Abstract
Vulvovaginal candidiasis (VVC) is an insidious infection that afflicts a large proportion of women of all ages, and 5 to 8% of affected women experience recurrent VVC (RVVC). The aim of this study was to explore the possible importance of vaginal bacterial communities in reducing the risk of RVVC. The species composition and diversity of microbial communities were evaluated for 42 women with and without frequent VVC based on profiles of terminal restriction fragment polymorphisms of 16S rRNA genes and phylogenetic analysis of cloned 16S rRNA gene sequences from the numerically dominant microbial populations. The data showed that there were no significant differences between the vaginal microbial communities of women in the two groups (likelihood score, 5.948; bootstrap P value, 0.26). Moreover, no novel bacteria were found in the communities of women with frequent VVC. The vaginal communities of most women in both groups (38/42; 90%) were dominated by species of Lactobacillus. The results of this study failed to provide evidence for the existence of altered or unusual vaginal bacterial communities in women who have frequent VVC compared to women who do not have frequent VVC. The findings suggest that commensal vaginal bacterial species may not be able to prevent VVC.
Vulvovaginal candidiasis (VVC) is a widespread and common disease affecting a large proportion of women of all ages. Classically, VVC is described as consisting of a white “cottage cheese” discharge with associated vulval and vaginal inflammation (31). In typical cases, the symptoms include vulva erythema, edema, excoriations, and fissures (33). Alternatively, women can test positive for Candida species during routine exams with no clinical evidence of infection (31). Several factors predispose women to VVC, including genetic factors (3), pregnancy (5), uncontrolled diabetes mellitus (6), and the use of high-estrogen contraceptives (11), steroids, and antibiotics (28). Approximately 75% of women experience at least one episode of VVC during their lives, most commonly when they are childbearing age (33). About 40 to 50% of these women experience a recurrence (17), and 5 to 8% of these women have recurrent VVC (RVVC), which is defined as four or more proven episodes of VVC in a 12-month period (35). The quality of life is greatly diminished for women who experience RVVC. In addition, the diagnosis and treatment of VVC cost an estimated $1 billion per year in the United States (12). The high incidence and healthcare costs associated with treatment of VVC highlight the need for understanding the pathogenesis of the infections and host defense mechanisms so that effective strategies can be developed to control and prevent this disease.
Relatively little is known about possible associations between the bacterial species found in the vagina and colonization by Candida species. While it is commonly thought that the normal vaginal microbiota plays an important role in the prevention of vaginal infections and transmission of pathogens responsible for sexually transmitted disease (20, 29), there is no consensus about the impact of vaginal bacteria on control of VVC and RVVC. Siegler (32) first suggested that vaginitis caused by Candida was associated with intermediate flora patterns (a Nugent score between 4 and 6 [23]), and then Hillier et al. (15) reported a similar association between VVC and intermediate or normal flora patterns. In contrast, Sobel and Chaim (34) compared the microbial flora of vaginal secretions in healthy, nonpregnant women of reproductive age with the microbial flora of vaginal secretions in similar women with acute RVVC and found that women with VVC did not have reduced numbers of Lactobacillus species. Further, there is no consensus on whether H2O2-producing lactobacilli in the vaginal microbial communities protect their hosts from VVC (16, 19, 39). It has also been observed that the use of antibiotics can lead to yeast infections (28), perhaps by killing or inhibiting bacterial populations that have antimycotic properties. This suggests that normal vaginal microbiota may have an important role in restricting yeast infections. Trials to evaluate the use of oral or vaginal probiotics containing Lactobacillus spp. to prevent postantibiotic VVC have been limited and have yielded inconsistent results (7, 27).
