a family of nine-carbon ketosugars that are ubiquitous on mammalian mucous membranes. However, sialic acids have a limited distribution among Bacteria and are confined mainly to pathogenic and commensal species. Vibrio pathogenicity island 2 (VPI-2), a 57-kb region found exclusively among pathogenic strains of Vibrio cholerae, contains a cluster of genes (nan-nag) putatively involved in the scavenging (nanH), transport (dctPQM), and catabolism (nanA, nanE, nanK, and nagA) of sialic acid. The capacity to utilize sialic acid as a carbon and energy source might confer an advantage to V. cholerae in the mucus-rich environment of the gut, where sialic acid availability is extensive. In this study, we show that V. cholerae can utilize sialic acid as a sole carbon source. We demonstrate that the genes involved in the utilization of sialic acid are located within the nan-nag region of VPI-2 by complementation of Escherichia coli mutants and gene knockouts in V. cholerae N16961. We show that nanH, dctP, nanA, and nanK are highly expressed in V. cholerae grown on sialic acid. By using the infant mouse model of infection, we show that V. cholerae ΔnanA strain SAM1776 is defective in early intestinal colonization stages. In addition, SAM1776 shows a decrease in the competitive index in colonization-competition assays comparing the mutant strain with both O1 El Tor and classical strains. Our data indicate an important relationship between the catabolism of sialic acid and bacterial pathogenesis, stressing the relevance of the utilization of the resources found in the host's environment.
Cholera, a severe diarrheal disease, is caused by the gram-negative bacterium Vibrio cholerae, a natural inhabitant of brackish and estuarine waters (17, 18, 39). Only two serogroups, O1 and O139, are known to cause epidemic cholera, and only the O1 serogroup is associated with the seven pandemics of the disease (22, 40, 60). In 1992 a new non-O1 serovar emerged, designated O139 Bengal, causing cholera outbreaks in India and Bangladesh (16, 59). Between 1994 and 1996 El Tor O1 serogroups reemerged as the predominant cause of cholera on the Indian subcontinent; the O139 serogroup has remained relatively quiescent since, except for an outbreak in Dhaka, Bangladesh, in 2002 (1, 22). There are a number of virulence factors required for V. cholerae to cause the disease. These include cholera toxin, which is the main cause of the profuse diarrhea and is encoded within the CTXphi phage, and the toxin-coregulated pilus (TCP), which is an essential colonization factor and is encoded within the Vibrio pathogenicity island (VPI) (also called the TCP island) (41, 43, 53, 70, 79). Another virulence factor of V. cholerae is neuraminidase (NanH), an extracellular enzyme that unmasks the receptors of cholera toxin and is also encoded within an integrative element named VPI-2 (24, 31, 34, 68).
VPI-2 is a 57-kb integrative element identified in V. cholerae O1 serogroup isolates (34, 35). The canonical VPI-2 region is present only in V. cholerae O1 strains, and the majority of the region has been deleted from O139 serogroup isolates (34). VPI-2 is inserted at a tRNA serine locus (VC1757.1) and is known to excise from the chromosome at a low rate to form a circular intermediate (48). VPI-2 encompasses loci VC1758 to VC1809, containing genes putatively involved in the transport and catabolism of sialic acid (nan-nag region), a restriction modification system, an integrase, and a Mu-phage like region (34). The nanH gene, encoding neuraminidase, is located directly downstream of the nan-nag region within VPI-2. NanH has the ability to cleave two sialic acid groups from the trisialogangliosides found in the intestinal mucus, releasing sialic acid into the environment (24, 69).
Sialic acids are a family of nine carbon ketosugars primarily found at terminal positions of numerous glycoconjugates on the surfaces of mammalian and avian cells, where they mediate a diverse range of cell-cell and cell-molecule interactions (61, 71, 75). Originally, it was thought that sialic acid was absent from prokaryotic cells (6). More recent data demonstrate that several bacterial pathogens, such as Campylobacter jejuni, enterohemorrhagic Escherichia coli, Haemophilus influenzae, Haemophilus ducreyi, Neisseria gonorrhoeae, Neisseria meningitidis, Pasteurella multocida, and Streptococcus agalactiae, can put sialic acid residues on their cell surface (sialylate) as a method of masking the bacterial cell from the host immune system (8, 10, 12, 13, 26, 27, 29, 32, 37, 38, 44, 52, 58, 63, 66, 67, 73, 74, 78). These pathogens use different mechanisms to acquired sialic acid, which include de novo biosynthesis (E. coli and N. meningitidis), scavenging (N. gonorrhoeae), or precursor scavenging (H. influenzae) (52, 73, 74, 78). Pathogenic bacteria such as Clostridium perfringens, E. coli O157:H7, H. influenzae, and P. multocida also use sialic acid as a carbon and nitrogen source by scavenging it from the surrounding environment (15, 45, 62, 66, 67, 75). Evolutionary analysis of nearly 2,000 bacterial genome sequences found that the genes required for sialic acid catabolism are confined to commensal or pathogenic species. Specifically, most of these species colonize sialic acid-rich areas such as the gut and lungs, which suggests that sialic acid catabolism might play an important role in survival in vivo (3).
The catabolic pathway of sialic acid consists of three core enzymes (the Nan cluster): N-acetylneuraminic acid lyase, encoded by nanA, which cleaves sialic acid to release N-acetylmannosamine (NAM) and pyruvate; NAM kinase, encoded by nanK, which adds a phosphate group to NAM; and NAM-6-phosphate epimerase, encoded by nanE, which converts NAM-6-phosphate into N-acetylglucosamine-6-phosphate. Two additional enzymes, NagA and NagB, complete the catabolic pathway of sialic acid in bacteria (19, 45, 56, 75-77). The uptake of sialic acid can be performed by three well-characterized transporters: NanT, a single-component system, which belongs to the major facilitator superfamily; an ATP binding cassette (ABC) transporter; or a tripartite ATP-independent periplasmic C4-dicarboxylate (TRAP) transport system, which consists of a periplasmic binding receptor (DctP) and two integral membrane proteins (DctM and DctQ) (57, 64, 75). To date, there is a lack of information regarding the possible role of sialic acid catabolism and pathogenicity (64). Also, the ability of V. cholerae to catabolize sialic acid has not been investigated yet. Here, we hypothesize that in the mucus-rich environment of the gut, V. cholerae strains with the ability to degrade sialic acid encounter a competitive advantage over those isolates that cannot utilize the aminosugar as a carbon source.
