Abstract
The Candida albicans plasma membrane plays critical roles in growth and virulence and as a target for antifungal drugs. Three C. albicans genes that encode Bin-Amphiphysin-Rvs homology domain proteins were mutated to define their roles in plasma membrane function. The deletion of RVS161 and RVS167, but not RVS162, caused strong defects. The rvs161Δ mutant was more defective in endocytosis and morphogenesis than rvs167Δ, but both were strongly defective in polarizing actin patches. Other plasma membrane constituents were still properly localized, including a filipin-stained domain at the hyphal tips. An analysis of growth under different in vitro conditions showed that the rvs161Δ and rvs167Δ mutants grew less invasively in agar and also suggested that they have defects in cell wall synthesis and Rim101 pathway signaling. These mutants were also more resistant to the antimicrobial peptide histatin 5 but showed essentially normal responses to the drugs caspofungin and amphotericin. Surprisingly, the rvs161Δ mutant was more sensitive to fluconazole, whereas the rvs167Δ mutant was more resistant, indicating that these mutations cause overlapping but distinct effects on cells. The rvs161Δ and rvs167Δ mutants both showed greatly reduced virulence in mice. However, the mutants were capable of growing to high levels in kidneys. Histological analyses of infected kidneys revealed that these rvsΔ mutants grew in a large fungal mass that was walled off by leukocytes, rather than forming disseminated microabscesses as seen for the wild type. The diminished virulence is likely due to a combination of the morphogenesis defects that reduce invasive growth and altered cell wall construction that exposes proinflammatory components to the host immune system.
Candida albicans is the most common human fungal pathogen and causes a wide range of infections in susceptible individuals (31). One characteristic of C. albicans is that the host milieu stimulates it to grow in different morphologies, including rounded buds, elongated pseudohyphae, and long filamentous hyphae (39). The induction of the hyphal form also correlates with the increased expression of virulence factors, such as adhesins that promote attachment to human cells and biofilm formation, secreted hydrolytic enzymes that may liberate nutrients and facilitate invasive growth, and antioxidant enzymes that counter the attack of the immune system (5, 7, 21, 47). The C. albicans plasma membrane plays special roles during these important morphological transitions. In addition to forming an important protective barrier, the plasma membrane contains components that sense the extracellular environment, induce changes in morphogenesis, and mediate virulence factor production. The plasma membrane is also involved in dynamic cellular processes, such as endocytosis and cell wall biogenesis. Consistent with its key role in infection, the plasma membrane is a target of most of the commonly used antifungal drugs (32).
Studies of Saccharomyces cerevisiae have shown that endocytosis contributes to environmental sensing and morphogenesis. The plasma membrane is not static, but rather it is continuously remodeled by the endocytic internalization of membrane constituents (13). Endocytosed lipids and proteins can be recycled back to the plasma membrane or sent to the vacuole for degradation. The stability of many nutrient receptors and transporters in the plasma membrane is regulated by endocytosis in response to changes in nutrient levels (18). Other signal transduction components can also be regulated by endocytosis, such as the ligand-induced internalization of mating pheromone receptors. Endocytosis is also thought to contribute to polarized morphogenesis by internalizing components and reinserting them at the site of cell polarization (40).
A large number of proteins contribute to the complex process of endocytosis. The roles of these proteins have been most well studied for S. cerevisiae, for which four different protein modules involved in the internalization process have been defined (19). Endocytic site assembly begins with the localization of clathrin and the formation of a coat complex in a module that also includes Pan1, End3, Sla1, and Sla2. The WASP/MYO module (Las17 and later Myo5) then interacts with the actin module ARP2/ARP3 complex to initiate actin polymerization, which drives the internalization of the actin-bound coat and attached membrane and cargo. The fourth module is comprised of the Rvs161 and Rvs167 amphiphysin homologs that bind to the invaginating tubule and promote scission to separate the vesicle from the basal plasma membrane. The trafficking of the released vesicle to its final destination then continues along actin cables within the cytoplasm.
In order to study the roles of plasma membrane organization and endocytosis in C. albicans, we analyzed the orthologs of the S. cerevisiae RVS161 and RVS167 (ScRVS161 and ScRVS167, respectively) genes. The deletion of RVS161 and RVS167 in S. cerevisiae causes a defect in the internalization phase of endocytosis with less impairment of actin function and morphogenesis than was seen with many other endocytic mutants (24, 28). The S. cerevisiae RVS genes were initially identified because their mutation caused a reduced viability upon starvation (10). Subsequent studies revealed that the Rvs proteins contain a BAR (Bin-Amphiphysin-Rvs) homology domain, which has been shown in other organisms to sense and promote membrane curvature (12, 52). The S. cerevisiae rvs mutants also display other phenotypes that may be directly or indirectly linked to their endocytosis defect, including a defect in vesicle trafficking, increased sensitivity to high salt, and decreased cell wall integrity (4, 6). The Rvs161 and Rvs167 proteins have been isolated in a detergent-resistant membrane fraction, which suggests that they localize in ergosterol- and sphingolipid-enriched plasma membrane domains that are similar to lipid rafts (16). We therefore mutated RVS161 and RVS167 in C. albicans (CaRVS161 and CaRVS167, respectively), along with a third uncharacterized BAR domain gene exhibiting similarity to S. cerevisiae RVS161, which we have termed RVS162. The results demonstrate that CaRVS161 and CaRVS167, but not CaRVS162, are important for efficient endocytosis, true hyphal filamentation, and virulence in a mouse model of hematogenously disseminated candidiasis.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains used in this study (Table 1) were grown in rich yeast extract-peptone-dextrose (YPD) medium with uridine (80 mg/liter) or in synthetic medium lacking the indicated nutrients (38). Homozygous mutant strains were constructed from the parental BWP17 strain by the sequential deletion of both copies of the targeted gene (C. albicans is diploid). Gene deletion cassettes were generated by PCR amplification of the ARG4 or HIS1 selectable marker genes using primers that also included 60 bp of DNA sequence homologous to the upstream or downstream regions of the targeted open reading frame (49). Cells which had undergone homologous recombination to delete the targeted gene were identified by PCR analysis using a combination of primers flanking sites of cassette integration and internal primers. Reconstituted strains in which the deletion mutations were complemented by the reintroduction of a wild-type copy of the targeted gene were constructed by integrating a plasmid containing the gene of interest and the URA3 selectable marker into the 5′ region of the deleted gene. Briefly, the entire open reading frame plus ∼1,000 bp upstream and 300 bp downstream were amplified by PCR and inserted into the SacI and SacII restriction sites in the plasmid pDDB57 (48). Prior to transformation into C. albicans deletion strains, complementing plasmids were linearized by restriction digestion in the promoter region for targeted integration.