Studies done previously to examine the importance of vaginal bacterial communities in determining susceptibility to RVVC have used cultivation-dependent methods. These traditional, “cultivation-dependent” methods have historically been used in clinical microbiology laboratories and in research to characterize the microbial populations associated with the human body. They are tedious and labor-intensive, and so their use for the analysis of large numbers of samples is impractical and costly. They are further limited by their reliance on selective media, and many bacterial populations are refractory to cultivation (37). Hence, they provide an incomplete assessment of prokaryotic diversity. In recent years, cultivation-independent methods based on analysis of 16S and 18S rRNA gene sequences directly extracted from samples have been used to overcome these limitations and are widely employed to explore microbial diversity in various habitats (10, 22, 24). The use of these approaches not only obviates the need to cultivate organisms but also permits high-throughput analysis of samples and provides precise and detailed information about the populations present. This has become the favored approach for characterizing microbial populations residing on and in the human body because the species (phylotype) composition of communities can be readily determined and similarities and differences among microbial communities can be discerned (13, 14, 18, 21, 25, 43). In this study, we used these methods to characterize the composition and structure of the vaginal microbiota of healthy women and women with frequent VVC to test the null hypothesis that the risk of frequent VVC is not related to the species composition of vaginal bacterial communities.
MATERIALS AND METHODS
Study design and subjects.
In this cross-sectional case-control study vaginal swabs were collected from 42 subjects who were between 18 and 40 years old in a clinical study conducted at the University of Iowa (Iowa City, IA). One half the samples were collected from patients with frequent VVC, while the other half were obtained from a control group. Subjects who had had at least four vaginal yeast infections during the past 2 years, at least one of which was diagnosed by a health care practitioner, were classified as having frequent VVC. Subjects in the control group were required to self-report that they had had no yeast infections in the past 2 years.
Subjects enrolled in the study were required to refrain from douching, vaginal medications (e.g., Vagisil), and sexual intercourse for 48 h and to refrain from bathing or showering for at least 2 h prior to the scheduled visit. Each participant had regular menstrual cycles (minimum, 21 days; maximum, 35 days) and completed questionnaires on habits and practices, medical history, and vaginitis symptoms. Individuals who were currently participating in another clinical study, were pregnant, or were menstruating at the time of a scheduled visit were excluded from the study, as were individuals diagnosed with vaginal infections such as trichomoniasis, chlamydial infection, vulval skin disease, or bacterial vaginosis at the time of a scheduled visit. Likewise, individuals currently using any of the following forms of birth control were excluded: an intrauterine device, spermicides, Depo-Provera, NuvaRing, and Seasonale. Individuals with preexisting systemic diseases or chronic conditions, such as diabetes or an immunological disease (human immunodeficiency virus or systemic lupus erythematosus), were excluded, as were individuals currently using immunosuppressive drugs, undergoing chemotherapy, or taking prescription medication of the following kinds: systemic antimicrobial or antifungal drugs or antifungals or antimicrobials to treat a vaginal infection within the previous 30 days. The subjects were not asked whether they had used vaginal probiotics in the past. The study protocol and informed consent document were reviewed and approved by the University of Iowa Institutional Review Board. Documented informed consent was obtained from all subjects prior to participation in this study.
At each scheduled visit the practitioner did a pelvic exam to assess possible clinical signs of infection. After this, a speculum (lubricated with sterile saline if necessary) was used to collect three vaginal samples from the left side of the upper half of the vaginal canal. One cotton swab was placed in a vial containing ∼1 ml of sterile saline and sent to a clinical laboratory to be evaluated microscopically for the presence of Candida sp., trichomoniasis, and “clue” cells. The second and third cotton swabs were placed in separate vials and stored at −70 ± 10°C. One of these swabs was shipped to the University of Idaho, while the other was retained as an archived specimen. A standardized protocol was utilized to determine the vaginal pH at each visit of a subject using pH paper with range from pH 3.0 to 5.5 (pHydrion pH papers; MicroEssential Laboratory, Brooklyn, NY). The practitioner obtained a sample of vaginal secretions from the vaginal side wall with a cotton-tipped applicator after placement of the vaginal speculum. The color of the pH paper was compared with the pH paper chart and recorded. Quality control was performed weekly to ensure the accuracy of the pH determinations within 0.5 pH unit.
The study was done in a blind fashion; the histories of frequent VVC in the women sampled were unknown to the investigators who analyzed the microbial communities. Once these analyses were completed, the blind was broken.
Genomic DNA isolation.
