Abstract
Streptococcus pneumoniae remains a major cause of bacteremia, pneumonia, and otitis media despite vaccines and effective antibiotics. The neuraminidase of S. pneumoniae, which catalyzes the release of terminal sialic acid residues from glycoconjugates, is involved in host colonization in animal models of infection and may provide a novel target for preventing pneumococcal infection. We demonstrate that the S. pneumoniae neuraminidase (NanA) cleaves sialic acid and show that it is involved in biofilm formation, suggesting an additional role in pathogenesis, and that it shares this property with the neuraminidase of Pseudomonas aeruginosa even though we show that the two enzymes are phylogenetically divergent. Using an in vitro model of biofilm formation incorporating human airway epithelial cells, we demonstrate that small-molecule inhibitors of NanA block biofilm formation and may provide a novel target for preventative therapy. This work highlights the role played by the neuraminidase in pathogenesis and represents an important step in drug development for prevention of colonization of the respiratory tract by this important pathogen.
Neuraminidases are widespread in animals and microorganisms and catalyze the release of terminal sialic acid residues from glycoconjugates (51). The best-characterized neuraminidase is the influenza virus neuraminidase, which is required to facilitate spread of this virus. Not only is the influenza virus neuraminidase a key antigen for the highly successful influenza vaccine, but it is also the target of the drugs zanamivir and oseltamivir, which have been useful for preventing and ameliorating influenza virus infection (57). Streptococcus pneumoniae produces at least three distinct neuraminidases (41); NanA is the neuraminidase that is most active and most highly expressed at the transcriptional level (5, 31), and it is conserved in all strains (21, 24, 41). Production of NanA can be detected in vivo, and its expression is upregulated upon interaction with host cells (27, 39, 46, 58). The pneumococcal neuraminidase modifies host glycoconjugates, including immune defense proteins (22, 23), and exposes potential binding receptors (3, 26, 28, 54, 55). Pneumococcal neuraminidase activity also provides a source of carbohydrates for bacterial metabolism, cleaving sugars from the mucosal surface (8, 23, 61), but whether this significantly contributes to bacterial growth in vivo has not been clearly established. Several studies have suggested that nanA mutants colonize the rodent respiratory tract less efficiently than wild-type strains (31, 40, 52), and vaccination with purified NanA affords some protection against nasopharyngeal colonization and otitis media (29, 30, 53). However, the differences can be mouse strain and animal model dependent (6, 13, 22, 23).
In addition to targeting host glycoconjugates, some bacterial neuraminidases have a role in biofilm formation, presumably targeting sialylated bacterial exopolysaccharides (47). S. pneumoniae biofilms have been characterized (1, 34, 36) and have been observed directly in the middle-ear mucosa from children with chronic otitis media (15), contributing to the colonization process (36). It is noteworthy that expression of nanA is upregulated when S. pneumoniae is grown under biofilm conditions (39). There is a need for new therapeutic strategies as the prevalence of serotypes not covered by available vaccines is increasing due to genetic recombination and strain replacement and these serotypes are increasingly associated with invasive disease (7, 20, 45, 59). We postulated that the neuraminidase of S. pneumoniae is involved in biofilm formation and sought to identify compounds that inhibit its activity in vitro.
MATERIALS AND METHODS
Bacterial strains and media.
S. pneumoniae strains D39 (4), D39 nanA (22), R6 (18), and R6 nanA (23) were grown on Trypticase soy (TS) agar or broth supplemented with 200 U/ml of catalase (Worthington) and 1 μg/ml of chloramphenicol for nanA strains. Plate cultures were grown at 37°C in the presence of carbon dioxide (5%). All chemicals were purchased from Sigma unless otherwise stated.
Epithelial cell culture.
16HBE14o− human bronchial epithelial cells and 1HAEo− human airway cells (originally obtained from D. Gruenert, California Pacific Medical Center Research Institute, San Francisco) were grown in minimum essential medium with Earle's salts (Cellgro and Gibco, respectively) supplemented with 10% fetal bovine serum (Cambrex and Gibco, respectively), 100 U/ml penicillin, and 100 μg/ml streptomycin. The medium used for 16HBE14o− cells was also supplemented with 2 mM glutamine (Invitrogen). The cells were grown at 37°C with 5% CO2 in a humidified incubator.