Toward that end, we demonstrate that V. cholerae can utilize sialic acid as a carbon and energy source. We determined that the genes involved in sialic acid catabolism are encoded within VPI-2 by using complementation analysis of E. coli mutants and knockout mutation analysis in V. cholerae. We found a significant increase in the expression of nanH, nanA, and nanK, as well as dctP, which is part of a putative TRAP transporter, and rpiR, a putative regulator, when sialic acid is utilized as a sole carbon source. We examined the infection dynamics of wild-type V. cholerae N16961 and SAM1776, a ΔnanA mutant strain, and demonstrate a significant growth disadvantage in the early stages of intestinal colonization for SAM1776. We performed competition assays using the infant mouse model and show that SAM1776 has a competitive index of 0.06 compared to the wild-type strain and over a fivefold decrease in the competitive index compared to an O1 classical strain. Overall, our data document, in vitro and in vivo, the importance of the ability of V. cholerae to utilize sialic acid as a carbon and energy source, suggesting a significant role in increasing survival and fitness in the host.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. All genetic manipulations utilized Escherichia coli strains DH5α λpir, the β2155 λpir diaminopimelic acid (DAP) auxotroph, and S171-1 λpir. Unless otherwise stated, bacteria were grown on Luria-Bertani broth (LB) or M9 minimal medium (M9) at 37°C with aeration. The E. coli β2155 DAP auxotroph was cultured on medium containing 1 mM DAP (Fluka). The antibiotics added to LB had the following concentrations: streptomycin (Sm), 100 μg/ml; chloramphenicol (Cm), 25 μg/ml; and ampicillin, 25 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotype | Reference |
|---|---|---|
| V. cholerae strains | ||
| N16961 | O1 El Tor, VPI-2+, Smr | 30 |
| O395 | O1 classical, VPI-2+, Smr | 46 |
| MO10 | O139, VPI-2−, Smr | 80 |
| LAC1 | O395, ΔlacZ, Smr | 25 |
| SAM2338 | N16961, ΔlacZ, gfp, Smr | This study |
| SAM1776 | N16961, ΔVC1776, Smr | This study |
| SAM1781 | N16961, ΔVC1781, Smr | This study |
| SAM1782 | N16961, ΔVC1782, Smr | This study |
| SAM1776C | SAM1776, pSAM1776C, Smr, Cmr | This study |
| SAM1781C | SAM1781, pSAM1781C, Smr, Cmr | This study |
| SAM1782C | SAM1782, pSAM1782C, Smr, Cmr | This study |
| SAM1781C2 | SAM1781, pSAM1781-82C, Smr, Cmr | This study |
| E. coli strains | ||
| BW25113 | Wild-type strain | 20 |
| JW3194 | BW25113, ΔnanA, Knr | 9 |
| JW3192 | BW25113, ΔnanE, Knr | 9 |
| JW5538 | BW25113, ΔnanK, Knr | 9 |
| JW3194C | JW3194, pSAM1776C, Knr, Cmr | This study |
| JW3192C | JW3192, pSAM1781C, Knr, Cmr | This study |
| JW5538C | JW5538, pSAM1782C, Knr, Cmr | This study |
| JW3192C2 | JW3192, pSAM1781-82C, Knr, Cmr | This study |
| Plasmids | ||
| pDS132 | Suicide plasmid, Cmr, SacB | 55 |
| pBBR1MCS | Expression plasmid, Cmr | 42 |
| pJZ111 | pCVD442, plac::gfp::lacZ, Cmr | J. Zhu |
| pSAM1776 | ΔVC1776 cloned into pDS132 | This study |
| pSAM1781 | ΔVC1781 cloned into pDS132 | This study |
| pSAM1782 | ΔVC1782 cloned into pDS132 | This study |
| pSAM1776C | VC1776 cloned into pBBR1MCS | This study |
| pSAM1781C | VC1781 cloned into pBBR1MCS | This study |
| pSAM1782C | VC1782 cloned into pBBR1MCS | This study |
| pSAM1781-82C | VC1781 and VC1782 cloned into pBBR1MCS | This study |
Growth analysis.
Precultures of either wild-type or mutant strains were grown to stationary phase at 37°C in LB, and 100 μl of these overnight cultures was added to 5 ml of fresh M9 minimal medium supplemented with N-acetylneuraminic acid (1 mg/ml) or d-glucose (1 mg/ml) as indicated (Sigma). Growth assays were done in triplicate on at least three occasions by adding 200 μl of the inoculated medium per well to a 96-well microtiter plate and incubating at 37°C under shaking conditions. Optical densities were measured at various time points using a Genios microplate reader and Magellan plate reader software (Tecan). Sigmaplot software was used to construct graphs based on the data obtained.
In silico analysis.
We performed a BLAST search (blastp) against the sequenced genome of V. cholerae N16961 using as seeds the sequences of proteins known to be involved in the metabolism of sialic acid in other bacteria (4, 5, 30). The cutoff value for our search was 1e−20 except for those genes that were already annotated as the seed we were using.
RNA extraction and quantitative real-time PCR.
Total RNA from V. cholerae N16961 was extracted at 1 h and 3 h postinoculation on M9 supplemented with sialic acid or glucose using RNAprotect Bacteria reagent (Qiagen) and an RNeasy mini kit (Qiagen); all samples were tested in triplicate. After confirmation of RNA quality, samples were DNase treated (DNA-free; Ambion) and then underwent a round of reverse transcription using SuperScript II reverse transcriptase (Invitrogen). The cDNA samples were diluted to a concentration of 4 ng/μl to perform quantitative PCR using SYBR green PCR Master Mix on an ABI Prism 7000 real-time PCR apparatus (Applied Biosystems); all assays were performed in triplicate at least twice. The gene-specific primers were designed using Primer3 software according to the real-time PCR guidelines and are listed in Table 2. The data were analyzed using ABI Prism 7000 SDS software (Applied Biosystems), and differences in the ratio of expression were extrapolated using the ΔΔCT method (54). Plots were made using Sigma Plot.
TABLE 2.
Oligonucleotides used in this study
| Function and oligonucleotide | Sequence (5′→3′) | Annealing temp | Size (bp) |
|---|---|---|---|
| Splice overlap extension PCR | |||
| VC1776A | GTTCGGTATTCGAGCGCAATCG | 63 | 319 |
| VC1776B | GTGTTGCGGCAGGTAAAGTTGC | 63 | |
| VC1776C | GCAACTTTACCTGCCGCAACACCATAAGCACCTTTCACTCC | 63 | 368 |
| VC1776D | ACCTAATGATTGGCATTCTTACCC | 63 | |
| VC1781A | CGTCGCAGCATTGACAGAAGC | 61 | 332 |
| VC1781B | TACAGGTTGTATGGAAACGACAG | 61 | |
| VC1781C | CTGTCGTTTCCATACAACCTGTAGCGGAAGGTCGATAC | 61 | 318 |
| VC1781D | CCTTCTGGCGTTACATAACCTG | 61 | |
| VC1782A | TGGCGGAAGGTCGATACAATAC | 61 | 313 |
| VC1782B | CCTTCTGGCGTTACATAACCTG | 61 | |
| VC1782C | CAGGTTATGTAACGCCAGAAGGGAATTGGTCTCGCAGAG | 61 | 345 |
| VC1782D | CAATACCCGCAAGAGTCATGTC | 61 | |
| Flanking | |||
| VC1776FF | ATAGCGACCGACGATACTGG | 57 | 2075 |
| VC1776FR | CAACTGAAGCGGCTGCTGTT | 57 | |
| VC1781FF | GCACGCCAGTTGAACTTCTG | 57 | 1878 |
| VC1781FR | CGTTACCTGAAGCCATGGAC | 57 | |
| VC1782FF | AGCGTGACTTGCCTGATAGC | 57 | 1853 |
| VC1782FR | CCGCGACCGTTACAAGAACT | 57 | |
| Complements | |||
| VC1776CF | CTCGAGACGGACAGTAGTTGAACTA | 49 | 971 |
| VC1776CR | GAGGCTCGATGAATATTCCTCCCTAG | 49 | |
| VC1781CF | CTCGAGGTTCATCAAGTCAGGAATTA | 49 | 785 |
| VC1781CR | GAGGCTCTGTTCCGCCGATATCGAT | 49 | |
| VC1782CF | CTCGAGCTCAATGGTTCAATAACGC | 49 | 926 |
| VC1782CR | GAGGCTCTTGCCTTTAATGCCATCGT | 49 | |
| Real-time PCR | |||
| VC1775QF | GTAGAACCTGAGCTCGATATTG | 62 | 152 |
| VC1775QR | CGACCGACGATACTGGATGC | 62 | |
| VC1784QF | CGTCCATTGTAGCAAGTAGCGTAA | 65 | 148 |
| VC1784QR | TCGGTATCCCAAGTTATACCGCC | 65 | |
| VC1779QF | TGATGATCGTGCCATGCTTCAGC | 65 | 123 |
| VC1779QR | TTCGCGACATAAGGGAGCATGAC | 65 | |
| VC1776QF | AGGAGTGAAAGGTGCTTATGTCTG | 65 | 114 |
| VC1776QR | CATCTAACTTCCCATCAACGGCTT | 65 | |
| VC1782QF | AACAGGTTATGTAACGCCAGAAGG | 65 | 128 |
| VC1782QR | TGCATCATTAAGAATGGAGACTGGT | 65 | |
| VC1781QF | ACTGTCGTTTCCATACAACCTGTAA | 65 | 124 |
| VC1781QR | ACGTTATTCACACCTTCAATGCGC | 65 | |
| VC0328QF | ATCGAGCGTAACGTAGCGGTTGA | 65 | 153 |
| VC0328QR | AGTCAGGTTGTAGATGTCGATACC | 65 |
Complementation of E. coli JW3194, JW3192, and JW5538.