The swab samples were thawed on ice and mixed vigorously by vortexing to dislodge cells from the swab. Prokaryotic genomic DNA was isolated from a 0.5-ml aliquot of each cell suspension using a two-step cell lysis procedure (42). First, bacterial cell walls were disrupted enzymatically by addition of mutanolysin (50 μg) and lysozyme (500 μg), followed by incubation for 1 h at 37°C. Second, the cells were mechanically disrupted by six freeze-thaw cycles. Each cycle consisted of 2 min of incubation at 100°C that was immediately followed by 2 min in a dry ice-ethanol bath. Between freeze-thaw cycles, the cell suspensions were incubated for 1 min in an ultrasonic cleaning bath (FS60; Fisher Scientific, Pittsburgh, PA). Proteins in the disrupted cell suspension were digested with proteinase K (Qiagen, Hilden, Germany) during a 1-h incubation at 55°C. Further isolation and purification of the total DNA extract were performed using a Wizard DNA purification kit (Promega, Madison, WI).
T-RFLP analysis of 16S rRNA genes.
The species composition and diversity of the numerically dominant populations in vaginal microbial communities were evaluated based on profiles of terminal restriction fragment polymorphisms (T-RFLP) of 16S rRNA genes. To examine the internal regions of 16S rRNA genes, each sample was amplified in two separate reactions using fluorescently labeled primer pairs 8fm-926r and 49f-926r (based on the position of the Escherichia coli 16S rRNA sequence). Primers 8fm, 49f, and 926r were labeled with VIC, NED, and 6-carboxyfluorescein, respectively (Applied Biosystems, Foster City, CA). The PCR protocol used was the protocol described previously (43). After amplification, the two fluorescently labeled amplicons were combined. This mixture was divided equally and separately digested with MspI and HaeIII, and then the digested products were recombined. The resulting mixture contained six fluorescently labeled terminal restriction fragments that resulted from the use of three fluorophores and two restriction enzymes, and this allowed high resolution of microbial communities. The T-RFLP profiles were determined using an ABI PRISM 3100 DNA analyzer with GeneScan software (Applied Biosystems) and with CST ROX 25-1000 (BioVentures, Inc., Murfreesboro, TN) as an internal standard.
A cluster analysis of community profiles was done to identify similar communities, and the numbers of clusters (kinds of communities) were independently assessed using statistical algorithms, as described by Abdo et al. (1). First, true peaks were identified once a threshold (baseline) had been defined. Second, hierarchical clustering was done to identify the fragments whose lengths were similar enough to group them in the same length category. Third, the Euclidean distances between T-RFLP profiles were calculated, and these distances were hierarchically clustered based on average linkage (unweighted-pair group method using average linkages) and a dendrogram was constructed. Finally, three clustering criteria were employed to identify a statistically meaningful number of groups in the data (1).
Clone library construction and 16S rRNA gene sequence analysis.
The samples used to construct clone libraries were chosen using a “coverage sampling approach” (1). This approach provided a way to identify the fewest samples necessary to describe 85% of the phylotype diversity within each cluster. The 16S rRNA genes in each sample identified using this approach were amplified using primers 8fm and 926r without fluorescent labels and cloned as previously described (42, 43). Approximately 100 clones from each sample were randomly chosen from each library, and the cloned inserts were partially sequenced using an ABI 3730 Prism DNA analyzer.
A phylogenetic analysis of cloned 16S rRNA gene sequences from the numerically dominant microbial populations was done to determine the composition of bacterial communities in each sample. High-quality sequences that contained less than 3% uncalled bases and that were more than 500 bp long were analyzed using high-throughput methods (2) to identify similar sequences in the eubacterial type strains in the Ribosomal Database Project and GenBank databases.
Comparison of women with and without frequent VVC.
A likelihood ratio test and a bootstrap analysis described by Zhou et al. (43) were conducted to evaluate differences between the microbial community structures for women with frequent VVC and the microbial community structures for women without frequent VVC based on profiles generated by T-RFLP analysis. Student's t test was used to assess the difference in vaginal pH between women with frequent VVC and women without frequent VVC. A difference was considered significant when the P value was <0.05.
RESULTS AND DISCUSSION
Classification of vaginal microbial communities from the women sampled based on T-RFLP data.