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Genotype |
|---|---|---|
| BWP17 | Sc5314 | ura3Δ::λimm434 ura3Δ::λimm434 his1::hisG his1::hisG arg4::hisG arg4::hisG |
| DIC185 | BWP17 | ura3Δ::λimm434 URA3 his1::hisG HIS1 arg4::hisG ARG4 |
| YSM42 | BWP17 | BWP17, except rvs161Δ::ARG4 rvs161Δ::HIS1 |
| YLD14 | YSM42 | BWP17, except rvs161Δ::ARG4 rvs161Δ::HIS1 ura3Δ::λimm434 URA3 |
| YLD11 | YSM42 | BWP17, except rvs161Δ::ARG4 rvs161Δ::HIS1 RVS161::URA3 |
| YSM37 | BWP17 | BWP17, except RVS161-HA::URA3 |
| YLD26 | YSM42 | BWP17, except rvs161Δ::ARG4 rvs161Δ::HIS1 CDC12-GFP::URA3 |
| YLD29 | YSM42 | BWP17, except rvs161Δ::ARG4 rvs161Δ::HIS1 SUR7-GFP::URA3 |
| YLD21 | BWP17 | BWP17, except rvs162Δ::ARG4 rvs162Δ::HIS1 |
| YLD22 | YLD21 | BWP17, except rvs162Δ::ARG4 rvs162Δ::HIS1 ura3Δ::λimm434/URA3 |
| YLD25 | YLD21 | BWP17, except rvs162Δ::ARG4 rvs162Δ::HIS1 RVS162::URA3 |
| YLD19 | BWP17 | BWP17, except RVS162-HA::URA3 |
| YLD28 | YLD21 | BWP17, except rvs162Δ::ARG4 rvs162Δ::HIS1 CDC12-GFP::URA3 |
| YLD31 | YLD21 | BWP17, except rvs162Δ::ARG4 rvs162Δ::HIS1 SUR7-GFP::URA3 |
| YLD3 | BWP17 | BWP17, except rvs167Δ::ARG4 rvs161Δ::HIS1 |
| YLD16 | YLD3 | BWP17, except rvs167Δ::ARG4 rvs161Δ::HIS1 ura3Δ::λimm434 URA3 |
| YLD12 | YLD3 | BWP17, except rvs167Δ::ARG4 rvs161Δ::HIS1 RVS167::URA3 |
| YSM54 | BWP17 | BWP17, except RVS167-HA::URA3 |
| YLD27 | YLD3 | BWP17, except rvs167Δ::ARG4 rvs161Δ::HIS1 CDC12-GFP::URA3 |
| YLD30 | YLD3 | BWP17, except rvs167Δ::ARG4 rvs161Δ::HIS1 SUR7-GFP::URA3 |
| YJA15 | BWP17 | BWP17, except SUR7-GFP::URA3 |
The rvs deletion strains used in virulence assays were transformed with a URA3-containing fragment to complement the remaining auxotrophy. The plasmid pBSK-URA3 (33) was digested with restriction enzymes PstI and NotI to liberate the URA3-IRO1 sequence, which was then transformed into the mutant strains for integration by homologous recombination to restore URA3 at its native locus.
The RVS genes were tagged by the insertion of the hemagglutinin (HA) epitope via homologous recombination at the C-terminal coding region of each gene with a HA-URA3 cassette. The HA-URA3 cassette was created by PCR amplification using primers directed to the p3xHA plasmid (26) that were flanked by 60 to 80 bp of sequence homologous to the regions surrounding the stop codon. RVS deletion strains containing the SUR7-GFP fusion gene (where GFP is the green fluorescent protein gene) were derived by transformation with a DNA fragment containing SUR7-GFP and the URA3 selectable marker that was created by PCR amplification of the corresponding DNA using strain YJA15 as a template and 23-bp primers designed to anneal ∼500 bp upstream and downstream of the SUR7 open reading frame stop codon. Plasmid pAW-CDC12-GFP (45) was linearized with restriction enzyme BsaB1 and transformed into the strains for the targeted integration of GFP at the C terminus of CDC12.
Western blotting.
Log-phase cells were lysed by agitation with glass beads for 1 min in the presence of lysis buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA) and protease inhibitors (1 μg/ml pepstatin, 1 mM benzamidine, 0.5 mM phenylmethylsulfonyl fluoride). Protein levels in the extracts were quantified by a modified Bradford assay (Pierce Biotechnology, Inc.), and 100 μg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane (Whatman International Limited). The filters were incubated in blocking solution (5% powdered milk, 10 mM Tris [pH 7.5], 150 mM NaCl) and then incubated overnight at 4°C with anti-HA rabbit polyclonal antibody (Bethyl Laboratories, Montgomery, TX), followed by incubation in anti-rabbit horseradish peroxidase-conjugated antibody. The chemiluminescent detection of the cross-reacting bands was performed using a SuperSignal West Dura kit (Pierce Biotechnology, Inc.).
Fluorescence microscopy.
For the assessment of endocytosis, log-phase cells were incubated for 15 min on ice with the lipophilic fluorescent dye FM4-64 (Invitrogen, Carlsbad, CA) at a concentration of 20 μM (42). The cells were then washed, resuspended in fresh medium, and evaluated by fluorescence microscopy at 15, 30, and 60 min for dye internalization and staining of the vacuolar membrane. For actin localization (1), log-phase cells were initially fixed in 5% formaldehyde for 75 min at 30°C, followed by incubation at room temperature for 30 min in 0.1 M potassium phosphate buffer (pH 7.5) containing 0.1% Triton X-100. After the cells were washed two times in 0.1 M potassium phosphate buffer (pH 7.5), five units of rhodamine-phalloidin (Invitrogen, Carlsbad, CA) were added and the cells incubated at 4°C overnight in the dark. The cells were washed three times in 0.1 M potassium phosphate buffer (pH 7.5) and then resuspended in mounting medium containing 10 μg/ml phenylenediamine (Sigma) and examined microscopically. Fluorescent staining of cell wall chitin was carried out by incubating the cells in 100 to 500 μl of 20 ng/ml calcofluor white (Fluorescent Brightener 28; Sigma) for ∼10 min at room temperature and then viewing by fluorescence microscopy (34). Ergosterol was stained by incubating cells with 10 to 20 μg/ml filipin complex (Sigma) for 5 min, and then the cells were examined immediately by fluorescence microscopy using a DAPI (4′-6-diamidino-2-phenylindole) filter set (2, 25).
Virulence assays.
C. albicans strains were grown overnight at 30°C in YPD medium with 80 mg/liter uridine, reinoculated into fresh medium the next day, and incubated again overnight at 30°C. Mutant strain cultures were sonicated for 5 s to break up aggregates, and then the cells were washed twice in phosphate-buffered saline (PBS), counted in a hemocytometer, and diluted to 1 × 107 cells/ml with PBS. Dilutions of cells were plated onto solid YPD medium plates to confirm the quantitation. Eight female BALB/c mice were inoculated per each C. albicans strain with 1 × 106 cells into the lateral tail vein of each mouse. The mice were monitored for 28 days, during which time a mouse was considered moribund if food and water could no longer be accessed. Humane euthanasia was then performed. For an analysis of the early stages of infection, infected mice were euthanized after 2 or 3 days, as indicated in Fig. 8. Graphing and statistical analysis of survival after infection was carried out using a log-rank test (Mantel-Haenszel test) with Prism 4 (GraphPad Software, Inc., La Jolla, CA). Kidneys excised for fungal burden assessment were weighed, placed in 5 ml PBS, and homogenized for 30 s with a tissue homogenizer (Pro Scientific, Inc., Oxford, CT). Serial dilutions of the homogenate were plated onto YPD medium plates and incubated at 30°C for 2 days to determine the number of CFU per gram of kidney tissue. Statistical analysis of the CFU data was carried out with Prism software using one-way analysis of variance with a nonparametric Kruskal-Wallis test and a Dunn's posttest. Periodic acid-Schiff (PAS) and hematoxylin-eosin staining for histological analysis were performed by a commercial laboratory (McLain Laboratories, Smithtown, NY). Calcofluor white staining was carried out on kidney tissue by pelleting 500 μl of homogenate and then resuspending it in 100 to 500 μl of 20 ng/ml calcofluor white for ∼10 min. The stained tissue was then resuspended in 1% KOH, incubated for 5 min at room temperature, and examined by fluorescence microscopy.
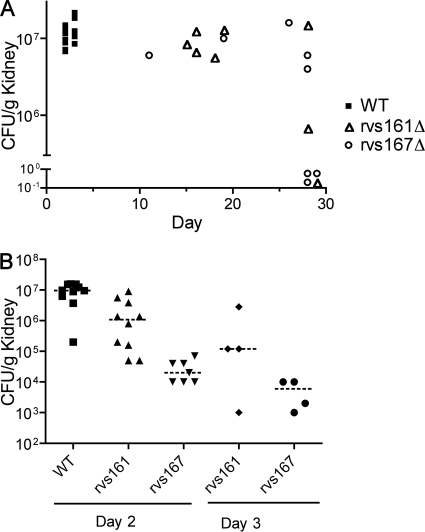
FIG. 8.