E. coli JW3194, a ΔnanA mutant, was complemented with the nanA gene (VC1776) from V. cholerae N16961 amplified by PCR using primer pairs listed in Table 2 and cloned into pBBR1MCS using standard procedures, resulting in pSAM1776C (Table 2). The transformed E. coli JW3194/pSAM1776C strain (JW3194C) was grown on M9 medium supplemented with sialic acid. To complement E. coli JW3192, a ΔnanE mutant, and JW5538, a ΔnanK mutant, the nanE (VC1781) and nanK (VC1782) genes from V. cholerae N16961 were amplified from N16961 using primer pairs listed in Table 2 and cloned into these strains following the same procedure. Growth was measured as described in “Growth analysis” above.
Construction of V. cholerae N16961 ΔnanA, ΔnanK, ΔnanE, and ΔlacZ mutants.
Using the V. cholerae N16961 genome sequence as a template, primers were designed to perform splice overlap extension PCR and obtain single-knockout mutants for VC1776 (nanA), VC1781 (nanE), and VC1782 (nanK). We constructed 267-bp, 354-bp, and 251-bp truncated versions of the three genes, respectively, as previously described (Table 2) (33, 48). Briefly, the ΔnanA construct was cloned into pDS132 to construct pSAM1776, which was then electroporated into E. coli DH5α and subsequently into E. coli β2155 λpir. E. coli β2155 λpir bearing pSAM1776 was cross-streaked with V. cholerae N16961 on LB plates supplemented with DAP to allow conjugation to occur. Exconjugants were replated on LB containing Sm and Cm. Single colonies were screened for single-crossover mutants using flanking primers and primers A and D from each of the genes (Table 2). Single-crossover mutants were inoculated on LB without antibiotics. Serial dilutions of the overnight culture were plated on LB-10% sucrose without NaCl and incubated at 30°C (47). Single colonies were screened by PCR, and products were purified and confirmed by sequencing. The same procedure was applied to construct V. cholerae SAM1781, a ΔnanE mutant, and SAM1782, a ΔnanK mutant. In order to complement V. cholerae SAM1776, pSAM1776C was electroporated into E. coli S17-1 λpir, and the cells were plated on LB containing Cm. The positive clones were cross-streaked with V. cholerae SAM1776 (ΔnanA). Cells with the plasmids were selected by plating on LB containing Sm and Cm. The same procedure was used for complementation of V. cholerae SAM1781 and SAM1782. SAM2338, a lacZ mutant of N16961, was constructed by using pJZ111 (a kind gift from J. Zhu), which exchanges lacZ for a gene encoding green fluorescent protein as described previously (11). Growth was measured as described in “Growth analysis” above.
Mouse infection and competition studies.
Stationary-phase cells were prepared by overnight growth at 37°C for V. cholerae N16961, SAM2338, and SAM1776. V. cholerae LAC1, an O1 classical lacZ-negative strain, was incubated on LB broth at 30°C. For the in vitro competition assays, 1:1,000 dilutions from N16961, SAM1776, SAM2338, and LAC1 were mixed 1:1 according to their optical densities (SAM1776 and SAM2338, N16961 and LAC1, and SAM1776 and LAC1), and 100 μl was added to 5 ml of LB. The cultures were grown overnight at 37°C with aeration. Serial dilutions were plated on LB containing Sm and supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml; Fischer), and recovered CFU were counted for the in vitro competitive index (see below). The experiments were performed in triplicate at least twice. For the in vivo assays, 3- to 5-day-old CD-1 mice were orogastrically infected with V. cholerae as previously done by others (7). We used a total of six mice per group and performed the experiments at least twice. For the infection assay, CD-1 mice were inoculated with 50 μl of a 1:1,000 dilution of either N16961 or SAM1776. Mice were sacrificed at 1, 3, 6, 9, 12, and 24 h postinfection, and their small intestines were removed and homogenized and serial dilutions plated on LB containing Sm. Next, we performed in vivo competition assays. For these experiments, we infected each mouse using 50 μl of strain mixtures of SAM1776 and SAM2338, N16961 and LAC1, and SAM1776 and LAC1. We used a total of eight mice per group and performed the experiments at least twice. Mice were sacrificed at 24 h postinfection, and their small intestines were removed and homogenized and serial dilutions plated on LB containing Sm and supplemented with X-Gal (40 μg/ml; Fischer). Viable CFU were counted for the in vivo competitive index. The competitive indices were calculated as described by Osorio et al. (51).
RESULTS AND DISCUSSION
V. cholerae N16961 loci involved in sialic acid catabolism.
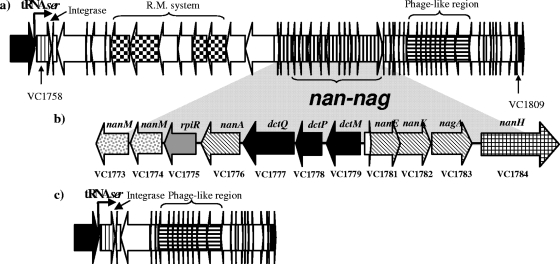
Initially sialic acid metabolism was thought to be confined to higher eukaryotes and metazoans. However, more recent data have shown that several bacterial human pathogens coat their surfaces with sialic acid to mask them from the host immune systems (75). Bacteria such as Haemophilus influenzae, Neisseria meningitidis, and E. coli K1 can either synthesize sialic acid de novo or scavenge sialic acid from the mammalian host (64, 75). We performed a BLAST search against the published genome of V. cholerae N16961 in order to identify putative homologues of previously characterized genes that are known to be involved in the metabolism of sialic acid in Bacteria (Table 3) (4, 5, 30). We did not find homologues of neuC, neuB, and neuA from E. coli, which are required for the synthesis of sialic acid and the addition of CMP to sialic acid before transfer by sialyltransferases to the bacterial cell surface (75). No homologues of the two known bacterial sialyltransferases (Lst and LsgB) was present within V. cholerae N16961 genome (Table 3) (23, 28, 82). However, a small portion at the 3′ end of Lst, between amino acids 498 and 664, showed high similarity with VC0727, a 236-amino-acid protein annotated as transcriptional regulator PhoU; the possible relationship between these two proteins remains to be elucidated (Table 3). As is well known, V. cholerae pathogenic isolates encode a neuraminidase (NanH), which unmasks the receptors of the cholera toxin (24, 31). NanH (VC1784) is located within VPI-2, and adjacent to this gene are four genes encoding enzymes in the sialic acid catabolic pathway: nanA (VC1776), nanK (VC1782), nanE (VC1781), and nagA (VC1783) (Fig. 1, Table 3) (34, 35). Homologues of the NanT and NanR proteins from E. coli, which function as a sialic acid transporter and a regulator, respectively (56, 76, 77), were not identified. However, a gene for a putative TRAP transporter was identified in the BLAST search; this clustered with the catabolic genes VC1778 (dctQ), VC1779 (dctP), and VC1777 (dctM) and shared similarity to a sialic acid TRAP transporter from H. influenzae (2, 66, 67). In addition, a putative homologue of rpiR (VC1775), which in H. influenzae is a negative regulator of the sialic acid catabolism gene cluster, was present within the nan-nag region (75). Finally, two genes putatively encoding two sialic acid mutarotases, which are involved in the epimerization of α-N-acetylneuraminic acid into β-N-acetylneuraminic acid, were found adjacent to VC1775 (65) (Table 3).