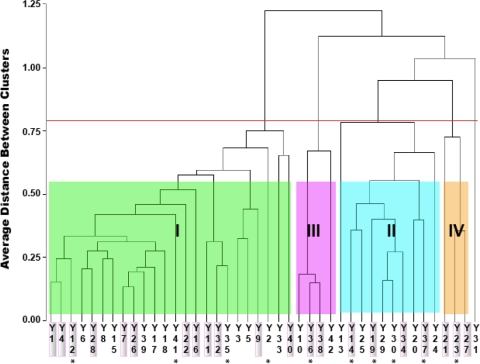
The vaginal microbial communities of the 42 women sampled were characterized on the basis of the T-RFLP of 16S rRNA genes derived from the numerically dominant populations present in each community. The data were subjected to cluster analysis to identify similar communities, and the number of clusters was evaluated. The communities were placed into clusters on the basis of differences in the sizes and abundances of terminal restriction fragments derived from 16S rRNA, providing a means for distinguish the communities in terms of the species (phylotypes) present and their abundance. In the 42 women, there were five different clusters of bacterial communities (Fig. 1), four of which were found in three or more women (clusters I, II, III, and IV) and one of which was found in a single individual (cluster V).
FIG. 1.
Clustering of vaginal microbial communities in women with and without frequent VVC based on the data from T-RFLP analysis of 16S rRNA genes. The red line indicates the clustering threshold determined by using three statistical cluster criteria (1). Samples from patients with frequent VVC are indicated by a shaded background. Clusters for more than three women are indicated by different colors of shading and roman numerals. Cluster V is not indicated because it was a “singleton” and was obtained for only one subject (branch on the right). Asterisks indicate the samples used to construct libraries of cloned 16S rRNA genes.
Differences between the women with and without frequent VVC.
The frequencies of vaginal microbial community types in women with and without frequent VVC are shown in Table 1. A likelihood ratio test and bootstrap analysis (43) showed that there were no significant differences in the distribution of women with and without frequent VVC among the various community types (likelihood score, 5.948; bootstrap P value, 0.26). This suggests that women who have frequent VVC do not have altered or unusual vaginal bacterial communities and that the risk of recurrent yeast infections is not correlated with the composition of vaginal bacterial communities. This finding is consistent with the of findings of Sobel and Chaim (34), who also showed, using culture-dependent methods, that the development of RVVC was not correlated with an abnormal vaginal microbiota. However, the possibility that rare bacterial populations are correlated with risk of frequent VVC cannot be excluded. It should also be pointed out that only one of the women with frequent VVC had acute VVC at the time of sampling, and thus nothing can be concluded about shifts in community composition that may accompany an acute infection.
TABLE 1.
Kinds of vaginal microbial communities in women with and without frequent VVC
| Cluster | No. of women with frequent VVC | No. of women without frequent VVC | Total |
|---|---|---|---|
| I | 12 | 12 | 24 |
| II | 4 | 6 | 10 |
| III | 2 | 2 | 4 |
| IV | 3 | 0 | 3 |
| V | 0 | 1 | 1 |
| Total | 21 | 21 | 42 |
Compositions of the vaginal microbiota in women with and without frequent VVC.
The species compositions of vaginal microbial communities in women who had frequent VVC and women who did not have frequent VVC were determined by phylogenetic analysis of 16S rRNA gene sequences that had been cloned from samples representing each major cluster. A total of 10 clone libraries were prepared, and ∼100 clones from each library were sequenced. The data showed that about 90% (38/42) of the vaginal microbial communities were dominated by species of Lactobacillus that were phylogenetically related to L. crispatus, L. iners, and L. gasseri (clusters I, II, and III, respectively, in Fig. 1) (Table 2). Among the Lactobacillus-dominated communities, those that included L. crispatus (cluster I) were the most common and accounted for 57% of the women sampled (24/42). In this cluster, L. crispatus constituted over 85% of the sequenced clones in each library, and there were low numbers of other lactobacilli, including L. iners, L. jensenii, L. vaginalis, L. gasseri, Lactobacillus sp. strain KC38 (26), and L. kitasatonis. The second most common type of communities (cluster II) was found in 24% (10/42) of the women sampled, and these communities were dominated by L. iners, but L. crispatus, L. jensenii, and Lactobacillus sp. strain KC38 were also common. Lactobacillus sp. strain KC38 is phylogenetically related to L. jensenii (26) and is likely to be homofermentative, as are the other lactobacilli in these communities. The communities in cluster III were dominated by L. gasseri, while those in cluster IV contained Atopobium vaginae, several species of lactobacilli, and various other microbial species at low levels. Communities in clusters III and IV were recovered from 9.5% and 7% of the women sampled, respectively. It should be noted that all three subjects with communities in cluster IV, which was dominated by A. vaginae and L. iners, had frequent VVC. The potential relationship between the risk of frequent VVC and communities of this sort needs to be substantiated in future studies that include larger numbers of women and that also take into account differences in innate and adaptive immunity systems. It should also be noted that no novel bacteria were found in the vaginal communities of women with frequent VVC and there was no significant difference between the vaginal pH of women with frequent VVC and the vaginal pH of the control group (data not shown) (P = 0.0713, Student's t test). Over 90% women sampled (38/42) had a low vaginal pH (pH 3.5 to 4.5).