Fungal load in infected kidneys. (A) Determination of CFU per gram of kidney tissue in mice that became moribund on the indicated day of infection or after euthanasia on day 28 of infection. (B) Determination of CFU per gram of kidney tissue in mice that were euthanized after 2 or 3 days of infection, as indicated. Strains used were YLD14 (rvs161Δ), YLD16 (rvs167Δ), and wild-type DIC185.
Antifungal susceptibility.
Etest gradient strips (AB Biodisk, Solna, Sweden) were used to assess the sensitivity of the C. albicans strains to amphotericin B, fluconazole, and caspofungin essentially as recommended by the manufacturer. Strains were grown overnight in YPD medium plus 80 mg/liter uridine at 30°C and then diluted to 1 × 106 cells/ml. A total of 250 μl of cells was then spread onto solid RPMI 1640 medium containing glutamine and buffered to pH 7.0 with 165 mM morpholinepropanesulfonic acid. The surface of each plate was allowed to dry, and then Etest strips were applied. The plates were incubated at 30°C, and the MIC was recorded at 24 and 48 h as the lowest level at which the elliptical zone of growth inhibition intersected with the strip. Susceptibility to histatin 5 (Hst 5) was tested with strains grown overnight in YPD medium at 30°C, washed two times in 10 mM sodium phosphate buffer (pH 7.4), and then diluted to 2.5 × 105 cells/ml (22). A total of 20 μl (5,000 cells) were then added to 20 μl of the indicated concentration of Hst 5 (Sigma-Aldrich) in low-retention tubes. The reactions were incubated for 90 min at 37°C with shaking at 250 rpm, followed by dilution with 360 μl of 10 mM sodium phosphate buffer. A total of 30 μl was then spread onto a YPD medium plate and incubated at 30°C for 48 h. The number of CFU was then assessed and the percentage of loss of viability calculated as 1 − (Hst 5 CFU/control CFU) (22).
RESULTS
Three Rvs proteins produced in C. albicans.
Protein similarity searches revealed that C. albicans encodes three BAR domain family proteins (Fig. 1). Two are similar to S. cerevisiae Rvs161 and Rvs167, and the third protein lacks an ortholog in S. cerevisiae (orf19.6349). We named this new protein Rvs162, since it is more closely related to Rvs161 (∼35% identity). Rvs162 is also similar to Rvs161 in that it lacks the GPA and SH3 domains present in Rvs167, which are thought to mediate interaction with other proteins (14, 15). RVS162 orthologs were detected in other fungal genomes that are more closely related to C. albicans (e.g., Debaryomyces hansenii).
FIG. 1.
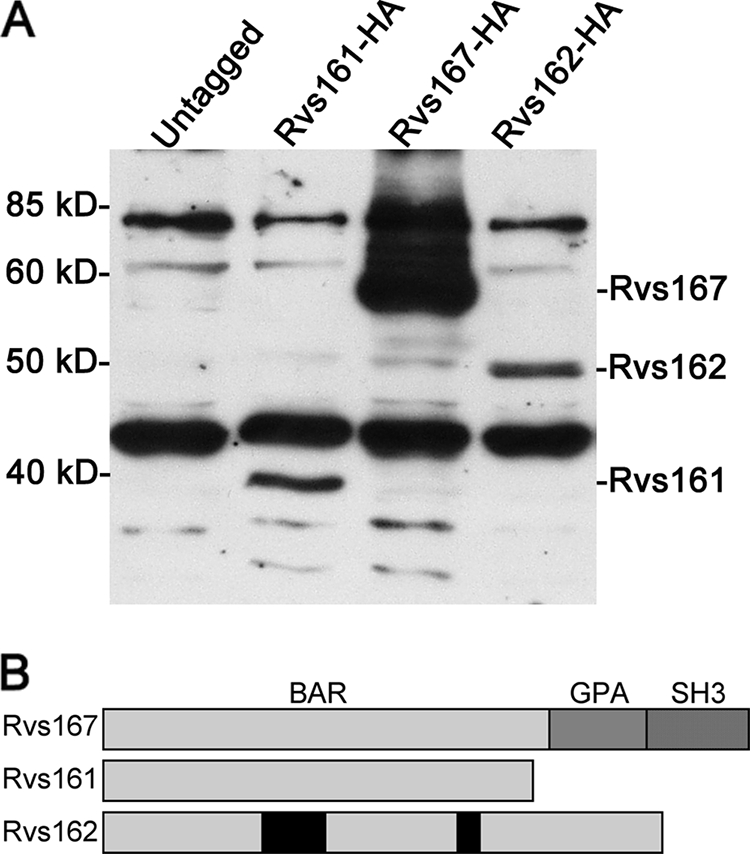
The Rvs161, Rvs167, and Rvs162 proteins are produced by C. albicans. (A) Western blot analysis of an untagged control C. albicans strain (DIC185) and the following strains in which the indicated RVS gene was tagged with a triple HA epitope at the 3′ end of the open reading frame: YSM37 (RVS161-HA), YLD19 (RVS162-HA), and YSM54 (RVS167-HA). (B) Diagram of Rvs protein structures. Rvs161, Rvs167, and Rvs162 each contain an N-terminal BAR domain. Rvs167 also has an internal glycine-, proline-, and alanine-rich sequence (GPA), followed by a C-terminal SH3 domain.
To confirm the production of the Rvs proteins in C. albicans, one allele of each gene was tagged with a triple HA epitope. One of the two RVS162 alleles contains numerous mutations (orf19.13706 genome sequence and our unpublished data), including a frameshift that causes a premature truncation of the open reading frame, so we tagged the other allele (orf19.6349), which contains an open reading frame across the whole BAR domain. Western blots revealed that all three Rvs proteins were produced by budding cells (Fig. 1A) and by cells induced with serum to form hyphae (data not shown). The gel mobilities of the proteins were similar to the sizes of the predicted proteins (Rvs161, 34 kDa; Rvs162, 39.4 kDa; Rvs167, 53.3 kDa) plus the additional mass of the 3× HA tag. Rvs167 was consistently produced at higher levels than Rvs161 or Rvs162. Rvs167 also exhibited additional forms with lower gel mobility, possibly due to the phosphorylation of sites within the putative GPA domain as has been reported for S. cerevisiae Rvs167 (Fig. 1B) (15).
Altered morphogenesis and growth properties of rvs161Δ and rvs167Δ mutants.
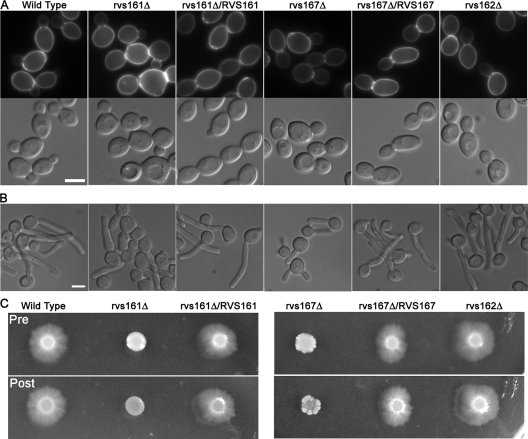
Homozygous deletion mutants of the three RVS genes were constructed for C. albicans strain BWP17 (see Materials and Methods). The rvs161Δ and rvs167Δ cells displayed slight defects in bud morphogenesis, forming somewhat larger, rounder buds than the wild type and an increase in the random budding pattern (Fig. 2A). The defects were complemented by the reintroduction of the corresponding wild-type RVS gene. The rvs161Δ and rvs167Δ mutants displayed increased sensitivity to calcofluor white and Congo red, which indicates they have altered cell walls and suggests they may have a defect in chitin synthesis (Table 2). However, the mutants were not generally defective in cell wall synthesis, since they were not more sensitive to growth in the presence of high salt (1.5 M NaCl), 0.5 M CaCl2, or 0.06% sodium dodecyl sulfate. The Carvs161Δ and Carvs167Δ mutants were also very sensitive to 150 mM LiCl and weakly sensitive to growth under an alkaline pH, suggesting that they may have defects in the activation of the RIM101 pathway similar to those of other endocytosis mutants (11, 20). Both the Carvs161Δ and Carvs167Δ mutants were more sensitive to 5 mM H2O2, with Carvs167Δ being the most sensitive. In contrast, the deletion of CaRVS162 did not cause detectable effects. Surprisingly, the CarvsΔ mutants differed from the ScrvsΔ mutants (10) in that they did not display reduced viability upon starvation, even after incubation for 8 weeks in water.
