Abstract
Trypanosoma congolense is a protozoan parasite that causes severe diseases in livestock. Three major quantative trait loci (QTL), Tir1, Tir2, and Tir3, control the survival time of mice after infection with T. congolense. Congenic mice carrying the C57BL/6 resistance alleles on the A/J background were developed for each of these loci. The congenic mice were used to physically map the regions containing the QTL gene(s) and to investigate the physiological effect of each locus. Clinical chemistry data for infected A/J, C57BL/6, and BALB/c mice were obtained for 15 analytes at five time points. Congenic mice were assessed for survival, parasitemia, and anemia as well as seven clinical-chemical analytes. The survival times were significantly increased in the Tir1 and Tir2 mice but not Tir3 congenic mice. The survival time of the parental inbred mice correlated negatively with parasitemia but positively with alanine aminotransferase activities in serum, suggesting that inflammatory reactions in the liver had a beneficial effect possibly associated with reduced parasitemia. However, there was no difference in parasitemia or liver enzyme activities of Tir1 and Tir2 congenic mice relative to their controls, showing that survival, parasitemia, and degree of liver damage are not associated with each other, despite the correlation in the parental lines. These data suggest that the congenic loci affect survival but do not affect control of parasite number. They may therefore act by limiting the pathological consequences of T. congolense infection.
African trypanosomes are extracellular protozoan parasites that multiply in the bloodstream and cause severe diseases in humans and livestock with fatal consequences unless treated. While trypanosomiasis due to Trypanosoma brucei causes significant morbidity and mortality in humans, trypanosomiasis caused by Trypanosoma congolense and Trypanosoma vivax is one of the most significant constraints on cattle production in Africa and the cause of major economic losses with serious effects on human health and welfare (13). Anemia is the key feature of disease in cattle and persists after the first wave of parasitemia when parasite numbers have declined to low or undetectable levels. Anemia rather than parasitemia is best correlated with productivity and is used as the primary indicator of when to treat the infection; treatment to clear the parasites usually resolves the anemia.
African trypanosomes also infect laboratory mice, and the survival time after infection varies significantly between inbred strains of mice. A/J, BALB/c, and C57BL/6 mice have survival times of ∼16, 50, and 110 days after infection with T. congolense strain 5E-12 (17a), and similar survival times have been observed with T. congolense IL1180 used in this study (8-10). However differences in survival times between C57BL/6, A/J, and BALB/c mice are not dependent on the degree of anemia developed (17, 24), suggesting that effects other than anemia are the major cause of death in mice. Nevertheless any insight into the mechanisms regulating anemia in mice may be of relevance to the anemia of cattle.
Three quantitative trait loci (QTL) affecting survival time after infection have been mapped in independent crosses between C57BL/6 and A/J mice and between C57BL/6 and BALB/c mice (10). The three loci located on proximal chromosome 17, near the center of chromosome 5, and distal chromosome 1, respectively, were designated Tir1, Tir2, and Tir3 for “Trypanosoma infection response.” The genomic positions of these large loci were refined to five smaller regions in a study of the same crosses that were extended to the F6 generation (8). The QTL contributed 36, 22, and 32 days, respectively, to the difference in survival in the C57BL/6 × BALB/c F2 cross and 31 and 22 days for Tir1 and Tir2 in the A/J × C57BL/6 F2 cross. The Tir3 locus was nonsignificant in the F2 A/J × C57BL/6 cross but highly significant in a subsequent F6 cross. These regions contain hundreds of genes; a combination of strategies is therefore required to identify plausible candidate genes for detailed evaluation. We have therefore collected extensive clinical chemistry data on these animals in order to help identify candidate genes that might plausibly control aspects of phenotype that correlate with survival.
Our previous mapping studies have shown that A/J and BALB/c mice appear to carry the same susceptibility alleles at the Tir1, Tir2, and Tir3 loci (8, 10), although BALB/c mice have a significantly longer survival time than A/J mice (50 versus 16 days). Therefore, phenotypes regulated by strain-specific genotypes of QTL affecting survival time are likely to exhibit similar profiles in A/J and BALB/c mice, with BALB/c mice possibly showing intermediate values for parameters that are correlated with survival. We do not expect to see intermediate values of any phenotype in C57BL/6 mice, and consequently, where we do, that trait is unlikely to be associated with survival. Hence, by including BALB/c mice in any experiment, we can increase our power to detect correlations between any given parameter and survival.
Congenic mice are a powerful tool for identifying the specific effect of a single locus on a phenotype. A panel of three congenic mouse lines for the Tir loci has been created previously and phenotyped at the N4 generation (12). We have now backcrossed the congenic lines to A/J for a further 4 generations to N8 to reduce the expected amount of residual non-target-donor DNA from 6% to <1%. And we have produced a specific control line for each congenic line, since it has been shown that even the small proportion of residual non-target-donor DNA that remains in the N8 genome after 7 generations of backcrossing can double the survival time of the mice. These controls carry the same nontarget regions derived from C57BL/6 mice as the congenic lines, and since these are present in test and control animals, they are not expected to give rise to observable differences in response to infection. Using these mice, it is possible to evaluate the effect of each QTL on the phenotype.
While the immune response to infection with T. congolense has been studied intensively (19, 31), we are not aware of any systematic studies of the clinical chemistry of mice following infection with African trypanosomes, with the exception of studies of the interaction between host and parasite glycolysis (3, 33). Clinical chemistry is routinely used in human medicine to identify pathological alterations of tissues, organ functions, or metabolic pathways that develop during the course of a disease, to estimate the severity of these alterations, and to decide on interventions that might be required.
In this article, we test the hypothesis that pathological consequences of trypanosomiasis, other than anemia, are responsible for differences in survival times of inbred mouse strains by reporting on 15 clinical chemical parameters for three inbred mouse strains and 8 parameters for six congenic mouse lines at five time points over the course of T. congolense infection.
MATERIALS AND METHODS
The founders of the inbred mouse lines C57BL/6J OlaHsd (C57BL/6), A/J OlaHsdnd (A/J), and BALB/cJ OlaHsdce (BALB/c) were purchased from Harlan UK, Ltd. (Bicester, Oxon, United Kingdom), and bred in the small animal unit at the International Livestock Research Unit, together with the congenic mice. All animals were treated in accordance with the Institute's Animal Care and Use Committee policies.
Congenic mouse lines are created by crossing two different lines, one of which is designated the donor line (C57BL/6 in this case) and the other the recipient strain (A/J). At each generation after F1, the offspring are genotyped to identify those animals that carry alleles from the donor strain (C57BL/6) in the region of interest. These are then backcrossed to the recipient line (A/J). The amount of donor genome present in the recipient line of the offspring halves at each generation, and by the N7 generation, it is reduced to <1% in addition to the target region.