TABLE 3.
Known gene products involved in sialic acid metabolism in bacteria and their homologues in V. cholerae
| Homologue | Sourcea | Accession no. | V. cholerae locus | % Identity | E value | Function |
|---|---|---|---|---|---|---|
| NeuA | A | YP_854393 | None | Neu5Ac CMP transferase | ||
| NeuB | A | YP_854394 | None | Neu5Ac synthase | ||
| NeuC | A | YP_854392 | None | UDP-GlcNAc 2-epimerase | ||
| Lst | B | BAA25316 | None | α2,6-Sialyltransferase | ||
| LsgB | C | YP_249415 | None | α2,3-Sialyltransferase | ||
| NanH | NP_231419 | VC1784 | 100 | 0.00E+00 | Neuraminidase | |
| NanA | A | YP_858833 | VC1776 | 29 | 5.00E−25 | Neu5Ac aldolase |
| NanK | A | YP_858830 | VC1782 | 40 | 8.00E−41 | ManNAc kinase |
| NanE | A | YP_858831 | VC1781 | 57 | 9.00E−50 | ManNAc-6-P 2-epimerase |
| NagA | A | YP_851790 | VC1783 | 33 | 1.00E−54 | GlcNAc-6-P deacetylase |
| NagA | A | YP_851790 | VC0994 | 61 | 1.00E−135 | GlcNAc-6-P deacetylase |
| NagB | A | YP_851791 | VCA1025 | 79 | 8.00E−119 | GlcN-6-P deaminase |
| DctQ | C | YP_247714 | VC1778 | 20 | 1.00E−06 | TRAP transporter small permease |
| DctP | C | YP_247715 | VC1779 | 48 | 6.00E−82 | TRAP transporter periplasmic component |
| DctM | C | YP_247713 | VC1777 | 37 | 3.00E−48 | TRAP transporter large permease |
| NanT | A | YP_858832 | None | MFS transporter | ||
| NanR | A | YP_858834 | None | Sialic acid metabolism regulator | ||
| RpiR | D | YP_718908 | VC1775 | 29 | 5.00E−26 | Sialic acid metabolism regulator |
| NanM | A | AP_004802 | VC1773 | 29 | 7E−46 | Neu5Ac mutarotase |
| NanM | A | AP_004802 | VC1774 | 35 | 6E−60 | Neu5Ac mutarotase |
FIG. 1.
Schematic diagram of VPI-2 and the nan-nag region in V. cholerae N16961. (a) VPI-2. Black arrows, core genes; bent black arrow, the tRNASer insertion site of VPI-2; thin vertically striped arrow, integrase; checkerboard arrow, restriction-modification system; bold vertically striped arrows, nan-nag region; horizontally striped arrows, phage-like region. (b) nan-nag region. Stippled arrows, mutarotases; light gray arrow, putative regulator; hatched arrows, putative genes involved in the catabolism of sialic acid; black arrows, putative genes involved in the transport of sialic acid; grid pattern arrow, neuraminidase. Above each arrow, the name of the putative gene is indicated. Below each arrow, the locus tag of the gene is indicated. (c) Structure of VPI-2 in V. cholerae MO10 (O139 strain). Black arrows, core genes; bent black arrow, the tRNASer insertion site of VPI-2; thin vertically striped arrow, integrase; horizontally striped arrows, phage-like region.
In sum, we found that V. cholerae N16961 is genetically equipped to scavenge (nanH), epimerize (nanM), transport (dctPQM), and catabolize (nanA, nanE, nanK, and nagAB) sialic acid. Interestingly, V. cholerae is the only sequenced member of the family Vibrionaceae that does not contain the genes required for the synthesis of sialic acid and also is the only species that encodes a neuraminidase.
Growth of V. cholerae N16961, O395, and MO10 on sialic acid as a sole carbon and energy source.
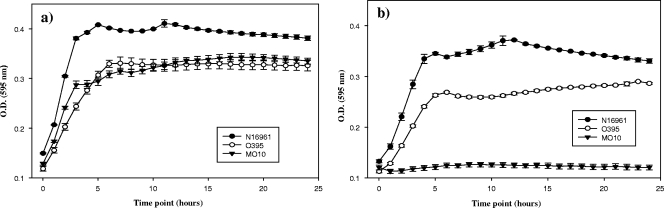
The in silico analysis indicates that V. cholerae N16961 carries the genes for the catabolism of sialic acid within VPI-2. Our next step was to determine whether V. cholerae was able to utilize sialic acid as a sole carbon and energy source. First we determined the ability of three V. cholerae strains to grow on M9 minimal medium supplemented with glucose or sialic acid (Fig. 2). V. cholerae N16961 (an O1 El Tor VPI-2-positive strain), O395 (an O1 classical VPI-2-positive strain), and MO10 (an O139 VPI-2-negative strain) all grew on M9 plus glucose, showing similar growth patterns and reaching final optical densities of between 0.33 and 0.4 (Fig. 2a). However, only N16961 and O395 grew on M9 plus sialic acid (Fig. 2b). This finding shows that V. cholerae VPI-2-positive strains are able to utilize sialic acid as a sole carbon and energy source, whereas the VPI-2-negative strain is not. Our data add to the limited list of bacterial pathogens and commensals that have been shown to utilize sialic acid as a carbon and energy source, such as C. perfringens, E. coli K-12, E. coli O157:H7, Haemophilus influenzae, and Pasteurella multocida (49, 66, 67, 76, 77).
FIG. 2.
Growth of different V. cholerae strains on minimal medium supplemented with glucose or sialic acid. The strains were incubated at 37°C under aerobic conditions on M9 minimal medium supplemented with glucose (a) or sialic acid (b). N16961 and O395 are VPI-2 positive, whereas MO10 is VPI-2 negative. Plots are represented on natural log scale. O.D., optical density. Error bars indicate standard deviations.
Differential expression of the genes within the nan-nag region.