TABLE 2.
Species compositions of vaginal communities in women with and without frequent VVC
| Phylotypea | % of phylotypes in communities
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster Ib
|
Cluster II
|
Cluster III (Y36; n = 96)c | Cluster IV (Y23; n = 96)c | |||||||
| Y12 (n = 96)c | Y41 (n = 95) | Y35 (n = 93) | Y2 (n = 96) | Y14 (n = 95)c | Y19 (n = 96)c | Y30 (n = 94) | Y37 (n = 96)c | |||
| L. crispatus | 86.5 | 89.5 | 94.6 | 99 | 24.2 | 0 | 0 | 16.7 | 0 | 0 |
| L. iners | 0 | 7.4 | 2.2 | 0 | 56.8 | 76 | 81.9 | 43.8 | 0 | 29.2 |
| L. gasseri | 13.5 | 0 | 0 | 0 | 0 | 4.2 | 0 | 0 | 100 | 0 |
| L. jensenii | 0 | 1.1 | 1.1 | 0 | 12.6 | 15.6 | 16 | 30.2 | 0 | 9.4 |
| L. galinarum | 0 | 0 | 0 | 0 | 1.1 | 0 | 0 | 0 | 0 | 0 |
| L. kitasatonis | 0 | 0 | 1.1 | 0 | 1.1 | 0 | 0 | 0 | 0 | 0 |
| L. vaginalis | 0 | 0 | 1.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lactobacillus sp. strain KC38d | 0 | 2.1 | 0 | 1 | 4.2 | 4.2 | 1.1 | 9.4 | 0 | 5.2 |
| A. vaginae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55.2 |
| Diaphorobacter sp. | 0 | 0 | 0 | 0 | 0 | 0 | 1.1 | 0 | 0 | 0 |
| Prevotella bivia | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Clones were classified by comparing their 16S rRNA gene sequences to those of known organisms. Genus and species names were used if the sequence similarity to a type species was >97%; only the genus only was used if the sequence similarity was <97% but >90%.
Community clusters are based on T-RFLP analysis of 16S rRNA genes. See Fig. 1. The numbers of women with the clusters were as follows: cluster I, 24; cluster II, 10; cluster III, 4; and cluster IV, 3.
Woman with frequent VVC. n is the number of clones in the library that were sequenced.
Lactobacillus sp. strain KC38 was first characterized by Pavlova et al. (26), and it is phylogenetically distinct from known species of Lactobacillus.
Several clinical trials have been conducted to determine the efficacy of probiotics for prevention of RVVC (7, 27, 30, 40). The results of these trials have not shown a clear benefit of taking lactobacilli orally or intravaginally for the prevention of RVVC. This is consistent with our findings that women who experienced frequent VVC did not have reduced numbers of Lactobacillus species or an increased vaginal pH, which suggests that Lactobacillus species may not play a major role in defense against overt candidiasis.
The pathogenic and immunological mechanisms that allow the recurrence of VVC are still obscure. It is well known that lactic acid bacteria metabolize glycogen and other components in vaginal secretions and produce lactic acid, which lowers the pH and creates a restrictive environment that precludes the growth of many pathogenic organisms. The results of this study showed that the bacterial communities of women who experience frequent VVC all include high numbers of lactic acid-producing bacteria, including various species of lactobacilli and Atopobium. This is consistent with the low vaginal pH observed in subjects sampled in this study, suggesting that a low vaginal pH per se might not be sufficient to prevent the recurrence of VVC and that other factors may be more important.