Three congenic lines were created corresponding to each of the predicted Trypanosoma infection response (Tir) loci: Tir1, Tir2, and Tir3 on chromosomes 17, 5, and 1, respectively. The progeny at each backcross generation were genotyped with microsatellite markers defining the genetic intervals containing the QTL using the following markers: D17Mit29, D17Mit16, and D17Mit11 for Tir1; D5Mit200, D5Mit157, and D5Mit58 for Tir2; and D1Mit60, D1Mit217, and D1Mit87 for Tir3. At the seventh generation of backcrossing, each line was typed with a series of markers at ∼2-cM intervals flanking the Tir loci. The individuals with the shortest donor haplotype extending beyond the QTL interval were used for breeding the next generation by intercrossing full- or half-sib male and female carriers of the C57BL/6 donor region. The progeny of these were genotyped, and those individuals homozygous for the alternative haplotypes were used as founders to propagate each line, which were denoted either “Tir1AA,” “Tir2AA,” or “Tir3AA” for homozygotes at the QTL for the recipient A/J haplotype or “Tir1CC,” “Tir2CC,” or “Tir3CC” for homozygotes at the QTL for the donor C57BL/6 haplotype. Thus, a total of six lines were produced: these are Tir1AA and Tir1CC, Tir2AA and Tir2CC, and Tir3AA and Tir3CC. The Tir1CC line is homozygous for a C57BL/6 haplotype spanning the 10-cM interval between markers D17Mit84 and D17Mit177 on Mmu17. The Tir2CC line is homozygous for a C57BL/6 haplotype spanning ∼30 cM between markers D5Mit184 and D5Mit136 on Mmu5. Tir3CC individuals have a C57BL/6 haplotype spanning ∼10 cM between markers D1Mit49 and D1Mit139 on Mmu1.
The recommended names for the test lines according to the Mouse Genome Informatics would be “A.B6-Tir1,” “A.B6-Tir2,” and “A.B6-Tir3.” However, there is no recommended nomenclature for the control lines, so for clarity, the style Tir1AA to -3AA and Tir1CC to -3CC will be used in the following description. Survival of congenic mouse lines was compared using the Kaplan-Meier log rank test implemented in SPSS.
The homozygous congenic mouse lines were genotyped at the Wellcome Trust Clinical Research Facility, Edinburgh, United Kingdom, using the Illumina mouse medium-density linkage panel on an Illumina BeadStation 500 instrument. This panel contains 1,449 single nucleotide polymorphism (SNP) markers, of which 959 were informative between A/J and C57BL/6, with a mean spacing of 2.61 Mb. Using these data, it was possible to identify the approximate boundaries of the introgressed regions and also to identify nontarget regions of C57BL/6 origin that had been carried through into the congenic lines.
Clinical chemistry data were collected from a total of nine lines: the three inbred strains A/J, BALB/c, and C57BL/6; each of the three congenic lines Tir1CC, Tir2CC, and Tir3CC; and each of their respective control lines, Tir1AA, Tir2AA, and Tir3AA. Mice were co-housed, and each cage was randomly allocated a place on the rack to minimize batch effects of the environment. Forty mice of each line were infected intraperitoneally (i.p.) with 104 T. congolense IL1180 parasites, and 10 mice of each line were inoculated i.p. with 0.01 M phosphate-NaCl (pH 8.0)-1% glucose buffer (PSG) as controls. All mice that were not being monitored for parasitemia were checked by microscopy for patent parasitemia at day 7 when a 3-μl blood sample was taken from the tail. Mice in which no parasites were seen were resampled after 48 h. Six of the 320 infected mice remained aparasitemic and were excluded from the study.
Mice were weighed and killed by CO2 anesthesia at different time points to collect serum samples.
Groups of 10 mice of each line were sampled immediately before infection and on each of days 3, 9, 17, and 35 following infection. On each occasion, processing started at 1:30 p.m. and was completed by 5:00 p.m. Food and water were not withdrawn before killing. In order to minimize hemolysis during postmortem sample collection, blood was collected by opening the thoracic cavity, removing the sternum, cutting the vena cava caudalis and the aorta cranial to the diaphragm, and collecting all the blood from the thoracic cavity using a pipette; the volume collected was 0.5 to 1 ml. Blood was left for 2 h at room temperature to clot and then stirred and centrifuged at 4,600 × g for 10 min, and the serum was collected.
Serum samples were stored at −80°C until all samples were collected. Then they were shipped to the German Mouse Clinic on dry ice and stored again at −80°C until they were analyzed. For the clinical chemistry analysis, samples were thawed, centrifuged, transferred to 1.5-ml Eppendorf tubes, and stored overnight at 4°C before analysis using an AU400 autoanalyzer (Olympus) and adapted test kits (Olympus). Samples were briefly vortexed and centrifuged again to remove clots. Uric acid concentrations were measured from undiluted samples, except for the samples from the three parental inbred strains, which were diluted for all parameters. For the analysis of all other parameters, samples were diluted 1:2 with deionized water, mixed, and centrifuged again, before being tested (see Table 2). Samples that contained <90 μl of serum were pooled with one other sample collected from another animal of the same strain or congenic line at the same time point, in order to get enough material to test all parameters chosen.
TABLE 2.
Clinical chemistry of inbred micea
| Analyte |
P value for:
|
Adjusted r2 | ||
|---|---|---|---|---|
| Strain | Day | Day × strain | ||
| ALAT | 0.04 | <0.001 | 0.028 | 0.335 |
| α-Amylase | 0.001 | <0.001 | <0.001 | 0.584 |
| Albumin | <0.001 | <0.001 | 0.035 | 0.711 |
| ALP | 0.007 | <0.001 | 0.041 | 0.597 |
| Chloride | <0.001 | <0.001 | 0.024 | 0.423 |
| Creatinine | NSb | 0.025 | 0.043 | 0.211 |
| Glucose | 0.001 | 0.003 | 0.052 | 0.498 |
| Phosphorus | NS | NS | NS | 0.011 |
| Lactate | 0.003 | NS | NS | 0.16 |
| Lipase | NS | NS | NS | 0.067 |
| Potassium | 0.001 | 0.009 | <0.001 | 0.388 |
| Sodium | 0.069 | 0.001 | 0.033 | 0.277 |
| Transferrin | <0.001 | <0.001 | <0.001 | 0.726 |
| Urea | 0.002 | <0.001 | 0.394 | 0.407 |
| Uric acid | 0.008 | <0.001 | 0.053 | 0.364 |
Values represent the probabilities of strain- and/or time-dependent differences in response to infection for the analytes tested. ALAT data were logn transformed before analysis; all other data were approximately normally distributed. There were highly significant effects detected for the levels of most analytes both over time (day) and between strains. No multiple-testing correction has been applied, but if Bonferroni's correction was used, then 0.0033 would be equivalent to the 0.05 significance threshold used here. Plots of transferrin levels have been published previously but are included here for completeness (25).
NS, not significant.
Hemoglobin levels.