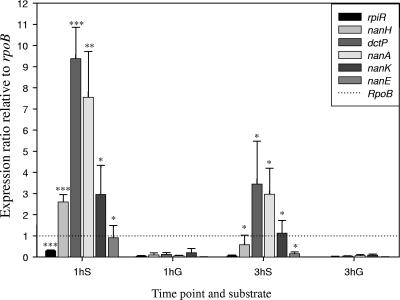
The in silico BLAST search and in vitro growth assay strongly indicated that the genes involved in the degradation of sialic acid are carried within the nan-nag region of VPI-2. We examined whether the genes in the nan-nag region are expressed in the presence of glucose or sialic acid to determine if their expression was constitutive or induced. We investigated the expression of six genes, all within VPI-2: rpiR, encoding a putative regulator; nanH, encoding neuraminidase; dctP, encoding a putative transporter protein; and nanA, nanK, and nanE, encoding homologues of sialic acid catabolism proteins (Table 3). We isolated RNA from cultures of V. cholerae N16961 at 37°C with aeration on M9 supplemented with glucose or M9 supplemented with sialic acid. Real-time PCR was performed, and the six genes showed little expression on M9 supplemented with glucose at both 1 h and 3 h postinoculation compared to rpoB (Fig. 3). In contrast, N16961 cultured for 1 h and 3 h on M9 plus sialic acid showed markedly increased levels of expression of all six genes compared with the expression levels on M9 supplemented with glucose (Fig. 3). Two of the most noticeable differences in the expression ratios are those of dctP, a component of the putative TRAP transporter, with over an 85-fold increase in expression on M9 supplemented with sialic acid compared to expression on M9 supplemented with glucose, and nanA, the first gene in the catabolic pathway, with a 150-fold difference in expression on M9 supplemented with sialic acid at 1 hour postinoculation compared to expression on M9 supplemented with glucose. Again, after 3 h of growth, the expression levels of both dctP and nanA on M9 supplemented with sialic acid are significantly higher than those on M9 supplemented with glucose, with 114-fold and 49-fold increases in their expression, respectively (Fig. 3). These results indicate that the levels of expression of the genes within the nan-nag region are strongly induced in the presence of sialic acid in V. cholerae. Interestingly, differential gene expression analysis of V. cholerae collected from the luminal fluid of the ligated ileal loop at 12 h postinoculation and from stationary-phase bacteria grown in LB broth also shows significantly increased expression of the nan-nag genes within the rabbit intestine (50). It was demonstrated in Arthrobacter sialophilus and C. perfringens that the expression of neuraminidase is increased by the presence of sialic acid, which is also seen in our study of V. cholerae (49, 81). It could be argued that sialic acid may act as a signaling molecule for entrance into the small intestine and that the putative TRAP transporter acts to transmit the signal from the host milieu to the bacterial cell interior.
FIG. 3.
Transcriptional analysis of the nan-nag genes. The bars represent expression ratio of the nan-nag genes relative to the expression of rpoB as determined using real-time PCR. V. cholerae N16961 was inoculated on M9 plus sialic acid (S) and M9 plus glucose (G). The RNA was extracted at 1 h and 3 h postinoculation. The genes under study were VC1775 (rpiR), VC1784 (nanH), VC1779 (dctP), VC1776 (nanA), VC1782 (nanK), and VC1781 (nanE). The expression of rpoB was used to normalize our test genes. The paired t test was used to infer statistical significance of the differences in expression of the genes between the two substrates. *, 0.01 < P ≤ 0.1; **, 0.001 < P ≤ 0.01; ***, P ≤ 0.001. Error bars indicate standard deviations.
Complementation of E. coli JW3194, JW3192, and JW5538.
To determine further the roles of nanA (VC1776), nanE (VC1781), and nanK (VC1782), we cloned each gene from V. cholerae N16961 into the broad-host-range vector pBBR1MCS (Table 1). We electroporated each plasmid into previously characterized E. coli deletion mutants for these genes (9). E. coli JW3194 (ΔnanA) was transformed with pSAM1776C, JW5538 (ΔnanK) with pSAM1782C, and JW3192 (ΔnanE) with pSAM1781C (Table 1) (9). First, we examined growth on LB and M9 supplemented with glucose as a sole carbon source (Table 4). E. coli JW3194, JW5538, and JW3192 showed growth patterns similar to that of strain BW25113 (Table 4). Subsequently, we studied their growth on M9 supplemented with sialic acid as a sole carbon source. As expected, BW25113 is able to utilize sialic acid as a sole carbon source and therefore grows on this medium (76). Both nanA (VC1776) and nanK (VC1782) from V. cholerae N16961 complemented the E. coli mutant strains JW3194 and JW5538, showing growth on M9 supplemented with sialic acid (Table 4). It is a note of interest that E. coli JW5538, the ΔnanK mutant, does not show a complete defect in growth compared to E. coli JW3194 (9, 20, 56, 76, 77). In addition, we were unable to complement E. coli JW3192, the ΔnanE mutant strain, with nanE (VC1781) from V. cholerae, and no growth was shown on M9 supplemented with sialic acid (Table 4). A total of three different VC1781 constructs were designed and tested in E. coli JW3192, but all failed to complement (data not shown). This behavior is often shown when the mutation causes downstream effects in an adjacent gene; in this case the mutation in nanE might have affected nanK, which is likely due to overlapping of the two genes. In order to study whether the inability to complement E. coli JW3192 was due to a polar effect caused by the mutation, we cloned into pBBR1MCS the nanE and nanK operon using the primer pair VC1781F and VC1782R, generating pSAM1781-82C (Tables 1 and 2). In this case pSAM1781-82C complemented E. coli JW3192. Overall, our data indicate that the proteins encoded by VC1776, VC1781, and VC1782 have the same functions as NanA, NanE, and NanK in E. coli, respectively.
TABLE 4.
Growth of E. coli and V. cholerae strains on different media
| Species and strain | Growth ona:
|
||
|---|---|---|---|
| LB | M9+G | M9+S | |
| E. coli | |||
| BW25113 | *** | ** | ** |
| JW3194 | *** | ** | |
| JW3194C | *** | ** | ** |
| JW3192 | *** | ** | |
| JW3192C | *** | ** | |
| JW5538 | *** | ** | * |
| JW5538C | *** | ** | ** |
| JW3192C2 | *** | ** | ** |
| V. cholerae | |||
| N16961 | *** | ** | ** |
| SAM1776 | *** | ** | |
| SAM1776C | *** | ** | ** |
| SAM1781 | *** | ** | |
| SAM1781C | *** | ** | |
| SAM1782 | *** | ** | * |
| SAM1782C | *** | ** | ** |
| SAM1781C2 | *** | ** | |
M9+G, M9 supplemented with glucose; M9+S, M9 supplemented with sialic acid; ***, final optical density of >0.8; **, final optical density of <0.8; *, lagged growth.
V. cholerae SAM1776 (ΔnanA), SAM1781 (ΔnanE), and SAM1782 (ΔnanK) mutant strains.
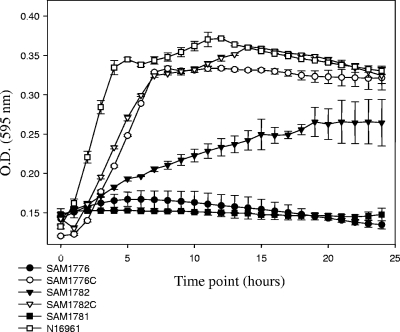
We constructed three isogenic knockout strains of V. cholerae N16961, with truncated versions of nanA, nanE, and nanK (Table 2). The three mutant strains SAM1776 (ΔnanA), SAM1781 (ΔnanE), and SAM1782 (ΔnanK) were inoculated into LB or M9 supplemented with glucose or sialic acid (Table 2; Fig. 4). All three strains grew similarly on LB and M9 supplemented with glucose (Table 4). However, neither SAM1776 nor SAM1781 grew on M9 supplemented with sialic acid, as expected (Fig. 4). V. cholerae SAM1782 (ΔnanK) showed impaired growth on M9 supplemented with sialic acid, similar to the growth pattern for E. coli JW5538 (ΔnanK), suggesting that both E. coli and V. cholerae have an additional kinase within the cell that can function similarly to NanK (Table 4).
FIG. 4.
Growth of V. cholerae sialic acid catabolism mutant strains. The constructed mutants and complements for the genes involved in the catabolism of sialic acid were incubated at 37°C under aerobic conditions. Growth was on M9 minimal medium supplemented with sialic acid. O.D., optical density. Error bars indicate standard deviations.