The evidence available suggests that VVC may often reflect a resurgence of Candida rather than reinfection (31, 33). It has been shown that strains isolated before and after treatment are identical in more than two-thirds of candidiasis recurrences, indicating the persistence of some strains of yeast that colonize the vagina (4, 36, 38). The transition from a quiescent (silent) phase to an acute phase in a woman with frequent VVC is associated with increased numbers of Candida organisms. However, the host response to the presence of greater numbers of Candida in the vagina may determine whether a woman is symptomatic in the acute frequent VVC phase. It has been documented that host hypersensitivity or immune mechanisms are probably involved in this symptomatic process (33, 41). Fidel et al. proposed that the symptoms related to the presence of Candida are due to infiltration of polymorphonuclear neutrophils (9); that is, symptoms of Candida infection are not the result of a failure of a woman's immune system but rather are a result of an overly aggressive innate immune response (8).
It should be noted that our study, along with other previous studies, characterized the composition of the vaginal microbiota of women with frequent VVC in a cross-sectional fashion. Moreover, most subjects in our study were in the quiescent (silent) phase and were not experiencing acute frequent VVC at the time of sampling. Thus, we cannot exclude the possibility that the composition of the vaginal communities of these subjects may vary over time (even over very short time scales). It could be that the vaginal communities of women with frequent VVC vary more often or more extensively than those of women who do not have frequent VVC. It might be interesting and potentially important to set up longitudinal studies for assessment of the composition of the vaginal microbiota of women with frequent VVC. This may help us to better understand the dynamics of vaginal microbial communities in women with frequent VVC and to establish whether fluctuations in these communities are linked to the onset of acute frequent VVC.
The results of this study failed to provide evidence of altered or unusual vaginal bacterial communities in women with frequent VVC compared with women without frequent VVC. This suggests that the risk of recurrent yeast infections is not correlated with the composition of vaginal bacterial communities. Specifically, the results indicate that the vaginal communities of women with frequent VVC did not have reduced proportions of lactobacilli. These findings suggest that indigenous vaginal bacterial species, such as Lactobacillus species, may not be key players in the defense against Candida infections.
Acknowledgments
We thank Ursel Schütte, Jacob D. Pierson, and Maria G. Schneider for their technical assistance; Celeste Brown and Zaid Abdo for their valuable advice and help with analyses of T-RFLP and sequence data; and Linda Rogers and Ivan Kuletz for editing the manuscript.
This project was supported by grants P20RR16448 and P20 RR016454 from the National Center for Research Resources, a component of the National Institutes of Health. Financial support from the Procter & Gamble Company, Cincinnati, OH, was used to design and conduct the study.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Editor: A. Casadevall
Footnotes
Published ahead of print on 15 June 2009.
REFERENCES
- 1.Abdo, Z., U. Schuette, S. Bent, C. Williams, L. J. Forney, and P. Joyce. 2006. Statistical methods for characterizing diversity in microbial communities by analysis of terminal restriction fragment length polymorphism of 16S rRNA. Environ. Microbiol. 8929-938. [DOI] [PubMed] [Google Scholar]
- 2.Brown, C. J., M. Wong, C. C. Davis, A. Kanti, X. Zhou, and L. J. Forney. 2007. Preliminary characterization of the normal microbiota of the human vulva using cultivation-independent methods. J. Med. Microbiol. 56271-276. [DOI] [PubMed] [Google Scholar]
- 3.Calderon, L., R. Williams, M. Martinez, K. V. Clemons, and D. A. Stevens. 2003. Genetic susceptibility to vaginal candidiasis. Med. Mycol. 41143-147. [DOI] [PubMed] [Google Scholar]