Before the mice were killed, three samples of 2 μl of whole blood were collected from the tail (after discarding the first drop). The relative hemoglobin concentrations were measured spectrophotometrically at 540 nm using Drabkin's method (2a). Each of the triplicate samples of 2 μl of tail blood was diluted in 200 μl of Drabkin's solution mixed with Brij detergent (Sigma) in a 96-well round-bottom plate (Costar 3799; Corning, Inc., Corning, NY). After 30 min at room temperature, the optical density was measured at 540 nm in an enzyme-linked immunosorbent assay plate reader (Multiscan MCC/340; Titertek Instruments, Huntsville, AL).
Parasitemia.
At days 4, 6, 8, 10, and 13, 3 μl of blood was collected from the tails of five mice per strain or congenic line. The blood was diluted 100:1 in two stages, and parasitemia was determined with a hemocytometer.
Statistics.
Data were explored by analysis of variance using the general linear model univariate procedure in SPSS. The inbred mice and each of the pairs of congenic lines and their controls were analyzed independently, and no comparison was made between different congenic lines or between congenic lines and inbred mice. Consequently there were a maximum of three strains and five time points for each analysis. Time points were considered as an explanatory variable (i.e., independent) as mice were sacrificed at each time point. Duncan's multiple comparison test was used to identify strains or time points that differed significantly from each other. No multiple-testing protection was applied to the results since the primary objective was to generate hypotheses by screening a large panel of analytes for any that might show an association with breed. Histograms of standardized residuals and plots of fitted values versus residuals were used to check the assumptions of the model, and natural log transformations were used where necessary.
RESULTS
Positions of donor haplotypes within recipient genome.
SNP genotypes of the mice obtained with the Illumina 1,536-SNP panel are shown in supplemental material for genotypes with positions relative to mouse genome assembly NCBI36. These data showed that the Tir1CC congenic mice carried the expected region of C57BL/6 origin between 26.0 and 43.9 Mb on Mmu17 in the A/J background. The Tir2CC mice carry four regions of C57BL/6 origin on chromosome 5: 27.4 to 32.9 Mb, 35.8 to 37.4 Mb, around 78.5 Mb, and 99.5 to 118.2 Mb. The Tir3CC mice harbor a region of C57BL/6 origin between 93.3 and 123.6 Mb on Mmu1. In addition, all congenic lines carried nontarget C57BL/6 sequences on some other chromosomes: 70.7 to 82.6 Mb on Mmu15 in 4/5 Tir1CC mice; 52.8 to 63.7 Mb on Mmu14 in 5/5 Tir1CC mice; around 98.8, 102.2, and 121.5 Mb on Mmu8 in 1/3 Tir2CC mice; 108.5 to 117.8 Mb on Mmu9 in 1/3 Tir2CC mice; 51.6 to 86.4 Mb on Mmu8 in 3/4 Tir3CC mice; and 0 to 21.7 Mb on Mmu6 in 3/4 Tir3CC mice. In most cases, these C57BL/6 regions were also present in the littermate control mice and were therefore unlikely to account for genotype-specific differences found between carriers of the congenic region and control mice from the same line. Complete data are shown in the supplemental material table for genotypes.
Effect of congenic loci on survival.
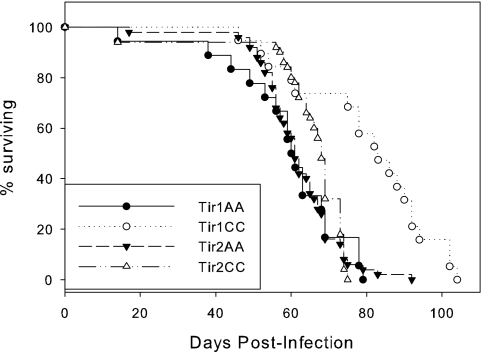
Survival times postinfection are shown in Table 1 and Fig. 1. The Tir1CC mice carrying the C57BL/6 allele at the Tir1 locus survived 21 days longer than their littermate controls carrying the A/J allele at this locus. Similarly, the Tir2CC animals carrying the C57BL/6 allele at the Tir2 locus survived 10 days longer than their littermate controls carrying the A/J allele. There was no significant difference in survival between the Tir3CC and Tir3AA mice.
TABLE 1.
Mean number of days that carriers of each congenic region and littermate controls survived after infection
| Strain | No. of mice | Mean survival (days) | SE | χ2P valuea |
|---|---|---|---|---|
| Tir1AA | 18 | 60.33 | 3.75 | <0.001 |
| Tir1CC | 21 | 81.24 | 3.56 | |
| Tir2AA | 52 | 63.67 | 1.55 | <0.001 |
| Tir2CC | 94 | 73.99 | 1.47 | |
| Tir3AA | 19 | 57.84 | 1.95 | 0.905 |
| Tir3CC | 66 | 57.35 | 1.14 |
The χ2 P values are for the significance of the Kaplan-Meier function for difference in survival.
FIG. 1.
Survival of Tir1CC and Tir2CC congenic mice and their control lines (Tir1AA and Tir2AA) after infection with T. congolense showing the difference in survival between lines. In each case, the Tir1CC or Tir2CC line survived longer than the Tir1AA or Tir2AA line.
Clinical chemistry of inbred mice.
Clinical chemistry data were obtained in two steps. In step 1, parental A/J, BALB/c, and C57BL/6 inbred mice were screened for 15 parameters prior to infection and at four time points (3, 9, 17, and 35 days postinfection). In step 2, the congenic mice were screened for seven parameters that showed differences between the inbred strains or that varied over the period of infection. The complete results are available in the supplemental material, under “Clinical chemistry.” P values indicating the probability of a significant influence of strain, day postinfection, and the interaction strain × day postinfection on each parameter measured are shown in Table 2.
Electrolytes.
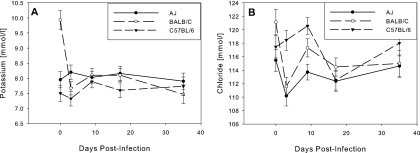
Levels of potassium and chloride in preinfection samples from BALB/c mice were relatively high for unknown reasons, but by day 3 postinfection, these had dropped to within the range for A/J and C57BL/6 mice. Thereafter, there were only small differences in potassium and chloride levels between the inbred parental mouse strains (Fig. 2). C57BL/6 mice had a mean K+ of 7% less than A/J and BALB/c mice; chloride in A/J mice was significantly lower than in BALB/c or C57BL/6 mice, but only by 1 to 3%. There were no significant differences between strains in sodium and inorganic phosphorus (not shown). The very small absolute variations in potassium and chloride levels between strains were statistically significant (P < 0.001), but this may be a reflection of the highly reproducible measurements of these analytes rather than any clinical significance.
FIG. 2.
Levels of electrolytes over the course of infection. (A) Potassium; (B) chloride. Means ± standard errors are shown. BALB/c mice had high initial levels of both potassium and chloride, but these dropped by day 3 to within the range of the other strains. From day 3 onwards, all values for all strains remained within a tight band.