Previous studies have shown that the TRAP transporter encoded by dctPMQ adjacent to the nanA, nanE, and nanK genes from H. influenzae is a high-affinity transporter for sialic acid and that this can rapidly increase the concentration inside the cell, causing a toxic effect (36, 76). To determine whether the presence of sialic acid impairs the growth of V. cholerae SAM1776, our ΔnanA mutant, due to internal accumulation, we compared the growth of the wild type and SAM1776 on LB or LB supplemented with 100 μM sialic acid and found no differences in growth on both media (data not shown). These data suggest that accumulation of sialic acid is not detrimental to the cell or that V. cholerae avoids accumulation of sialic acid in a yet-unknown manner.
Complementation of the V. cholerae SAM1776 and SAM1782 transformed with pSAM1776C and pSAM1782C, respectively, was shown by growth on M9 supplemented with sialic acid (Fig. 4). Surprisingly, we were unable to complement V. cholerae SAM1781 (ΔnanE) using several different constructs, including pSAM1781-82C (data not shown). It is possible that the regulation of the Nan cluster in V. cholerae is more complex than that in E. coli and requires the presence of certain regulatory sequences for optimal expression. Overall, our data show that VC1776, VC1781, and VC1782 are required for the catabolism of sialic acid.
Sialic acid catabolism in the infection and competition dynamics of V. cholerae.
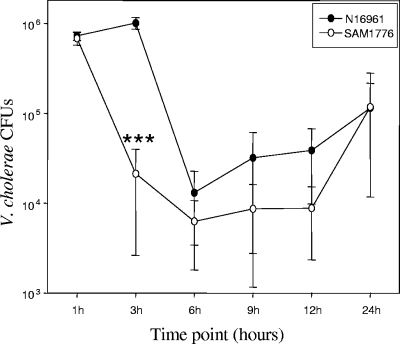
V. cholerae colonizes the heavily sialylated mucus of the human gut. The ability of V. cholerae to catabolize sialic acid as a carbon and energy source should give the organism a growth advantage compared to strains unable to utilize sialic acid. We performed single-infection assays in the infant mouse model to compare the wild type and ΔnanA mutant SAM1776. We inoculated, using the orogastric route, 3- to 5-day-old CD-1 mice with similar numbers of V. cholerae N16961 or SAM1776 organisms (∼5 × 105). We inoculated six mice per group. We extracted the small intestines at 1, 3, 6, 9, 12, and 24 h postinfection and calculated the number of viable CFU for each time point for each strain (Fig. 5). For the wild-type strain, N16961, we found numbers equivalent to those shown by Angelichio et al. (7). At 1 hour postinoculation, both strains showed very similar numbers (∼8 × 105); however, after 3 h, there was a noticeable decrease in the number of SAM1776 CFU compared to that at the 1-h time point and compared to the wild type (Fig.5). After 6 h postinoculation, the wild-type N16961 did decrease in numbers between 4.5 × 103 and 2.3 × 104. The number of CFU recovered at 9 h postinoculation increased from the previous time point, with the number of V. cholerae N16961 cells ranging between 6.7 × 103 and 2.5 × 104 and the number of SAM1776 cells ranging between 1 × 102 and 1.7 × 104 (Fig. 5). No difference were found between the counts obtained after 9 and 12 h postinoculation (Fig. 5). Finally, at 24 h postinoculation, both strains achieved similar numbers of CFU. The data show that SAM1776 is unable to maintain the same high cell density as the wild-type strain in early stages of infection. This suggests that the ability to utilize sialic acid as a carbon source can increase the likelihood of successfully colonizing a heavily populated environment such as the human gut.
FIG. 5.
Infection dynamics of V. cholerae N16961 and a sialic acid catabolism-deficient strain in the suckling mouse intestine. Three- to 5-day-old CD-1 mice were inoculated with V. cholerae N16961 or V. cholerae SAM1776 (ΔnanA). Six mice were used per strain and time point. The intestines were removed after 1, 3, 6, 9, 12, and 24 h. Serial dilutions of the homogenates were plated on LB containing Sm and the CFU counted. We used the paired t test to compare the numbers of CFU of N16961 and SΑΜ1776 at the same time point. ***, P < 0.0005. Error bars indicate standard deviations.
To test this further, we performed in vitro and in vivo competition assays between V. cholerae SAM2338 (N16961 ΔlacZ) and SAM1776 (ΔnanA). We also compared the competition indices of N16961 and SAM1776 with that of LAC1, a ΔlacZ derivative of O395, a classical strain, in order to see whether the inability of SAM1776 to utilize sialic acid had an effect in comparison to other V. cholerae strains rather than V. cholerae O1 El Tor. We inoculated orogastrically a total of eight CD-1 infant mice per group with mixtures of SAM1776 and SAM2338, N16961 and LAC1, or SAM1776 and LAC1. After overnight incubation, mice were sacrificed and intestinal numbers calculated. We found that the competitive index of V. cholerae SAM1776 versus SAM2338 was 0.06 (1/16.6), showing a 17-fold decrease; that of N16961 versus LAC1 was 0.17 (1/5.9), which is a 6-fold decrease; and that of SAM1776 versus LAC1 was 0.03 (1/33.3), indicating a 33-fold decrease. These data support the hypothesis that the ability of V. cholerae pathogenic strains to utilize sialic acid as a carbon source gives them a competitive advantage in the sialic acid-rich environment of the gut.
Conclusions.
The diversity in the ways by which different bacterial pathogens utilize sialic acid as a means to increase their fitness is fascinating, ranging from avoiding phagocytosis or opsonization to preventing serum killing by impeding the insertion of the complement attack complex (64, 72, 75). However, to date, no direct relationship between sialic acid catabolism and bacterial pathogenesis has been established (64, 67, 73). Nonetheless, when studying carbon nutrition of commensal E. coli strains, Chang et al. found that a strain lacking the ability to take up and degrade sialic acid yielded fewer CFU in the feces than the wild type, strongly suggesting a role for sialic acid metabolism in bacterial colonization (15). However, Fabich et al. showed that a ΔnanAT strain of pathogenic E. coli EDL933 did not show the same disadvantage as the commensal strain (21). In this study, we show for the first time that sialic acid catabolism is important in the early infection stages of V. cholerae. The ability to utilize sialic acid as a carbon source confers a competitive advantage in the mouse intestine to strains of V. cholerae that carry a functional sialic acid gene cluster. The utilization of sialic acid as a carbon source might act as a “jump start” for V. cholerae when it is trying to colonize the intestine, where competition for resources is high.
The presence of the nanA gene, encoding the key enzyme in sialic acid degradation, has been documented for several bacterial pathogens, such as H. influenzae, Mycoplasma spp., Salmonella enterica, Shigella boydii, Staphylococcus aureus, Streptococcus pneumoniae, V. vulnificus, Yersinia enterocolitica, and Yersinia pestis, as well as for a wide group of human commensals, including the numerically important groups Bacteroidetes and clostridia (3). All these bacterial species colonize heavily sialylated mucous surfaces of the host, such as the lungs and the gut, suggesting that the ability to utilize this nine-carbon amino sugar may convey a fitness advantage. It is a note of interest that in most V. cholerae O139 serogroup strains, only open reading frames VC1758, VC1759, and VC1789 to VC1809 are present, whereas neuraminidase and the sialic acid transport and catabolism region are deleted from all O139 strains recovered after 1992 (34, 35). The O139 serogroup emerged in 1992 to cause numerous severe outbreaks of cholera in the Bengal delta region, surpassing El Tor isolates as the major cause of cholera in the region; however, by the mid-1990s the El Tor biotype reemerged as the predominant cause of cholera. We suspect that the deletion of VPI-2 from O139 isolates has a detrimental effect on the fitness of these strains. Among V. cholerae non-O1 serogroup isolates that cause gastroenteritis, neuraminidase and the sialic acid transport and catabolism region are present, indicating that this may be a significant determinant in their intestinal survival (48).