FIG. 2.
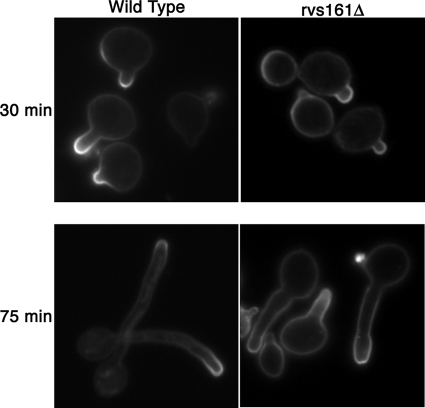
Carvs161Δ and Carvs167Δ exhibit defects in morphology. (A) Morphology of log-phase yeast-form cells grown in YPD medium plus uridine at 30°C. Calcofluor white-stained cells are shown in the top panels and DIC images in the bottom panels. The rvs161Δ and rvs167Δ cultures contained a greater level of large, round mother cells than the wild-type or complemented strains, to which a single copy of the corresponding RVS open reading frame was restored. (B) Morphology of cells grown under hyphal-inducing conditions in liquid medium (YPD plus 10% bovine calf serum for 75 min at 37°C). rvs161Δ and rvs167Δ, but not rvs162Δ, exhibited defects in hyphal growth. rvs161Δ formed predominantly pseudohyphae, while rvs167Δ produced both pseudohyphae and true hyphal filaments. However, the development of rvs167Δ filaments was slower than that of the wild-type and complemented strain filaments. (C) Defects of the rvs161Δ and rvs167Δ mutants when grown on solid-medium agar plates containing 4% bovine calf serum to stimulate invasive hyphal growth. Plates were incubated for 7 days at 37°C. The top and bottom panels show the plate before and after washing to remove the noninvasive cells. Strains used were YLD14 (rvs161Δ), YLD22 (rvs162Δ), YLD16 (rvs167Δ), YLD11 (rvs161Δ/rvs161Δ/RVS161), YLD12 (rvs167Δ/rvs167Δ/RVS167), and wild-type DIC185.
TABLE 2.
Growth phenotypes of C. albicans rvsΔ mutants under different conditionsa
| Condition | Wild type | rvs161Δ | rvs162Δ | rvs167Δ |
|---|---|---|---|---|
| NaCl (1.5 M) | +++ | +++ | +++ | +++ |
| CaCl2 (0.5 M) | +++ | +++ | +++ | +++ |
| LiCl (0.15 M) | +++ | − | +++ | − |
| Calcofluor white (40 μg/ml) | +++ | + | +++ | ++ |
| Congo red (100 μg/ml) | +++ | + | +++ | + |
| Sodium dodecyl sulfate (0.06%) | +++ | ++ | +++ | +++ |
| HEPES (pH 8.5; 150 mM) | +++ | ++ | +++ | ++/+++ |
| H2O2 (5 mM) | +++ | ++ | +++ | + |
| 8-wk starvationb | +++ | +++ | +++ | +++ |
Dilutions of cells were spotted on the indicated plates, and then relative growth was recorded. Growth relative to the wild-type strain under each condition is indicated as follows: +++, >30% of that of the wild type; ++, 3% to 30% of that of the wild type; +, 0.3% to 3% of that of the wild type; and −, no growth.
wk, week. rvs161Δ grew more slowly coming out of the stationary phase but did not display reduced viability.
Hyphal morphogenesis was examined by inducing cells with 10% bovine calf serum at 37°C (Fig. 2B). Interestingly, the rvs161Δ mutant was strongly defective and developed chiefly pseudohyphal-type forms. The rvs167Δ mutant formed hyphae that appeared more similar to those of the wild type, but the hyphal growth was slower to develop. The rvs162Δ mutant formed hyphae similar to those of the wild type (Fig. 2B). Thus, the different RVS genes make distinct contributions to hyphal morphogenesis. Consistent with their altered morphogenesis, the rvs161Δ and rvs167Δ mutants grew less invasively in agar (Fig. 2C).
Endocytosis defects of the C. albicans rvs161Δ and rvs167Δ mutants.
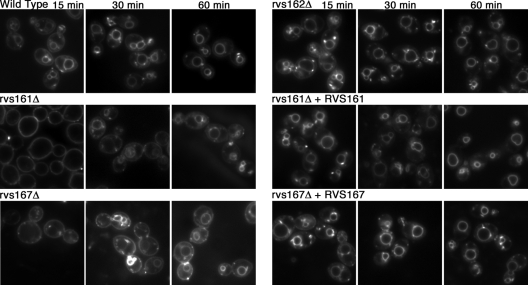
The endocytic capacity of the Carvs mutants was examined by assaying their ability to internalize the lipophilic dye FM4-64. As expected for the wild-type control strain (DIC185), the plasma membrane was no longer stained after 15 min of incubation, and instead, a ring pattern of intracellular staining appeared, indicating that FM4-64 was efficiently endocytosed and localized in the vacuolar membrane (Fig. 3). Similar results were observed for the rvs162Δ mutant. In contrast, FM4-64 staining was still confined to the plasma membrane of the rvs161Δ strain after 15 min. The rvs167Δ mutant gave an intermediate phenotype, with faint staining of the vacuole and endocytic intermediates in addition to plasma membrane staining. By 60 min, both mutants demonstrated increased vacuolar staining, indicating that trafficking to the vacuole still occurred, albeit at a slower rate (Fig. 3). Similar defects were observed in mutant strains induced for hyphal growth (data not shown). The endocytosis defects were complemented by the reintroduction of a wild-type copy of the corresponding gene.
FIG. 3.
Carvs161Δ and Carvs167Δ, but not Carvs162Δ, exhibit defects in endocytosis. Log-phase cells were incubated in 20 μM FM4-64 on ice for 15 min and monitored for internalization of the stain after incubation at 30°C for 15, 30, and 60 min. rvs161Δ exhibited the greatest defect, with little or no internalization of the stain after 15 min, while rvs167Δ demonstrated both surface staining and some staining of endocytic intermediates and the vacuolar membrane. However, after 60 min, both rvs161Δ and rvs167Δ cells displayed an increased ring pattern indicative of vacuolar membrane staining. The deletion of RVS162 resulted in endocytic internalization similar to that of the wild-type strain. The strains used were wild-type DIC185, YLD14 (rvs161Δ), YLD22 (rvs162Δ), YLD16 (rvs167Δ), YLD11 (rvs161Δ/rvs161Δ/RVS161), and YLD12 (rvs167Δ/rvs167Δ/RVS167).
Actin polarization defects in the rvs161Δ and rvs167Δ mutants.