Serum enzyme activities.
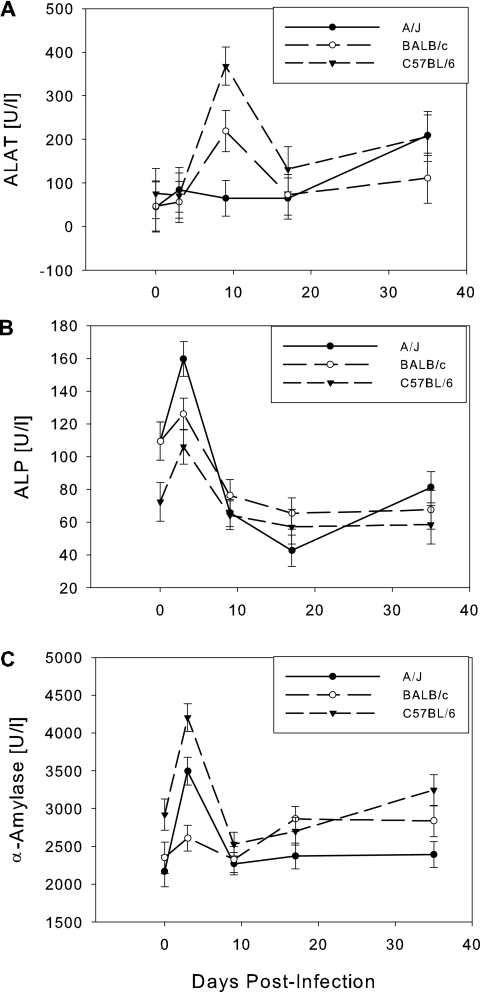
Alanine aminotransferase (ALAT) activity (Fig. 3) reached a maximum around the first peak of parasitemia at day 9 in BALB/c and C57BL/6 mice, but did not respond until day 35 in A/J mice. A/J is the most susceptible strain, and by day 35 postinfection, many of the A/J mice began to show visible symptoms, such as swollen spleen, spiky coat, hunched posture, and reduced mobility.
FIG. 3.
Serum enzymes. (A) ALAT; (B) ALP; (C) α-amylase. Means ± standard errors are shown. ALP and α-amylase levels spiked in most mice at day 3. ALAT levels spiked at day 9 when levels were directly correlated with survival time.
Alkaline phosphatase (ALP) activity (Fig. 3B) showed a marked increase at day 3 postinfection in all strains and then returned to baseline levels.
α-Amylase activity peaked at day 3 in all three mouse strains and then returned to baseline levels (Fig. 3C). The strongest reaction was seen in C57BL/6 mice.
Markers of kidney function.
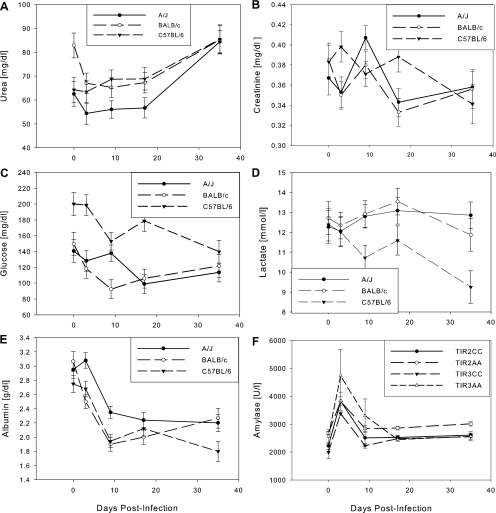
Kidney diseases lead to the failure of the urinary excretion of metabolites, thereby resulting in increased serum levels of creatinine and urea. Creatinine levels did not differ during the observation period between strains and changed only slightly over time, similar to urea levels, which remained constant in all strains until they increased by 21 to 47% at day 35 (Fig. 4A and B).
FIG. 4.
Levels of urea (A), creatinine (B), glucose (C), lactate (D), and serum albumin (E) in inbred mice and α-amylase (F) in congenic mice. Means ± standard errors are shown. Urea declined in BALB/c mice from day 0 to day 3; thereafter, all strains remained stable until day 17, after which they increased to day 35. A/J mice had lower levels at all time points. Creatinine remained within normal ranges at all time points. Both glucose and lactate declined in C57BL/6 mice over the course of the infection, but levels remained constant in the other two strains. The inverse relationship between glucose and lactate in C57BL/6 mice, compared to the other two strains, might indicate reduced glucose flux through the erythrocyte glycolysis pathway or increased gluconeogenesis in this strain. Albumin declined consistent with an acute-phase response; transferrin increased, presumably in response to the developing anemia. The differences in α-amylase between Tir2CC and Tir2AA mice and between Tir3CC and Tir3AA mice were significant. The general trend was very similar to that of the A/J parental strain (Fig. 3C).
Uric acid.
Uric acid is an intermediate in purine catabolism, and levels can be increased as a consequence of infections and massive cell death. Uric acid metabolism has been implicated in the control of malaria and also T. congolense infection in cape buffalo (6, 34). However, in the present study uric acid levels remained reasonably constant during the course of infection in A/J and BALB/c mice, while they fluctuated between 2 and 4 mg/dl in C57BL/6 mice (not shown). However, these values are within the ranges seen in wild-type mice of different strains.
Albumin concentration.
Albumin is known as a negative acute-phase protein since levels decline during the acute-phase response. Albumin declined throughout the infection in all mouse strains (Fig. 4F), probably indicating a decline in liver function, while total plasma protein remained constant (data not shown).
Glucose and lactate concentrations.
Glucose levels were 19 to 75% higher in C57BL/6 mice than in A/J and BALB/c mice at all time points, and glucose concentrations declined in all strains over the course of infection (Fig. 4C). Lactate levels remained constant in A/J and BALB/c mice but declined in C57BL/6 mice. Lactate levels were significantly lower in C57BL/6 mice than in A/J or BALB/c mice, particularly after day 3 (Fig. 4D).
Clinical chemistry findings in congenic mice.
The data for each congenic/control pair were compared, and the resulting probabilities are shown in Table 3. There were significant changes of all analytes over time in at least one congenic-control comparison, but only one, α-amylase, differed significantly between congenic mice and their respective control groups, and in this case the differences were small relative to the differences between parental strains. The Tir2 and Tir3 regions showed effects on α-amylase activity, and the Tir1 region showed a significant interaction between day and strain. All strains showed a spike in amylase activity at day 3 postinfection. Carriers of the C57BL/6 allele at the Tir2 and Tir3 loci displayed increased levels at day 3 and at days 3 and 9, respectively, compared to the corresponding control mice. This was in the same direction as the observed effect in the parental mice, suggesting that both loci might have an influence on amylase levels (Fig. 4F).
TABLE 3.