Acknowledgments
We thank the reviewers for their insightful comments on the manuscript. We also thank those who kindly provided us with the V. cholerae strains used in this study. We especially thank F. Jerry Reen for technical assistance and advice. We thank Michelle Parent and Seth Blumerman, Department of Biological Sciences, University of Delaware, for help and assistance with mouse studies and analysis.
This study was supported in part by a University of Delaware Research Foundation grant (UDRF2008-2009) and a Science Foundation Ireland student fellowship (2005 to 2008) to S.A.M.
Editor: A. Camilli
Footnotes
Published ahead of print on 29 June 2009.
REFERENCES
- 1.Alam, M., N. A. Hasan, A. Sadique, N. A. Bhuiyan, K. U. Ahmed, S. Nusrin, G. B. Nair, A. K. Siddique, R. B. Sack, D. A. Sack, A. Huq, and R. R. Colwell. 2006. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl. Environ. Microbiol. 724096-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, S., A. Zaleski, J. W. Johnston, B. W. Gibson, and M. A. Apicella. 2005. Novel sialic acid transporter of Haemophilus influenzae. Infect. Immun. 735291-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almagro-Moreno, S., and E. F. Boyd. 2009. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol. Biol. 9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angata, T., and A. Varki. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102439-469. [DOI] [PubMed] [Google Scholar]
- 7.Angelichio, M. J., J. Spector, M. K. Waldor, and A. Camilli. 1999. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 673733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avril, T., E. R. Wagner, H. J. Willison, and P. R. Crocker. 2006. Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect. Immun. 744133-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer, S. H., M. Mansson, D. W. Hood, J. C. Richards, E. R. Moxon, and E. K. Schweda. 2001. A rapid and sensitive procedure for determination of 5-N-acetyl neuraminic acid in lipopolysaccharides of Haemophilus influenzae: a survey of 24 non-typeable H. influenzae strains. Carbohydr. Res. 335251-260. [DOI] [PubMed] [Google Scholar]
- 11.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 1851384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlin, A. F., A. L. Lewis, A. Varki, and V. Nizet. 2007. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J. Bacteriol. 1891231-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaffin, D. O., L. M. Mentele, and C. E. Rubens. 2005. Sialylation of group B streptococcal capsular polysaccharide is mediated by CpsK and is required for optimal capsule polymerization and expression. J. Bacteriol. 1874615-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challacombe, J. F., A. J. Duncan, T. S. Brettin, D. Bruce, O. Chertkov, J. C. Detter, C. S. Han, M. Misra, P. Richardson, R. Tapia, N. Thayer, G. Xie, and T. J. Inzana. 2007. Complete genome sequence of Haemophilus somnus (Histophilus somni) strain 129Pt and comparison to Haemophilus ducreyi 35000HP and Haemophilus influenzae Rd. J. Bacteriol. 1891890-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 1017427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cholera Working Group. 1993. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 342387-390. [PubMed] [Google Scholar]
- 17.Colwell, R. R., J. Kaper, and S. W. Joseph. 1977. Vibrio cholerae, Vibrio parahaemolyticus, and other vibrios: occurrence and distribution in Chesapeake Bay. Science 198394-396. [PubMed] [Google Scholar]
- 18.Colwell, R. R., and W. M. Spira. 1992. The ecology of Vibrio cholerae, p. 107-127. In D. Barua and W. B. Greenough III (ed.), Cholera. Plenum Medical Book Company, New York, NY.
- 19.Comb, D. G., and S. Roseman. 1960. The sialic acids. I. The structure and enzymatic synthesis of N-acetylneuraminic acid. J. Biol. Chem. 2352529-2537. [PubMed] [Google Scholar]
- 20.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabich, A. J., S. A. Jones, F. Z. Chowdhury, A. Cernosek, A. Anderson, D. Smalley, J. W. McHargue, G. A. Hightower, J. T. Smith, S. M. Autieri, M. P. Leatham, J. J. Lins, R. L. Allen, D. C. Laux, P. S. Cohen, and T. Conway. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 761143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faruque, S. M., D. A. Sack, R. B. Sack, R. R. Colwell, Y. Takeda, and G. B. Nair. 2003. Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. USA 1001304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269496-512. [DOI] [PubMed] [Google Scholar]
- 24.Galen, J. E., J. M. Ketley, A. Fasano, S. H. Richardson, S. S. Wasserman, and J. B. Kaper. 1992. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect. Immun. 60406-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 642246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert, M., J. R. Brisson, M. F. Karwaski, J. Michniewicz, A. M. Cunningham, Y. Wu, N. M. Young, and W. W. Wakarchuk. 2000. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. Identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-mhz (1)h and (13)c NMR analysis. J. Biol. Chem. 2753896-3906. [DOI] [PubMed] [Google Scholar]
- 27.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15192-198. [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 1874627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harvey, H. A., W. E. Swords, and M. A. Apicella. 2001. The mimicry of human glycolipids and glycosphingolipids by the lipooligosaccharides of pathogenic Neisseria and Haemophilus. J. Autoimmun. 16257-262. [DOI] [PubMed] [Google Scholar]
- 30.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmgren, J., I. Lonnroth, and L. Svennerholm. 1973. Fixation and inactivation of cholera toxin by GM1 ganglioside. Scand. J. Infect. Dis. 577-78. [DOI] [PubMed] [Google Scholar]
- 32.Hood, D. W., A. D. Cox, M. Gilbert, K. Makepeace, S. Walsh, M. E. Deadman, A. Cody, A. Martin, M. Mansson, E. K. Schweda, J. R. Brisson, J. C. Richards, E. R. Moxon, and W. W. Wakarchuk. 2001. Identification of a lipopolysaccharide alpha-2,3-sialyltransferase from Haemophilus influenzae. Mol. Microbiol. 39341-350. [DOI] [PubMed] [Google Scholar]
- 33.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 7761-68. [DOI] [PubMed] [Google Scholar]
- 34.Jermyn, W. S., and E. F. Boyd. 2002. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (NanH) among toxigenic Vibrio cholerae isolates. Microbiology 1483681-3693. [DOI] [PubMed] [Google Scholar]
- 35.Jermyn, W. S., and E. F. Boyd. 2005. Molecular evolution of Vibrio pathogenicity island-2 (VPI-2): mosaic structure among Vibrio cholerae and Vibrio mimicus natural isolates. Microbiology 151311-322. [DOI] [PubMed] [Google Scholar]
- 36.Johnston, J. W., A. Zaleski, S. Allen, J. M. Mootz, D. Armbruster, B. W. Gibson, M. A. Apicella, and R. S. Munson, Jr. 2007. Regulation of sialic acid transport and catabolism in Haemophilus influenzae. Mol. Microbiol. 6626-39. [DOI] [PubMed] [Google Scholar]
- 37.Jones, P. A., N. M. Samuels, N. J. Phillips, R. S. Munson, Jr., J. A. Bozue, J. A. Arseneau, W. A. Nichols, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2002. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 27714598-14611. [DOI] [PubMed] [Google Scholar]
- 38.Kahler, C. M., and D. S. Stephens. 1998. Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin). Crit. Rev. Microbiol. 24281-334. [DOI] [PubMed] [Google Scholar]