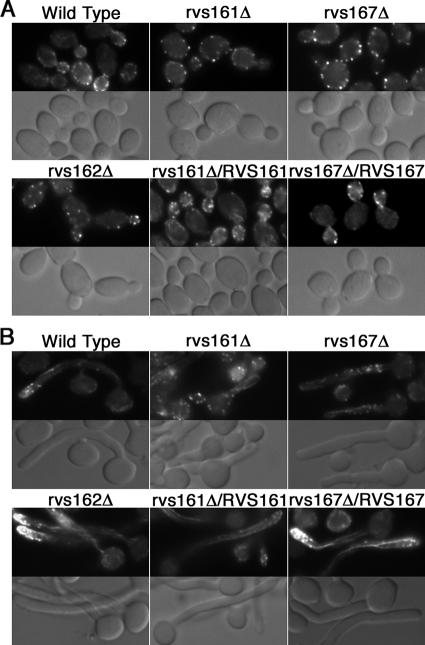
Rhodamine-phalloidin staining of budding cells to detect filamentous actin revealed the expected cortical actin patches localized to the growing buds in the control strain DIC185 and in rvs162Δ (Fig. 4A). In contrast, the rvs161Δ and rvs167Δ mutants showed extensive mislocalization of actin patches to the mother cells, similar to the corresponding S. cerevisiae mutants (28). Also, fewer actin cables were detected in the mutants. This correlates with a larger, rounder bud morphology that was most obvious in the Carvs161Δ cells (Fig. 2A). The actin localization defects were complemented by the reintroduction of a wild-type copy of the corresponding gene.
FIG. 4.
Actin polarization defects of rvs161Δ and rvs167Δ strains. Actin localization in budding cells (A) and cells induced to form hyphae (B) by growth in medium containing 10% bovine calf serum at 37°C. Wild-type strain DIC185 and the indicated rvsΔ mutant strains were analyzed by staining fixed cells with rhodamine-conjugated phalloidin (top panels) and by DIC light microscopy (bottom panels). The deletion of RVS161 and RVS167 resulted in the increased appearance of actin patches on the mother cell surface with few cables. The rvs162Δ strain appeared similar to the wild-type and complemented strains in which the corresponding wild-type RVS gene was reintroduced. Strains used were YLD14 (rvs161Δ), YLD22 (rvs162Δ), YLD16 (rvs167Δ), YLD11 (rvs161Δ/rvs161Δ/RVS161), and YLD12 (rvs167Δ/rvs167Δ/RVS167).
Wild-type cells induced with 10% serum showed strong polarization of actin patches to the hyphal tip. In contrast, actin patches were mislocalized throughout the rvs161Δ and rvs167Δ mutant hyphae (Fig. 4B). Interestingly, actin cables could still be detected projecting from the leading edge of growth in the mutant cells. Although there were fewer cables detected than in the wild-type hyphae, the actin cables presumably help promote the degree of hyphal growth that is detected in these mutants.
rvs161Δ cells exhibit polarized filipin staining of ergosterol at the leading edge of hyphal growth.
The ability of the rvsΔ mutants to undergo polarized hyphal growth was defined further by staining cells with filipin, an autofluorescent polyene antibiotic that binds ergosterol (2). Filipin stains the leading edge of hyphal growth, but not buds or pseudohyphae, suggesting that this distinct lipid composition contributes to highly polarized growth at the hyphal tips (25). Although rvs161Δ cells displayed the strongest endocytosis defect, they still exhibited increased filipin staining at polarized sites of growth after a short 30-min induction in 10% serum (Fig. 5). Similar results were observed for the rvs167Δ and rvs162Δ mutants (data not shown). After 75 min of incubation in serum, increased filipin staining was seen at the leading edge of growth in the rvs161Δ mutant similar to that of the wild type, even in cells with a pseudohyphal morphology. Thus, the formation of acute hyphal projections is not required for polarized filipin staining. Altogether, these assays indicate that efficient endocytosis is not essential for the altered plasma membrane structure of hyphal tips. Additional studies showed that the rvsΔ mutations did not affect the localization of plasma membrane proteins that are thought to be present in other specialized lipid domains (data not shown), including the localization of Sur7-GFP to punctate patches in the plasma membrane (3) or the localization of septin proteins to the bud neck (8).
FIG. 5.
Polarized filipin staining of ergosterol in rvs161Δ grown under hyphal-inducing conditions. Wild-type DIC185 and YLD14 (rvs161Δ) cells were stimulated with 10% bovine calf serum at 37°C for 30 and 75 min, stained with filipin, and then immediately examined by fluorescence microscopy. rvs161Δ cells exhibited increased filipin staining of ergosterol at the leading edges of growth similar to the wild type at both 30 and 75 min.
Altered sensitivity to fluconazole and Hst 5.
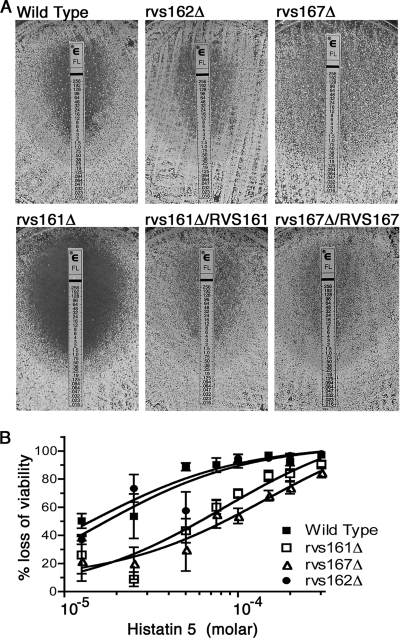
The rvsΔ mutants were tested for sensitivity to antifungal drugs that target plasma membrane functions (32) using Etest strips, which release a gradient of drug that diffuses into a solid medium agar plate (Fig. 6). The mutant strains showed essentially the wild-type response to caspofungin and amphotericin B (data not shown). In contrast, rvs161Δ was more sensitive to fluconazole, which blocks an intermediate step in ergosterol synthesis (32). The MIC was approximately fivefold lower for rvs161Δ. Similar results were observed for cells grown in liquid cultures (data not shown). Surprisingly, the rvs167Δ mutant was more resistant to fluconazole (Fig. 6). However, there was always lighter growth surrounding the Etest strip, indicating partial sensitivity to lower doses of fluconazole. Liquid broth micro- and macrodilution assays (37) confirmed that the partial inhibition of rvs167Δ occurred at lower doses than for the wild type but that the rvs167Δ mutant was also capable of growing, albeit more slowly, at doses of fluconazole that were ≥50-fold higher than the dose that inhibited the wild type. The different sensitivities of the rvs161Δ and rvs167Δ mutants to fluconazole indicate that these phenotypes do not correlate with the overall endocytosis defects and suggest that this may be due to another function of the Rvs proteins.
FIG. 6.
Altered responses of rvsΔ mutants to antifungal agents. (A) Increased susceptibility of rvs161Δ mutant to fluconazole. The indicated cells were spread onto solid RPMI medium, and then an Etest strip was applied to the agar surface to generate a gradient of fluconazole. rvs161Δ demonstrated increased susceptibility to fluconazole at 48 h, with no growth within the ellipse of inhibition, unlike the wild-type, rvs167Δ, or rvs162Δ strain or a strain in which rvs161Δ was complemented by the introduction of one copy of RVS161. Strains used were YLD14 (rvs161Δ), YLD22 (rvs162Δ), YLD16 (rvs167Δ), YLD11 (rvs161Δ/rvs161Δ/RVS161), and wild-type DIC185. (B) Cells were incubated at 37°C with the indicated concentration of Hst 5 for 90 min and then dilutions were plated to determine the loss of viability. rvs161Δ and rvs167Δ showed greater resistance to Hst 5 than either the wild-type or rvs162Δ strains at all concentrations tested. The half-maximal responses indicated a 5.9-fold resistance for rvs161Δ and a 10.3-fold resistance for rvs167Δ. Each data point represents the average of the results from three independent experiments. Bars indicate standard errors. Strains used were YLD14 (rvs161Δ), YLD22 (rvs162Δ), YLD16 (rvs167Δ), and wild-type DIC185.
The rvsΔ mutants were also tested for sensitivity to the antimicrobial peptide Hst 5, which is secreted by acinar cells of the human parotid glands into the oral cavity (36). Interestingly, both rvs161Δ and rvs167Δ exhibited increased resistance to Hst 5 compared with that of the wild-type and rvs162Δ strains (Fig. 6B). The half-maximal response (50% effective concentration) indicated that rvs161Δ was 5.9-fold more resistant to Hst 5 and rvs167Δ was 10.3-fold more resistant. Efficient endocytosis may play a direct role in the fungicidal activity of Hst 5 by promoting its internalization into cells. However, the role of the Rvs proteins could also be indirect, due to changes in lipid composition or the physical properties of the plasma membrane (27, 41).
rvs161Δ and rvs167Δ exhibit attenuated virulence.