Clinical chemistry of congenic micea
| QTL and analyte |
P value for:
|
Adjusted r2 | Model | ||
|---|---|---|---|---|---|
| Strain | Day | Day × strain | |||
| Tir1 | |||||
| ALAT | 0.675 | <0.001 | 0.262 | 0.425 | Logn |
| Albumin | 0.359 | <0.001 | 0.201 | 0.649 | Normal |
| ALP | 0.372 | <0.001 | 0.027 | 0.334 | Normal |
| Glucose | 0.073 | <0.001 | 0.275 | 0.379 | Normal |
| Lactate | 0.258 | 0.020 | 0.322 | 0.093 | Normal |
| Transferrin | 0.865 | 0.009 | 0.588 | 0.107 | Normal |
| α-Amylase | 0.501 | <0.001 | 0.005 | 0.597 | Normal |
| Tir2 | |||||
| ALAT | 0.670 | <0.001 | 0.691 | 0.407 | Logn |
| Albumin | 0.16 | <0.001 | 0.52 | 0.45 | Normal |
| ALP | 0.798 | <0.001 | 0.703 | 0.231 | Normal |
| Glucose | 0.734 | <0.001 | 0.279 | 0.347 | Normal |
| Lactate | 0.166 | 0.007 | 0.021 | 0.208 | Normal |
| Transferrin | 0.200 | 0.006 | 0.144 | 0.147 | Normal |
| α-Amylase | 0.015 | <0.001 | 0.762 | 0.441 | Normal |
| Tir3 | |||||
| ALAT | 0.17 | <0.001 | 0.33 | 0.29 | Logn |
| Albumin | 0.765 | <0.001 | 0.179 | 0.586 | Normal |
| ALP | 0.67 | <0.001 | 0.23 | 0.27 | Normal |
| Glucose | 0.291 | <0.001 | 0.773 | 0.334 | Normal |
| Lactate | 0.21 | 0.07 | 0.72 | 0.04 | Normal |
| Transferrin | 0.148 | <0.001 | 0.955 | 0.234 | Normal |
| α-Amylase | 0.02 | <0.001 | 0.28 | 0.33 | Logn |
Values represent the analysis of variance probabilities of strain- and/or time-dependent responses to infection. There was a highly significant response to infection over time (day) in the levels of most analytes, since the day postinfection had a significant effect on almost all analytes in all strains. However, significant strain effects (P < 0.05) were seen for amylase activity only.
Effect of congenic loci on anemia.
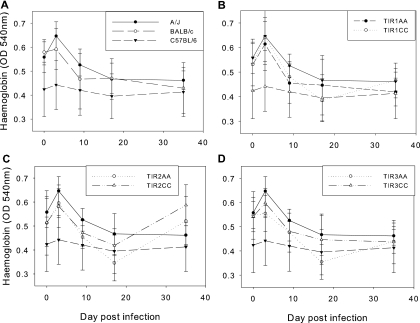
Anemia is an important component of the pathology in cattle, and the degree of anemia varies significantly between the parental mouse strains after infection. The hemoglobin levels of the congenic mice and their respective controls were measured in order to determine whether the genotype of the congenic regions affects hemoglobin levels and whether there was a significant association between anemia and survival (Fig. 5). The plots show that hemoglobin levels declined after infection, as expected. A/J mice had significantly higher hemoglobin than C57BL/6 mice; however, there was no evidence of significant differences in hemoglobin levels between any of the three congenic lines and their controls, suggesting that the congenic regions do not affect this phenotype.
FIG. 5.
Hemoglobin levels in congenic mice compared to their controls and the inbred mice. Means ± standard errors are shown. (A) Inbred strains; (B) Tir1 strains; (C) Tir2 strains; (D) Tir3 strains. Hemoglobin declined in all strains after infection, except in C57BL/6 mice, in which it started low and stayed low. Both the Tir2 strains appeared to make a recovery in hemoglobin levels, but since there was no difference between them, this is not likely to be due to the congenic region. The hemoglobins of A/J and C57BL/6 mice are shown on each of the congenic plots with the same symbols as in panel A.
Effect of congenic loci on parasitemia.
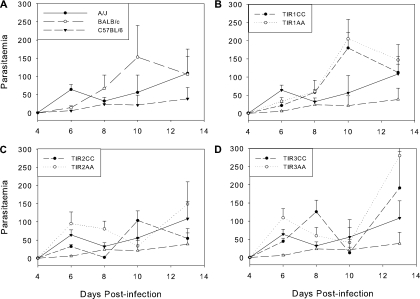
In mice, T. congolense parasitemia normally increases to a first peak about 7 to 9 days after infection with T. congolense strain IL1180. Although there is no direct correlation between parasitemia and survival, the intensity of parasitemia is both a consequence of the hosts' ability to control the parasite and a driver of many host responses. Parasitemia levels over the course of the infection are shown in Fig. 6. It is well established that BALB/c and A/J mice tend to have higher levels of parasitemia than the more resistant C57BL/6 mice, and this was confirmed again here. There was no evidence for control of parasitemia by any of the QTL in the congenic mice.
FIG. 6.
Parasitemia units in the three congenic strains compared to their controls and the inbred mice. Means ± standard errors are shown. OD, optical density. (A) Inbred strains; (B) Tir1 strains; (C) Tir2 strains; (D) Tir3 strains. The parasitemia of A/J and C57BL/6 mice is shown on each of the congenic plots with the same symbols as in panel A.
DISCUSSION
Survival time.
The Tir1CC and Tir2CC congenic mice survived 21 and 10 days longer after infection with T. congolense than their respective control strains. These differences in survival time are similar to those found in the earlier generation study (20 and 17 days, respectively) (12) and ∼10 days less than those predicted in the original F2 mapping experiment (31 and 22 days, respectively) (10). It is possible that combining lines would lead to a difference in survival that was greater than the sum of the differences found here. However, no evidence for interaction between loci was found in the original mapping studies (8, 10), and consequently any additional increase in survival in a combined line over the sum of the differences observed here is likely to be small.
The Tir3 QTL that was identified in the F2 study was found to consist of three QTL in the subsequent fine-mapping F6 study, which were designated Tir3a, Tir3b, and Tir3c (8). The Tir3 congenic line carried C57BL/6 alleles from 93.3 to 123.6 Mb on Mmu1, which corresponds to Tir3a, the most proximal of these QTL. It is possible that the map position of Tir3a has been confounded by the closely linked Tir3b and Tir3c QTL. The failure to detect a difference in this study, whereas a 28-day difference was found in the earlier congenic study (12), may be a consequence of additional recombination events in the introgressed region or the use of matched controls in this study rather than the common controls that were used in the earlier one. New congenic lines are being developed to test the effects of Tir3a, Tir3b, and Tir3c and might identify the number and location of these QTL.
Clinical chemistry.