- 4.Chong, P. P., Y. L. Lee, B. C. Tan, and K. P. Ng. 2003. Genetic relatedness of Candida strains isolated from women with vaginal candidiasis in Malaysia. J. Med. Microbiol. 52657-666. [DOI] [PubMed] [Google Scholar]
- 5.Cotch, M. F., S. L. Hillier, R. S. Gibbs, and D. A. Eschenbach. 1998. Epidemiology and outcomes associated with moderate to heavy Candida colonization during pregnancy. Vaginal Infections and Prematurity Study Group. Am. J. Obstet Gynecol. 178374-380. [DOI] [PubMed] [Google Scholar]
- 6.de Leon, E. M., S. J. Jacober, J. D. Sobel, and B. Foxman. 2002. Prevalence and risk factors for vaginal Candida colonization in women with type 1 and type 2 diabetes. BMC Infect. Dis. 21-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falagas, M. E., G. I. Betsi, and S. Athanasiou. 2006. Probiotics for prevention of recurrent vulvovaginal candidiasis: a review. J. Antimicrob. Chemother. 58266-272. [DOI] [PubMed] [Google Scholar]
- 8.Fidel, P. L., Jr. 2007. History and update on host defense against vaginal candidiasis. Am. J. Reprod. Immunol. 572-12. [DOI] [PubMed] [Google Scholar]
- 9.Fidel, P. L. J., M. Barousse, T. Espinosa, M. Ficarra, J. Sturtevant, D. H. Martin, A. J. Quayle, and K. Dunlap. 2004. An intravaginal live Candida challenge in humans leads to new hypotheses for the immunopathogenesis of vulvovaginal candidiasis. Infect. Immun. 722939-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7210-220. [DOI] [PubMed] [Google Scholar]
- 11.Foxman, B. 1990. The epidemiology of vulvovaginal candidiasis: risk factors. Am. J. Public Health 80329-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foxman, B., R. Barlow, H. D'Arcy, B. Gillespie, and J. D. Sobel. 2000. Candida vaginitis: self-reported incidence and associated costs. Sex. Transm. Dis. 27230-235. [DOI] [PubMed] [Google Scholar]
- 13.Fredricks, D. N., T. L. Fiedler, and J. M. Marrazzo. 2005. Molecular identification of bacterial vaginosis. N. Engl. J. Med. 3531899-1911. [DOI] [PubMed] [Google Scholar]
- 14.Gao, Z., C. H. Tseng, Z. Pei, and M. J. Blaser. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA 1042927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillier, S. L., M. A. Korhn, R. P. Nuget, and R. S. Gibbs. 1992. Characteristics of three vaginal flora patterns assessed by gram stain among pregnant women. Am. J. Obstet Gynecol. 166938-944. [DOI] [PubMed] [Google Scholar]
- 16.Hillier, S. L., M. A. Krohn, L. K. Rabe, S. J. Klebanoff, and D. A. Eschenbach. 1993. The normal vaginal flora, H2O2-producing lactobacilli, and bacterial vaginosis in pregnant women. Clin. Infect. Dis. 16S273-S821. [DOI] [PubMed] [Google Scholar]
- 17.Hurley, R., and J. De Louvois. 1979. Candida vaginitis. Postgrad. Med. J. 55645-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, P. S., E. J. Leys, J. M. Bryk, F. J. Martinez, M. L. Moeschberger, and A. L. Griffen. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 443665-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez, R. C., S. A. Franceschini, M. C. Patta, S. M. Quintana, A. C. Nunes, J. L. Moreira, K. C. Anukam, G. Reid, and E. C. De Martinis. 2008. Analysis of vaginal lactobacilli from healthy and infected Brazilian women. Appl. Environ. Microbiol. 744539-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClelland, R. S., B. A. Richardson, W. M. Hassan, V. Chohan, L. Lavreys, K. Mandaliya, J. Kiarie, W. Jaoko, J. O. Ndinya-Achola, J. M. Baeten, A. E. Kurth, and K. K. Holmes. 2008. Improvement of vaginal health for Kenyan women at risk for acquisition of human immunodeficiency virus type 1: results of a randomized trial. J. Infect. Dis. 1971361-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson, J. K., E. Holmes, and I. D. Wilson. 2005. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3431-438. [DOI] [PubMed] [Google Scholar]
- 22.Nocker, A., M. Burr, and A. K. Camper. 2007. Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54276-289. [DOI] [PubMed] [Google Scholar]