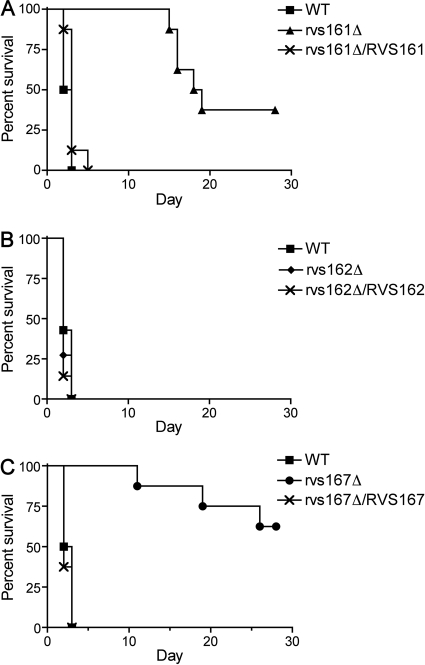
The pathogenicity of the rvs mutants was examined in a mouse model of hematogenously disseminated candidiasis. As expected, BALB/c mice injected with the wild-type control strain DIC185 into the lateral tail vein become moribund in 2 to 3 days. Similar results were observed for mice infected with the rvs162Δ mutant. In contrast, all mice infected with either the rvs161Δ or rvs167Δ mutant strains survived beyond 10 days, with some mice remaining viable at the completion of the assay at 28 days (Fig. 7). The defects were complemented by the reintroduction of a wild-type copy of the corresponding gene. The growth rate of the rvs161Δ mutant at 37°C in vitro was similar to that of the wild type (doubling time, 1.1 h versus 1 h), but the rvs167Δ mutant grew significantly slower (doubling time, 1.5 h), which could contribute to its virulence defect.
FIG. 7.
Attenuated virulence of rvs161Δ and rvs167Δ mutants in a mouse model of hematogenously disseminated candidiasis. Survival curves of mice infected with rvs161Δ (A), rvs162Δ (B), or rvs167Δ (C) compared with that of the wild-type strain DIC185. Note the reduced virulence of the rvs161Δ and rvs167Δ strains (P = 0.0001). The difference between the rvs161Δ and rvs167Δ strains was not statistically different. Strains used were YLD14 (rvs161Δ), YLD22 (rvs162Δ), YLD16 (rvs167Δ), YLD11 (rvs161Δ/rvs161Δ/RVS161), YLD12 (rvs167Δ/rvs167Δ/RVS167), and wild-type DIC185.
The fungal burden per gram of kidney tissue was assessed, because kidneys are the major target organs of hematogenously disseminated C. albicans in mice (Fig. 8A). Although the wild-type strain resulted in the highest level of CFU per gram, surprisingly, some rvs161Δ- and rvs167Δ-infected mice maintained comparatively high levels of CFU as well and still survived the entire 28-day period of the experiment. Other mice that also survived the assay to day 28 exhibited low CFU levels, apparently having cleared the majority of the fungal cells from the kidney.
To better understand the defects of the rvs161Δ and rvs167Δ mutants, the fungal load in the kidneys was analyzed after 2 and 3 days of infection. Mice infected with the wild-type strain succumbed to infection at day 2 with a median of 9.7 × 106 CFU/g kidney. Mice infected with rvs161Δ appeared to show a reduced fungal load (median CFU/g kidney was 1.1 × 106/g at day 2 and 0.12 × 106/g at day 3); however, these results were not statistically significant from those for the wild type (P > 0.05). In the case of the rvs167Δ mutant, the CFU/g kidney were 0.02 × 106/g after day 2 and 0.006 × 106/g after day 3, which were significantly different from those of the wild type (P < 0.001). Thus, the rvs167Δ mutants are defective in initiating an infection but can still grow to higher levels and cause a lethal infection. Although it appears that the CFU/g kidney in the mutants decreased between day 2 and day 3, this was not statistically significant.
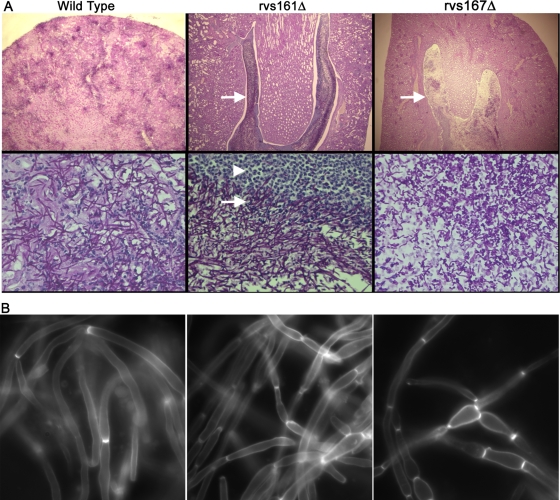
A histopathological analysis of formaldehyde-fixed kidney tissue sections using PAS staining revealed that the wild type and the complemented strains caused widely disseminated infections, with large numbers of hyphal filaments emanating from discrete foci that penetrated throughout the organ (Fig. 9A). In contrast, in mice surviving to day 28, the rvs161Δ and rvs167Δ mutants grew in the center of the kidney and were not disseminated to the outer cortex. These large zones of fungal growth for the rvs161Δ and rvs167Δ mutants were surrounded by layers of leukocytes, separating the areas of infection from the remaining healthy tissue, which presumably helps to contain the dissemination of the rvsΔ mutants. Both the rvs161Δ and rvs167Δ mutants were capable of forming filamentous cells in vivo in spite of their apparent defects in vitro. However, some kidney sections of mice infected for 28 days with rvs167Δ showed mainly shorter, pseudohyphal filaments rather than the longer true hyphae. This was mainly seen in zones adjacent to partially cleared areas of the kidney (Fig. 9A), and not throughout the kidney, suggesting that it may relate to an intermediate stage in which the infection is being cleared.
FIG. 9.
Microscopic analysis of infected kidney tissue. Kidneys were excised from mice infected with the wild-type strain (DIC185) for 2 days or with the indicated rvsΔ mutant strain that were euthanized after 28 days of infection. (A) Kidney sections were analyzed by PAS staining. Kidney tissue infected with the DIC185 wild-type strain demonstrated a high level of penetration by hyphal filaments emanating from multiple sites within the cortex. In contrast, tissue infected with the mutant YLD14 (rvs161Δ) and YLD16 (rvs167Δ) strains showed large internal zones of fungal cells (indicated by arrows) that were completely surrounded by leukocytes (indicated by the arrowhead). Note that the zone of clearing within the fungal mass of the YLD16 (rvs167Δ) strain contained cells with a bud or pseudohyphal morphology. Images in the top panels were captured through a 4× lens, and the bottom panels were captured through a 60× lens. (B) Kidney homogenates were stained with calcofluor, treated with 1% KOH to help dissolve host tissues, and then examined by fluorescence microscopy.
The morphogenesis of the mutants was examined in more detail by staining kidney homogenates with calcofluor white, which specifically stains fungal cell wall chitin but not human tissues which lack chitin. Wild-type C. albicans cells were present mainly in elongated filamentous growth forms, although pseudohyphal and round blastospore-type cells were also common. Interestingly, the rvs161Δ and rvs167Δ mutants also formed extensive filamentous growth in the kidney and even appeared to form some true hyphae. However, on close examination, it was obvious that the mutants formed a higher proportion of chains of pseudohyphal cells rather than true hyphae than the wild type. Thus, altered morphogenesis may contribute to the virulence defect of the rvs161Δ and rvs167Δ mutants in vivo.
DISCUSSION
Rvs proteins are important for virulence.