The dominant features of the clinical chemistry data are the successful maintenance of homeostasis of the major organ systems during the first 35 days of infection. Although there are responses to infection at specific time points, it is clear that the mice are not suffering serious loss of any organ functions despite the intensity of the infection. The stability of electrolyte levels indicates that the animals are maintaining homeostasis during the infection and that therefore the animal has some degree of control over its response. However, a common feature in all groups investigated were decreasing levels of glucose and albumin during the course of the disease, most likely reflecting developing cachexia.
Differences in glucose and lactate levels between the inbred strains might be caused by genetically fixed background effects, since C57BL/6J mice are known to show glucose intolerance due to impaired insulin production (31a). In the absence of differences in insulin resistance, the increased glucose and reduced lactate levels in the C57BL/6 mice relative to A/J and BALB/c mice could also be a consequence of one or more of the following three factors: reduced flux through the erythrocyte glycolysis pathway in the presence of more severe anemia, reduced flux through the parasite glycolysis pathway in the presence of lower parasitemia, and increased gluconeogenesis.
African trypanosomes have been estimated to consume twice their own weight in glucose per day, and hence high parasitemia could have a significant impact on glucose levels; however, trypanosomes actively excrete pyruvate rather than lactate, so the apparent inverse correlation between glucose and lactate levels is not readily explained by parasite glycolysis. It is not possible to choose between the other two hypotheses with the data presented here.
A recent nuclear magnetic resonance-based study found that lactate tended to increase and glucose tended to decrease in the NMRI strain of mice infected with T. brucei (GVR35) (35). This is not consistent with the data presented here. It is evident from this study that the levels of these analytes are dependent on mouse strain, and they may also be dependent on parasite strain as well. The differences in mouse and parasite strains could therefore account for the differences between the two studies.
Transferrin is also a negative acute-phase protein that normally declines during the acute-phase response (28), but it is upregulated in response to anemia and iron deficiency. The samples collected for this study were assayed for transferrin, and the data have been published separately (25): the level of transferrin increased significantly over the course of the infection, particularly in the A/J mice. The fact that transferrin levels rose rather than fell, as would be expected during a classical acute-phase response, is further evidence of the weakness of the inflammatory reaction. The relatively weak acute-phase response and the general maintenance of homeostasis are consistent with the behavior of mice during infection, which generally appeared normal upon visual inspection, even during peaks of parasitemia.
ALAT is a leakage enzyme (reflecting alterations in cell membrane function) and is found in the highest concentrations in the liver of mice. It is a useful analyte to measure as an index of hepatocellular damage (14). Increased activities of ALAT in serum of BALB/c and C57BL/6 mice most likely indicate liver cell damage due to inflammation around the peak of parasitemia at day 9. It has been shown that opsonized parasites are cleared from the blood via phagocytosis by Kupffer cells in the liver, which become greatly enlarged (29). Kupffer cells may also secrete inflammatory mediators, which could induce inflammatory reactions resulting in an increased ALAT level. It is interesting that the ALAT level did not rise in A/J mice or any of the congenic mice during the first peak of parasitemia, suggesting that these mice do not mount a significant inflammatory response to the first peak of parasitemia. Parasitemia could be a confounder of any measure of inflammation since it is not certain to what extent high parasitemia might induce more severe inflammation or conversely to what extent the inflammatory response might help to control parasitemia. In this case, ALAT levels were not ranked in the same order as parasitemia, suggesting that there was no direct correlation between the two. However, C57BL/6 mice had the highest ALAT levels and the lowest parasitemia, suggesting that a stronger inflammatory response might contribute to the reduction in parasitemia, although with only three mouse strains it is not possible to attach any statistical significance to this observation. A/J mice tend to have higher parasitemia than C57BL/6 mice (Fig. 6), and the lack of inflammation in the liver might be associated with reduced parasite clearance by Kupffer cells. The absence of an obvious acute phase or inflammatory response from the most susceptible mouse strain could suggest that this contributed to their early death after infection. However, all of the congenic mice also failed to exhibit an ALAT increase around the first peak of parasitemia, indicating that the individual congenic regions did not control the response resulting in increased ALAT activity on day 9. This is particularly surprising in the case of Tir1 since it includes the major histocompatibility complex region. Since there were significant differences in survival between congenic and control lines, it appears that the presence of an early response, as indicated by the clinical chemistry analytes measured here, does not contribute to prolonged survival after infection. However, plasma ALAT activities did rise in the late stage of infection in the congenic mice, although the levels were not significantly different between congenic and control mice, and urea levels were also increased on day 35 in the parental strains, suggesting that the latest time point chosen might have been too early to diagnose pathological consequences of the infection finally leading to the death of the animal.
Serum activity of ALP, an enzyme produced by hepatocytes lining the bile canicula and by osteoblasts, is typically increased in all liver diseases associated with cholestasis and in disorders of bone metabolism. Since ALP is also found in many other tissues (intestinal mucosa, kidneys, and blood vessels), increased activities sometimes can also be found under other pathological conditions such as peritonitis (1).
Increased activities of α-amylase are found in pancreatitis, peritonitis, diseases of the parotid glands, and kidney failure, while decreased activities are found in severe liver damage. α-Amylase was the only clinical chemistry analyte that differed between congenic lines and controls. The transient spikes in serum amylase and ALP in both inbred and congenic mice at day 3 could both be associated with a transient peritonitis caused by the i.p. injection of parasites. However, amylase also increases after feeding (26), so it is possible that the early stages of the infection are associated with increased appetance. Since food was not withdrawn before sampling, the differences in serum amylase between congenic mice and controls might be a consequence of greater increases in feeding in the Tir2 and Tir3 congenic mice than their controls or an interaction between the congenic regions and either appetite or digestion. Since Tir3 congenic mice did not survive longer than controls, it is unlikely that there is a direct correlation between serum amylase and survival.
The difference in survival between congenic and control mice is assumed to be a consequence of differences in the immune response and how it is initiated and subsequently controlled (2). The stronger inflammatory response of C57BL/6 mice as measured by ALAT levels may help control parasitemia and extend survival in this strain, consistent with the evidence that type 1 inflammatory immune responses are associated with lower parasitemia (11, 21). However, the absence of a difference in ALAT between congenic and control mice indicates that stronger inflammatory responses are not necessary for extended survival. Splenectomy and deletion of the proinflammatory cytokines tumor necrosis factor alpha and lymphotoxin-α increases survival after infection with T. brucei (16) but not T. congolense (11, 21). Deletion of proinflammatory T cells in mice and cattle also has no effect on survival (23, 30), but deletion of the antioxidant Sepp1 gene reduces survival (5). Deletion of both anti-inflammatory interleukin-10 and regulatory T cells reduces survival of T. congolense-infected mice (7), while overexpression of the antioxidant paraoxonase extends survival (4), suggesting that an effective anti-inflammatory response that controls the damage caused by infection is at least as important as effective parasite control.
Anemia and parasitemia.