- 23.Nugent, R. P., M. A. Krohn, and S. L. Hillier. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 29297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pace, N. R. 1997. A molecular view of microbial diversity and biosphere. Science 276734-740. [DOI] [PubMed] [Google Scholar]
- 25.Palmer, C., E. M. Bik, D. B. DiGiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlova, S. I., A. O. Kilic, S. S. Kilic, J. S. So, M. E. Nader-Macias, J. A. Simoes, and L. Tao. 2002. Genetic diversity of vaginal lactobacilli from women in different countries based on 16S rRNA gene sequences. J. Appl. Microbiol. 92451-459. [DOI] [PubMed] [Google Scholar]
- 27.Pirotta, M., J. Gunn, P. Chondros, S. Grover, P. O'Malley, S. Hurley, and S. Garland. 2004. Effect of Lactobacillus in preventing post-antibiotic vulvovaginal candidiasis: a randomised controlled trial. BMJ 329548-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pirotta, M. V., J. M. Gunn, and P. Chondros. 2003. “Not thrush again!” Women's experience of post-antibiotic vulvovaginitis. Med. J. Aust. 17943-46. [DOI] [PubMed] [Google Scholar]
- 29.Schwebke, J. R. 2005. Abnormal vaginal flora as a biological risk factor for acquisition of HIV infection and sexually transmitted diseases. J. Infect. Dis. 1921315-1317. [DOI] [PubMed] [Google Scholar]
- 30.Shalev, E., S. Battino, E. Weiner, R. Colodner, and Y. Keness. 1996. Ingestion of yogurt containing Lactobacillus acidophilus compared with pasteurized yogurt as prophylaxis for recurrent candidal vaginitis and bacterial vaginosis. Arch. Fam. Med. 5593-596. [DOI] [PubMed] [Google Scholar]
- 31.Sheary, B., and L. Dayan. 2005. Recurrent vulvovaginal candidiasis. Aust. Fam. Physician 34147-150. [PubMed] [Google Scholar]
- 32.Siegler, S. L. 1946. A new method for treatment of vaginitis and cervicitis. Am. J. Obstet. Gynecol. 521-13. [DOI] [PubMed] [Google Scholar]
- 33.Sobel, J. D. 2007. Vulvovaginal candidosis. Lancet 3691961-1971. [DOI] [PubMed] [Google Scholar]
- 34.Sobel, J. D., and W. Chaim. 1996. Vaginal microbiology of women with acute recurrent vulvovaginal candidiasis. J. Clin. Microbiol. 342497-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sobel, J. D., S. Faro, R. W. Force, B. Foxman, W. J. Ledger, P. R. Nyirjesy, B. D. Reed, and P. R. Summers. 1998. Vulvovaginal candidiasis: epidemiologic, diagnostic, and therapeutic considerations. Am. J. Obstet. Gynecol. 178203-211. [DOI] [PubMed] [Google Scholar]
- 36.Sobel, J. D. 1985. Epidemiology and pathogenesis of recurrent vulvovaginal candidiasis. Am. J. Obstet. Gynecol. 152924-935. [DOI] [PubMed] [Google Scholar]
- 37.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39321-346. [DOI] [PubMed] [Google Scholar]
- 38.Vazquez, J. A., J. D. Sobel, R. Demitriou, J. Vaishampayan, M. Lynch, and M. J. Zervos. 1994. Karyotyping of Candida albicans isolates obtained longitudinally in women with recurrent vulvovaginal candidiasis. J. Infect. Dis. 1701566-1569. [DOI] [PubMed] [Google Scholar]
- 39.Vitali, B., C. Pugliese, E. Biagi, M. Candela, S. Turroni, G. Bellen, G. G. Donders, and P. Brigidi. 2007. Dynamics of vaginal bacterial communities in women developing bacterial vaginosis, candidiasis, or no infection, analyzed by PCR-denaturing gradient gel electrophoresis and real-time PCR. Appl. Environ. Microbiol. 735731-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, A. B., C. Yu, K. Tashima, J. Burgess, and K. Danvers. 2001. Evaluation of two self-care treatments for prevention of vaginal candidiasis in women with HIV. J. Assoc. Nurses AIDS Care 1251-57. [DOI] [PubMed] [Google Scholar]
- 41.Witkin, S. S., I. M. Linhares, and P. Giraldo. 2007. Bacterial flora of the female genital tract: function and immune regulation. Best Pract. Res. Clin. Obstet. Gynaecol. 21347-354. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 1502565-2573. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, X., C. G. Brown, Z. Abdo, C. C. Davis, M. A. Hansmann, P. Joyce, J. A. Foster, and L. J. Forney. 2007. Difference in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1121-133. [DOI] [PubMed] [Google Scholar]