- 39.Kaper, J., H. Lockman, R. R. Colwell, and S. W. Joseph. 1979. Ecology, serology, and enterotoxin production of Vibrio cholerae in Chesapeake Bay. Appl. Environ. Microbiol. 3791-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 848-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 953134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16800-802. [PubMed] [Google Scholar]
- 43.Kovach, M. E., M. D. Shaffer, and K. M. Peterson. 1996. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology 1422165-2174. [DOI] [PubMed] [Google Scholar]
- 44.Lewis, A. L., M. E. Hensler, A. Varki, and V. Nizet. 2006. The group B streptococcal sialic acid O-acetyltransferase is encoded by neuD, a conserved component of bacterial sialic acid biosynthetic gene clusters. J. Biol. Chem. 28111186-11192. [DOI] [PubMed] [Google Scholar]
- 45.Martinez, J., S. Steenbergen, and E. Vimr. 1995. Derived structure of the putative sialic acid transporter from Escherichia coli predicts a novel sugar permease domain. J. Bacteriol. 1776005-6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mekalanos, J. J. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35253-263. [DOI] [PubMed] [Google Scholar]
- 47.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires ToxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy, R. A., and E. F. Boyd. 2008. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J. Bacteriol. 190636-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nees, S., R. Schauer, and F. Mayer. 1976. Purification and characterization of N-acetylneuraminate lyase from Clostridium perfringens. Hoppe Seyler's Z. Physiol. Chem. 357839-853. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osorio, C. G., J. A. Crawford, J. Michalski, H. Martinez-Wilson, J. B. Kaper, and A. Camilli. 2005. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsons, N. J., P. V. Patel, E. L. Tan, J. R. Andrade, C. A. Nairn, M. Goldner, J. A. Cole, and H. Smith. 1988. Cytidine 5′-monophospho-N-acetyl neuraminic acid and a low molecular weight factor from human blood cells induce lipopolysaccharide alteration in gonococci when conferring resistance to killing by human serum. Microb. Pathog. 5303-309. [DOI] [PubMed] [Google Scholar]
- 53.Pearson, G. D., A. Woods, S. L. Chiang, and J. J. Mekalanos. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. USA 903750-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Philippe, N., J. P. Alcaraz, E. Coursange, J. Geiselmann, and D. Schneider. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51246-255. [DOI] [PubMed] [Google Scholar]
- 56.Plumbridge, J., and E. Vimr. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 18147-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Post, D. M., R. Mungur, B. W. Gibson, and R. S. Munson, Jr. 2005. Identification of a novel sialic acid transporter in Haemophilus ducreyi. Infect. Immun. 736727-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ram, S., A. K. Sharma, S. D. Simpson, S. Gulati, D. P. McQuillen, M. K. Pangburn, and P. A. Rice. 1998. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J. Exp. Med. 187743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramamurthy, T., S. Garg, R. Sharma, S. K. Bhattacharya, G. B. Nair, T. Shimada, T. Takeda, T. Karasawa, H. Kurazano, A. Pal, et al. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341703-704. [DOI] [PubMed] [Google Scholar]
- 60.Sack, D. A., R. B. Sack, G. B. Nair, and A. K. Siddique. 2004. Cholera. Lancet 363223-233. [DOI] [PubMed] [Google Scholar]
- 61.Schauer, R. 2000. Achievements and challenges of sialic acid research. Glycoconj. J. 17485-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schauer, R., H. P. Buscher, and J. Casals-Stenzel. 1974. Sialic acids: their analysis and enzymic modification in relation to the synthesis of submandibular-gland glycoproteins. Biochem. Soc. Symp. 197487-116. [PubMed] [Google Scholar]
- 63.Schilling, B., S. Goon, N. M. Samuels, S. P. Gaucher, J. A. Leary, C. R. Bertozzi, and B. W. Gibson. 2001. Biosynthesis of sialylated lipooligosaccharides in Haemophilus ducreyi is dependent on exogenous sialic acid and not mannosamine. Incorporation studies using N-acylmannosamine analogues, N-glycolylneuraminic acid, and 13C-labeled N-acetylneuraminic acid. Biochemistry 4012666-12677. [DOI] [PubMed] [Google Scholar]
- 64.Severi, E., D. W. Hood, and G. H. Thomas. 2007. Sialic acid utilization by bacterial pathogens. Microbiology 1532817-2822. [DOI] [PubMed] [Google Scholar]
- 65.Severi, E., A. Muller, J. R. Potts, A. Leech, D. Williamson, K. S. Wilson, and G. H. Thomas. 2008. Sialic acid mutarotation is catalyzed by the Escherichia coli beta-propeller protein YjhT. J. Biol. Chem. 2834841-4849. [DOI] [PubMed] [Google Scholar]
- 66.Severi, E., G. Randle, P. Kivlin, K. Whitfield, R. Young, R. Moxon, D. Kelly, D. Hood, and G. H. Thomas. 2005. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 581173-1185. [DOI] [PubMed] [Google Scholar]
- 67.Steenbergen, S. M., C. A. Lichtensteiger, R. Caughlan, J. Garfinkle, T. E. Fuller, and E. R. Vimr. 2005. Sialic acid metabolism and systemic pasteurellosis. Infect. Immun. 731284-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svennerholm, L. 1964. The gangliosides. J. Lipid Res. 5145-155. [PubMed] [Google Scholar]
- 69.Taylor, G. 1996. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 6830-837. [DOI] [PubMed] [Google Scholar]
- 70.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 842833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varki, A. 1993. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 397-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vimr, E., and C. Lichtensteiger. 2002. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10254-257. [DOI] [PubMed] [Google Scholar]
- 73.Vimr, E., C. Lichtensteiger, and S. Steenbergen. 2000. Sialic acid metabolism's dual function in Haemophilus influenzae. Mol. Microbiol. 361113-1123. [DOI] [PubMed] [Google Scholar]
- 74.Vimr, E., S. Steenbergen, and M. Cieslewicz. 1995. Biosynthesis of the polysialic acid capsule in Escherichia coli K1. J. Ind. Microbiol. 15352-360. [DOI] [PubMed] [Google Scholar]
- 75.Vimr, E. R., K. A. Kalivoda, E. L. Deszo, and S. M. Steenbergen. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68132-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vimr, E. R., and F. A. Troy. 1985. Identification of an inducible catabolic system for sialic acids (Nan) in Escherichia coli. J. Bacteriol. 164845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vimr, E. R., and F. A. Troy. 1985. Regulation of sialic acid metabolism in Escherichia coli: role of N-acylneuraminate pyruvate-lyase. J. Bacteriol. 164854-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogel, U., H. Claus, G. Heinze, and M. Frosch. 1999. Role of lipopolysaccharide sialylation in serum resistance of serogroup B and C meningococcal disease isolates. Infect. Immun. 67954-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 2721910-1914. [DOI] [PubMed] [Google Scholar]
- 80.Waldor, M. K., and J. J. Mekalanos. 1994. Vibrio cholerae O139 specific gene sequences. Lancet 3431366. [DOI] [PubMed] [Google Scholar]
- 81.Wang, P., D. Schafer, C. A. Miller, S. W. Tanenbaum, and M. Flashner. 1978. Induction and regulation of neuraminidase synthesis in Arthrobacter sialophilus. J. Bacteriol. 136874-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamamoto, T., M. Nakashizuka, and I. Terada. 1998. Cloning and expression of a marine bacterial beta-galactoside alpha2,6-sialyltransferase gene from Photobacterium damsela JT0160. J. Biochem. 12394-100. [DOI] [PubMed] [Google Scholar]