Anemia, the most prominent feature of trypanosomiasis in cattle, is also seen in murine trypanosomiasis models (15, 17, 22, 24). A comparison of anemia and parasitemia between A/J mice and more resistant C57BL/6 mice revealed that anemia development was more severe in the C57BL/6 strain, despite the fact that this strain acquires lower parasitemia and survives longer after infection than strain A/J (24). The finding that none of the QTL regulated anemia is consistent with previous observations in infections with T. congolense and the closely related parasite T. brucei (17, 24). No correlation between either anemia or parasitemia and survival of C3H/HeN, BALB/c, C57BL/6, or CBA/Ca mice was found after infection with T. brucei (17). This is in contrast to the situation in cattle, where chronic anemia is associated with death, and anemia rather than parasitemia is used as the criterion to decide whether to treat the animal (32). In the mouse strains used in the present study, anemia is unrelated to survival, with the most resistant C57BL/6 mice developing the most severe anemia (24). The more severe anemia of C57BL/6 mice is likely to be associated with the stronger inflammatory response to infection of these mice as measured by Tnfa and Ifng expression. Nitric oxide levels have been correlated with anemia after T. brucei infection; however, deletion of T cells in cattle has no effect on anemia, nor does deletion of the Tnfa gene of mice or irradiation of rats (15, 18, 20, 21). Consequently, the more severe anemia of C57BL/6 mice may be due to differences in inflammation driven by the innate immune response.
Conclusion.
The congenic mice confirmed the physical location of two out of the three QTL tested and showed that these two loci did not regulate either parasitemia or anemia. Clinical chemical investigations identified differences in the early response to the parasite seen in the parental inbred strains, but with the possible exception of α-amylase, these were not seen in congenic lines and therefore are unlikely to represent a major contribution to differences in survival time. It has been proposed that the genetic response to parasites is comprised of “two conceptually different components: resistance (the ability to limit parasite burden) and tolerance (the ability to limit the disease severity induced by a given parasite burden)” (27). In this context, “tolerance” is used in the genetic sense and is similar to the term “resilience” used in veterinary helminthology and not “tolerance” as used by immunologists. The absence of an association between the QTL and parasitemia or proinflammatory response suggests that the Tir1 and Tir2 QTL regulate tolerance rather than resistance. Further experimental work will be required to test this hypothesis.
The congenic mice described here provide a powerful resource for discriminating between those factors that simply respond to infection and those that regulate survival after infection.
Supplementary Material
Acknowledgments
We thank the staff of the International Livestock Research Institute (ILRI) small animal unit and Elfi Holupirek of the German Mouse Clinic for expert technical assistance, Jane Poole of the ILRI for assistance with statistical analysis, and Kate Goodheart for preparation of the figures.
Funding to H.A.N., M.A., S.K., J.G., and A.B. was provided by the Wellcome Trust (GR066764MA to S.J.K.). The German Mouse Clinic Core (V.G.-D., H.F., and M.H.D.A.) and clinical chemistry laboratory (B.R. and E.W.) received funding from the EU (EUMODIC grant no. LSHG-2006-037188) and the Federal Ministry of Education and Research (National Genome Research Net NGFNplus grants 01GS0850 and 01GS0851).
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 13 July 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Andersson, R., H. E. Poulsen, and B. Ahren. 1991. Effect of bile on liver function tests in experimental E. coli peritonitis in the rat. Hepatogastroenterology 38388-390. [PubMed] [Google Scholar]
- 2.Antoine-Moussiaux, N., S. Magez, and D. Desmecht. 2008. Contributions of experimental mouse models to the understanding of African trypanosomiasis. Trends Parasitol. 24411-418. [DOI] [PubMed] [Google Scholar]
- 2a.Balasubramaniam, P., and A. Malathi. 1992. Comparative study of hemoglobin estimated by Drabkin's and Sahli's methods. J. Postgrad. Med. 388-9. [PubMed] [Google Scholar]
- 3.Barnard, J. P., B. Reynafarje, and P. L. Pedersen. 1993. Glucose catabolism in African trypanosomes. Evidence that the terminal step is catalyzed by a pyruvate transporter capable of facilitating uptake of toxic analogs. J. Biol. Chem. 2683654-3661. [PubMed] [Google Scholar]
- 4.Bhasin, K. K., J. M. Yu, A. Tward, D. Shih, D. A. Campbell, and A. J. Lusis. 2006. Trypanosoma congolense: paraoxonase 1 prolongs survival of infected mice. Exp. Parasitol. 114240-245. [DOI] [PubMed] [Google Scholar]
- 5.Bosschaerts, T., M. Guilliams, W. Noel, M. Herin, R. F. Burk, K. E. Hill, L. Brys, G. Raes, G. H. Ghassabeh, P. De Baetselier, and A. Beschin. 2008. Alternatively activated myeloid cells limit pathogenicity associated with African trypanosomiasis through the IL-10 inducible gene selenoprotein P. J. Immunol. 1806168-6175. [DOI] [PubMed] [Google Scholar]
- 6.Erdman, L. K., C. A. Finney, W. C. Liles, and K. C. Kain. 2008. Inflammatory pathways in malaria infection: TLRs share the stage with other components of innate immunity. Mol. Biochem. Parasitol. 162105-111. [DOI] [PubMed] [Google Scholar]
- 7.Guilliams, M., G. Oldenhove, W. Noel, M. Herin, L. Brys, P. Loi, V. Flamand, M. Moser, P. De Baetselier, and A. Beschin. 2007. African trypanosomiasis: naturally occurring regulatory T cells favor trypanotolerance by limiting pathology associated with sustained type 1 inflammation. J. Immunol. 1792748-2757. [DOI] [PubMed] [Google Scholar]
- 8.Iraqi, F., S. Clapcott, P. Kumari, C. Haley, S. Kemp, and A. Teale. 2000. Fine mapping of trypanosomiasis resistance loci in murine advanced intercross lines. Mamm. Genome 11645-648. [DOI] [PubMed] [Google Scholar]
- 9.Kemp, S. J., A. Darvasi, M. Soller, and A. J. Teale. 1996. Genetic control of resistance to trypanosomiasis. Vet. Immunol. Immunopathol. 54239-243. [DOI] [PubMed] [Google Scholar]
- 10.Kemp, S. J., F. Iraqi, A. Darvasi, M. Soller, and A. J. Teale. 1997. Localization of genes controlling resistance to trypanosomiasis in mice. Nat. Genet. 16194-196. [DOI] [PubMed] [Google Scholar]
- 11.Kitani, H., Y. Yagi, J. Naessens, K. Sekikawa, and F. Iraqi. 2004. The secretion of acute phase proteins and inflammatory cytokines during Trypanosoma congolense infection is not affected by the absence of the TNF-alpha gene. Acta Trop. 9235-42. [DOI] [PubMed] [Google Scholar]
- 12.Koudandé, O., J. Arendonk, and F. Iraqi. 2005. Marker-assisted introgression of trypanotolerance QTL in mice. Mamm. Genome 16112-119. [DOI] [PubMed] [Google Scholar]
- 13.Kristjanson, P. M., B. M. Swallow, G. J. Rowlands, R. L. Kruska, and P. N. de Leeuw. 1999. Measuring the costs of African animal trypanosomosis, the potential benefits of control and returns to research. Agric. Syst. 5979-98. [Google Scholar]
- 14.Loeb, W., and F. Quimby. 1999. The clinical chemistry of laboratory animals, 2nd ed. CRC Press, Boca Raton, FL.