The rvs161Δ and rvs167Δ mutants showed greatly reduced virulence in a mouse model of candidiasis (Fig. 7). For the rvs167Δ mutant, this is due in part to a decreased ability to colonize the kidney (Fig. 8B). This defect is interesting, given that rvs167Δ typically showed a milder phenotype than rvs161Δ in other assays. However, an exception is that rvs167Δ grew more poorly than rvs161Δ in the presence of 5 mM H2O2 (Table 2), suggesting that greater sensitivity to oxidative damage by the immune system may impair its ability to initiate an infection. Over time both the rvs161Δ and rvs167Δ mutants were capable of growing to high levels in the kidney, and about half the infected mice succumbed to infection by day 28. A histological analysis of kidney sections from mice that survived to day 28 showed that the rvs161Δ and rvs167Δ infections were confined to large internal regions surrounded by layers of leukocytes (Fig. 9A), which contrasts with the highly disseminated infection by wild-type C. albicans. This clustering presumably accounts for why mice with comparatively high fungal burdens of rvs161Δ or rvs167Δ cells still demonstrated normal behavior. It is surprising that the rvs161Δ and rvs167Δ mutants grow well in vivo, given that endocytosis is thought to influence a wide range of properties by regulating the plasma membrane composition and the stability of nutrient sensors and other components.
Relationship between morphogenesis and virulence.
The defects of the rvsΔ mutants in hyphal morphogenesis and invasive growth also appear to contribute to decreased virulence. Endocytosis and morphogenesis are interrelated; efficient endocytosis is thought to be critical for the appropriate localization of proteins required for polarized morphogenesis (35). Consistent with this, rvs161Δ was more defective than rvs167Δ in both endocytosis and hyphal morphogenesis, and a similar correlation was reported for mutations in other C. albicans genes, including WAL1, SLA2, and MYO5 (29, 30, 43). Interestingly, the rvs161Δ and rvs167Δ mutants formed extensive filamentous growth in the kidney, suggesting that stronger inducing signals in vivo improved filamentous growth (Fig. 9). However, the mutants more frequently formed chains of elongated pseudohyphae rather than true hyphae with parallel walls. The abnormal morphogenesis of the rvsΔ mutants also correlates with strong defects in invasive growth into agar (Fig. 2C). The reduced ability to grow invasively may account for why the rvs161Δ and rvs167Δ mutants were primarily detected in the central renal space and did not disseminate to the more solid cortex tissue. In agreement with this, other mutants that are defective in invasive growth in vitro, such as the cdc10Δ and cdc11Δ septin mutants, also grew as a large fungal mass in the central region of the kidney (44).
The impairment of endocytosis and morphogenesis in the rvsΔ mutants may also lead to the exposure of cell wall determinants recognized by the host immune system receptors. The ScRVS161 and ScRVS167 genes were reported to be members of a cell wall remodeling network that is essential for masking the proinflammatory inner β-glucan layer with a covering of mannan (46). The Carvs161Δ and Carvs167Δ mutants also displayed features of altered cell wall synthesis, since they were more sensitive to the cell wall-perturbing agents Congo red and calcofluor white (Table 2). Altered cell walls may help to recruit the leukocytes that were observed to surround the large mass of rvsΔ mutant growth in the kidney (Fig. 9). Similarly, the Cacdc10Δ and Cacdc11Δ septin mutants showed cell wall alterations and grew as large fungal masses surrounded by leukocytes in the kidney (44, 45). Thus, the cell wall defects of the rvsΔ and septin mutants may recruit leukocytes that help to prevent the dissemination of fungal growth.
It was suggested previously that endocytosis may also play a role in forming a special lipid domain at the hyphal tips (25, 30), which is thought to contribute to highly polarized hyphal morphogenesis and may influence the response to antifungal drugs (2). Surprisingly, the rvsΔ strains still exhibited enriched filipin staining of sterols at the hyphal tips (Fig. 5 and data not shown), indicating that efficient endocytosis is not required for lipid polarization. The failure of the previously described endocytosis-defective myo5Δ and sla2Δ mutants to show polarized filipin staining in response to hyphal inducers may be due to their more severe defect in actin organization (30). Actin filaments are important for polarized filipin staining (25). In contrast to the severely defective myo5Δ and sla2Δ mutants, the rvsΔ mutants showed detectable actin cables (Fig. 4) and were still able to undergo more highly polarized, albeit abnormal, hyphal growth.
RVS genes affect susceptibility to antifungal agents and environmental conditions.
Testing with antifungal drugs that alter plasma membrane functions (32) showed that rvs161Δ was more sensitive to fluconazole (Fig. 7A), which blocks an intermediate step in ergosterol synthesis and results in the accumulation of toxic sterol intermediates in the membrane. The mutation of C. albicans VPS28 and VPS32, vacuolar protein sorting (Vps) factors that are important in the late stages of endocytosis, also caused increased sensitivity to azoles (9). Endocytosis defects may increase sensitivity to fluconazole by altering membrane lipids or the function of multidrug transporters. However, the rvs167Δ mutant displayed a more complex phenotype. It was partially sensitive to low concentrations but still grew well with higher doses of fluconazole that inhibited the wild-type strain (Fig. 7). Thus, the different effects of the rvsΔ mutations on fluconazole sensitivity are likely due in part to other functions and not simply to endocytosis.
An analysis of the growth of the CarvsΔ mutants under different environmental conditions revealed additional phenotypes that likely influence the virulence of these strains (Table 2). Both rvs161Δ and rvs167Δ cells were more resistant to Hst 5, an antimicrobial peptide that acts in the oropharyngeal environment, which is consistent with a proposed role for endocytosis in its uptake (17, 23, 51). Both rvs161Δ and rvs167Δ were more sensitive to H2O2, which likely makes them more sensitive to attack by the immune system. The rvs161Δ and rvs167Δ mutants were also more sensitive to an alkaline pH (pH 8.5) and to 0.15 M LiCl, which are hallmarks of mutants in the RIM101 pH response pathway (11). Interestingly, ESCRT-III complex mutants that are defective in the later stages of endocytosis are also defective in the RIM101 pH response pathway (20, 50).
Surprisingly, although the CarvsΔ and ScrvsΔ mutants shared many similarities, they displayed some opposite phenotypes in that the Carvs161Δ and Carvs167Δ mutants were sensitive to calcofluor white and Congo red (6) but not to elevated NaCl (4). Thus, although the rvsΔ mutations result in altered cell wall function in both organisms, they do so in different ways. Furthermore, the CarvsΔ strains did not show the “reduced viability upon nutrient starvation” phenotype (Table 2) for which the Scrvs mutants were originally named (10).
Altogether, the analysis of the RVS genes in C. albicans indicates that endocytic mechanisms play diverse roles in promoting efficient virulence in mice. This is consistent with the fundamental role of endocytosis in regulating the plasma membrane, which is an important site for interfacing with the host environment and for regulating growth and morphogenesis. Although the rvs161Δ and rvs167Δ mutants could still grow well in vivo, their strong defects in virulence indicate that endocytosis could be targeted to diminish dissemination and to sensitize cells to antifungal therapy.
Acknowledgments
We thank Aaron Mitchell, Judith Berman, Amy Warenda, and Scott Filler for the plasmids and strains and James Bliska for advice. We also thank Kenneth Shroyer for advice on interpreting the histopathology studies.
This work was supported by grant RO1 AI47837 from the National Institutes of Health that was awarded to J.B.K.
Editor: A. Casadevall
Footnotes
Published ahead of print on 13 July 2009.