- 15.Mabbott, N., and J. Sternberg. 1995. Bone marrow nitric oxide production and development of anemia in Trypanosoma brucei-infected mice. Infect. Immun. 631563-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magez, S., B. Stijlemans, G. Caljon, H.-P. Eugster, and P. De Baetselier. 2002. Control of experimental Trypanosoma brucei infections occurs independently of lymphotoxin-α induction. Infect. Immun. 701342-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magez, S., C. Truyens, M. Merimi, M. Radwanska, B. Stijlemans, P. Brouckaert, F. Brombacher, E. Pays, and P. De Baetselier. 2004. P75 tumor necrosis factor-receptor shedding occurs as a protective host response during African trypanosomiasis. J. Infect. Dis. 189527-539. [DOI] [PubMed] [Google Scholar]
- 17a.Morrison, W., G. Roelants, K. Mayor-Withey, and M. Murray. 1978. Susceptibility of inbred strains of mice of Trypanosoma congolense: correlation with changes in spleen lymphocyte populations. Clin. Exp. Immunol. 3225-40. [PMC free article] [PubMed] [Google Scholar]
- 18.Murray, M., and T. M. Dexter. 1988. Anemia in bovine African trypanosomiasis. Acta Trop. 45389-432. [PubMed] [Google Scholar]
- 19.Naessens, J. 2006. Bovine trypanotolerance: a natural ability to prevent severe anaemia and haemophagocytic syndrome? Int. J. Parasitol. 36521-528. [DOI] [PubMed] [Google Scholar]
- 20.Naessens, J., H. Kitani, E. Momotani, K. Sekikawa, J. M. Nthale, and F. Iraqi. 2004. Susceptibility of TNF-alpha-deficient mice to Trypanosoma congolense is not due to a defective antibody response. Acta Trop. 92193-203. [DOI] [PubMed] [Google Scholar]
- 21.Naessens, J., H. Kitani, Y. Nakamura, Y. Yagi, K. Sekikawa, and F. Iraqi. 2005. TNF-alpha mediates the development of anaemia in a murine Trypanosoma brucei rhodesiense infection, but not the anaemia associated with a murine Trypanosoma congolense infection. Clin. Exp. Immunol. 139405-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naessens, J., S. G. Leak, D. J. Kennedy, S. J. Kemp, and A. J. Teale. 2003. Responses of bovine chimaeras combining trypanosomosis resistant and susceptible genotypes to experimental infection with Trypanosoma congolense. Vet. Parasitol. 111125-142. [DOI] [PubMed] [Google Scholar]
- 23.Naessens, J., A. J. Teale, and M. Sileghem. 2002. Identification of mechanisms of natural resistance to African trypanosomiasis in cattle. Vet. Immunol. Immunopathol. 87187-194. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, Y., J. Naessens, M. Takata, T. Taniguchi, K. Sekikawa, J. Gibson, and F. Iraqi. 2003. Susceptibility of heat shock protein 70.1-deficient C57BL/6 J, wild-type C57BL/6 J, and A/J mice to Trypanosoma congolense infection. Parasitol. Res. 90171-174. [DOI] [PubMed] [Google Scholar]
- 25.Noyes, H. A., M. H. Alimohammadian, M. Agaba, A. Brass, H. Fuchs, V. Gailus-Durner, H. Hulme, F. Iraqi, S. Kemp, B. Rathkolb, E. Wolf, M. H. de Angelis, D. Roshandel, and J. Naessens. 2009. Mechanisms controlling anaemia in Trypanosoma congolense infected mice. PLoS ONE 4e5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proctor, G. B., B. Asking, and J. R. Garrett. 1991. Serum amylase of non-parotid and non-pancreatic origin increases on feeding in rats and may originate from the liver. Comp. Biochem. Physiol. B 98631-635. [DOI] [PubMed] [Google Scholar]
- 27.Raberg, L., D. Sim, and A. Read. 2007. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science 318812-814. [DOI] [PubMed] [Google Scholar]
- 28.Ritchie, R., G. Palomaki, L. Neveux, O. Navolotskaia, T. Ledue, and W. Craig. 1999. Reference distributions for the negative acute-phase serum proteins, albumin, transferrin and transthyretin: a practical, simple and clinically relevant approach in a large cohort. J. Clin. Lab. Anal. 13273-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi, M., G. Wei, W. Pan, and H. Tabel. 2004. Trypanosoma congolense infections: antibody-mediated phagocytosis by Kupffer cells. J. Leukoc. Biol. 76399-405. [DOI] [PubMed] [Google Scholar]
- 30.Sileghem, M., and J. Naessens. 1995. Are Cd8 T-cells involved in control of African trypanosomiasis in a natural host environment. Eur. J. Immunol. 251965-1971. [DOI] [PubMed] [Google Scholar]
- 31.Tabel, H., R. S. Kaushik, and J. E. Uzonna. 2000. Susceptibility and resistance to Trypanosoma congolense infections. Microbes Infect. 21619-1629. [DOI] [PubMed] [Google Scholar]
- 31a.Toye, A., J. Lippiat, P. Proks, K. Shimomura, L. Bentley, A. Hugill, V. Mijat, M. Goldsworthy, L. Moir, A. Haynes, J. Quarterman, H. Freeman, F. Ashcroft, and R. Cox. 2005. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia 48675-686. [DOI] [PubMed] [Google Scholar]
- 32.Trail, J. C., G. D. d'Ieteren, A. Feron, O. Kakiese, M. Mulungo, and M. Pelo. 1990. Effect of trypanosome infection, control of parasitaemia and control of anaemia development on productivity of N′Dama cattle. Acta Trop. 4837-45. [DOI] [PubMed] [Google Scholar]
- 33.von Theodor, B. 1938. The metabolism of pathogenic trypanosomes and the carbohydrate metabolism of their hosts. Q. Rev. Biol. 1341-50. [Google Scholar]
- 34.Wang, J., A. Van Praagh, E. Hamilton, Q. Wang, B. Zou, M. Muranjan, N. B. Murphy, and S. J. Black. 2002. Serum xanthine oxidase: origin, regulation, and contribution to control of trypanosome parasitemia. Antioxid. Redox Signal. 4161-178. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Y., J. Utzinger, J. Saric, J. V. Li, J. Burckhardt, S. Dirnhofer, J. Nicholson, B. H. Singer, R. Burn, and E. Holmes. 2008. Global metabolic responses of mice to Trypanosoma brucei burcei infection. Proc. Natl. Acad. Sci. USA 1056127-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.