REFERENCES
- 1.Adams, A. E. M., and J. R. Pringle. 1991. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 194729-731. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, F. J., L. M. Douglas, and J. B. Konopka. 2007. Sterol-rich plasma membrane domains in fungi. Eukaryot. Cell 6755-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez, F. J., L. M. Douglas, A. Rosebrock, and J. B. Konopka. 2008. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol. Biol. Cell 195214-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balguerie, A., M. Bagnat, M. Bonneu, M. Aigle, and A. M. Breton. 2002. Rvs161p and sphingolipids are required for actin repolarization following salt stress. Eukaryot. Cell 11021-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blankenship, J. R., and A. P. Mitchell. 2006. How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 9588-594. [DOI] [PubMed] [Google Scholar]
- 6.Breton, A. M., J. Schaeffer, and M. Aigle. 2001. The yeast Rvs161 and Rvs167 proteins are involved in secretory vesicles targeting the plasma membrane and in cell integrity. Yeast 181053-1068. [DOI] [PubMed] [Google Scholar]
- 7.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9327-335. [DOI] [PubMed] [Google Scholar]
- 8.Casamayor, A., and M. Snyder. 2003. Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol. Cell. Biol. 232762-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornet, M., C. Gaillardin, and M. L. Richard. 2006. Deletions of the endocytic components VPS28 and VPS32 in Candida albicans lead to echinocandin and azole hypersensitivity. Antimicrob. Agents Chemother. 503492-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crouzet, M., M. Urdaci, L. Dulau, and M. Aigle. 1991. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast 7727-743. [DOI] [PubMed] [Google Scholar]
- 11.Davis, D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 441-7. [DOI] [PubMed] [Google Scholar]
- 12.Dawson, J. C., J. A. Legg, and L. M. Machesky. 2006. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 16493-498. [DOI] [PubMed] [Google Scholar]
- 13.Engqvist-Goldstein, A. E., and D. G. Drubin. 2003. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19287-332. [DOI] [PubMed] [Google Scholar]
- 14.Friesen, H., K. Colwill, K. Robertson, O. Schub, and B. Andrews. 2005. Interaction of the Saccharomyces cerevisiae cortical actin patch protein Rvs167p with proteins involved in ER to Golgi vesicle trafficking. Genetics 170555-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friesen, H., K. Murphy, A. Breitkreutz, M. Tyers, and B. Andrews. 2003. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Mol. Biol. Cell 143027-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germann, M., E. Swain, L. Bergman, and J. T. Nickels, Jr. 2005. Characterizing the sphingolipid signaling pathway that remediates defects associated with loss of the yeast amphiphysin-like orthologs, Rvs161p and Rvs167p. J. Biol. Chem. 2804270-4278. [DOI] [PubMed] [Google Scholar]
- 17.Helmerhorst, E. J., P. Breeuwer, W. van't Hof, E. Walgreen-Weterings, L. C. Oomen, E. C. Veerman, A. V. Amerongen, and T. Abee. 1999. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J. Biol. Chem. 2747286-7291. [DOI] [PubMed] [Google Scholar]
- 18.Hicke, L. 1999. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 9107-112. [DOI] [PubMed] [Google Scholar]
- 19.Kaksonen, M., C. P. Toret, and D. G. Drubin. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123305-320. [DOI] [PubMed] [Google Scholar]
- 20.Kullas, A. L., M. Li, and D. A. Davis. 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 31609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumamoto, C. A., and M. D. Vinces. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 71546-1554. [DOI] [PubMed] [Google Scholar]
- 22.Li, X. S., M. S. Reddy, D. Baev, and M. Edgerton. 2003. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J. Biol. Chem. 27828553-28561. [DOI] [PubMed] [Google Scholar]
- 23.Li, X. S., J. N. Sun, K. Okamoto-Shibayama, and M. Edgerton. 2006. Candida albicans cell wall ssa proteins bind and facilitate import of salivary histatin 5 required for toxicity. J. Biol. Chem. 28122453-22463. [DOI] [PubMed] [Google Scholar]
- 24.Lombardi, R., and H. Riezman. 2001. Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J. Biol. Chem. 2766016-6022. [DOI] [PubMed] [Google Scholar]
- 25.Martin, S. W., and J. B. Konopka. 2004. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot. Cell 3675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, S. W., and J. B. Konopka. 2004. SUMO modification of septin-interacting proteins in Candida albicans. J. Biol. Chem. 27940861-40867. [DOI] [PubMed] [Google Scholar]
- 27.Mochon, A. B., and H. Liu. 2008. The antimicrobial peptide histatin-5 causes a spatially restricted disruption on the Candida albicans surface, allowing rapid entry of the peptide into the cytoplasm. PLoS Pathog. 4e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munn, A. L., B. J. Stevenson, M. I. Geli, and H. Riezman. 1995. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 61721-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oberholzer, U., T. L. Iouk, D. Y. Thomas, and M. Whiteway. 2004. Functional characterization of myosin I tail regions in Candida albicans. Eukaryot. Cell 31272-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberholzer, U., A. Nantel, J. Berman, and M. Whiteway. 2006. Transcript profiles of Candida albicans cortical actin patch mutants reflect their cellular defects: contribution of the Hog1p and Mkc1p signaling pathways. Eukaryot. Cell 51252-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odds, F. C. 1988. Candida and candidosis. Bailliere Tindall, Philadelphia, PA.
- 32.Odds, F. C., A. J. Brown, and N. A. Gow. 2003. Antifungal agents: mechanisms of action. Trends Microbiol. 11272-279. [DOI] [PubMed] [Google Scholar]
- 33.Park, H., C. L. Myers, D. C. Sheppard, Q. T. Phan, A. A. Sanchez, J. E. Edwards, Jr., and S. G. Filler. 2005. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell. Microbiol. 7499-510. [DOI] [PubMed] [Google Scholar]
- 34.Pringle, J. R. 1991. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 194732-735. [DOI] [PubMed] [Google Scholar]
- 35.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. J. Cell Sci. 113571-585. [DOI] [PubMed] [Google Scholar]
- 36.Raj, P. A., M. Edgerton, and M. J. Levine. 1990. Salivary histatin 5: dependence of sequence, chain length, and helical conformation for candidacidal activity. J. Biol. Chem. 2653898-3905. [PubMed] [Google Scholar]
- 37.Sewell, D. L., M. A. Pfaller, and A. L. Barry. 1994. Comparison of broth macrodilution, broth microdilution, and E test antifungal susceptibility tests for fluconazole. J. Clin. Microbiol. 322099-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman, F. 2002. Getting started with yeast. Methods Enzymol. 3503-41. [DOI] [PubMed] [Google Scholar]
- 39.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12317-324. [DOI] [PubMed] [Google Scholar]
- 40.Valdez-Taubas, J., and H. R. Pelham. 2003. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 131636-1640. [DOI] [PubMed] [Google Scholar]
- 41.Veerman, E. C., M. Valentijn-Benz, K. Nazmi, A. L. Ruissen, E. Walgreen-Weterings, J. van Marle, A. B. Doust, W. van't Hof, J. G. Bolscher, and A. V. Amerongen. 2007. Energy depletion protects Candida albicans against antimicrobial peptides by rigidifying its cell membrane. J. Biol. Chem. 28218831-18841. [DOI] [PubMed] [Google Scholar]
- 42.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walther, A., and J. Wendland. 2004. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich syndrome protein homolog Wal1p. Eukaryot. Cell 3471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warenda, A. J., S. Kauffman, T. P. Sherrill, J. M. Becker, and J. B. Konopka. 2003. Candida albicans septin mutants are defective for invasive growth and virulence. Infect. Immun. 714045-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warenda, A. J., and J. B. Konopka. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 132732-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler, R. T., and G. R. Fink. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whiteway, M., and U. Oberholzer. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7350-357. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 1665-70. [DOI] [PubMed] [Google Scholar]
- 49.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1811868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, W., F. J. Smith, Jr., R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 155528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu, J., P. W. Luther, Q. Leng, and A. J. Mixson. 2006. Synthetic histidine-rich peptides inhibit Candida species and other fungi in vitro: role of endocytosis and treatment implications. Antimicrob. Agents Chemother. 502797-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmerberg, J., and S. McLaughlin. 2004. Membrane curvature: how BAR domains bend bilayers. Curr. Biol. 14R250-R252. [DOI] [PubMed] [Google Scholar]