Neuraminidase assay.
NanA was purified as previously described (19). Neuraminidase activity of NanA was detected using the fluorogenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (Sigma). The reaction mixtures contained 1.5 mM 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid and 1 nM NanA in 2.5 mM sodium phosphate buffer (pH 5). The reaction mixtures were incubated for 2 h at 37°C before the fluorescence intensity was measured using excitation and emission wavelengths of 360 nm and 465 nm, respectively, on a Tecan microplate reader (Männedorf, Switzerland). Compounds were obtained from a variety of sources (Otava, Kiev, Ukraine; Interbioscreen, Moscow, Russia; Chembridge, San Diego, CA; Maybridge, Cornwell, United Kingdom; Sigma, St. Louis, MO; Princeton Biomolecular Research, Princeton, NJ; Lifechem, Burlington, Canada; Enamine, Kiev, Ukraine). Neuraminidase assays with oseltamivir were performed using the hydrolyzed version of the compound. Briefly, a mixture of oseltamivir (300 mg) in methanol (10 ml) was added to 5 N NaOH (3 ml). The mixture was stirred overnight and evaporated under reduced pressure. The residue was dissolved in 10 ml H2O and then washed with 5 ml of ethyl acetate. The aqueous solution was acidified with 5 N HCl to pH 2 to 3. Evaporation of water under reduced pressure resulted in the hydrolyzed product. The structure of the product was confirmed by nuclear magnetic resonance and mass spectrometry. Divalent cations were supplied in the form of calcium, magnesium, ferric, and copper chlorides. All neuraminidase assays were performed at least in triplicate.
Quantification of asialo-GM1 exposure by flow cytometry.
16HBE14o− cells were grown in 24-well plates to confluence, exposed to bacterial supernatants for 3 h, and then washed three times with phosphate-buffered saline (PBS). Supernatants were concentrated approximately 30-fold (Amicon Ultra; Millipore), and the protein quantity was adjusted. As a control, medium alone was also concentrated. Cells were stained with rabbit polyclonal anti-asialo-GM1 antibody (Wako), followed by Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (Molecular Probes). Cells were detached from the plates using 0.02% EGTA in Hanks buffered saline solution, fixed with 1% paraformaldehyde, and analyzed with a FACSCalibur using CellQuest software (version 3.3; BD). Data were analyzed using WinMDI (version 2.8; Joseph Trotter).
Adherence assays.
Adherence assays were performed using 16HBE14o− cells. Bacterial strains were grown to mid-log phase and washed with PBS, and 0.7 × 107 to 2 × 107 CFU of bacterial cells were added to confluent monolayers in 24-well plates (multiplicity of infection, 30). Bacterial cells were allowed to adhere for 1 h at 37°C before three washes with PBS. Bacteria were dissociated from epithelial cells using TrypLE Express (Gibco) and were serially diluted before plating to determine the numbers of adherent bacteria. The assay was performed with three biological replicates and with duplicate technical replicates in two separate experiments.
Biofilm assay.
Bacterial strains were grown to mid-log phase before they were diluted 1:100 in TS broth with catalase. One hundred microliters of a diluted culture was added in triplicate to 96-well flat-bottom tissue culture-treated plates (Falcon) and incubated for 18 to 24 h at 37°C in the presence of 5% CO2. Plates were read at 600 nm to determine the levels of growth before they were washed in water. Adherent biofilm-forming cells were then stained with 125 μl of 1% crystal violet for 15 min before two washes in water and allowed to dry. The bound crystal violet was then suspended in 200 μl of ethanol and shaken for 15 min, and the results were read at 540 nm.
Biofilm formation after epithelial cell interaction.
Bacterial strains were grown and inoculated onto 1HAEo− cells as described above for the adherence assay. After the initial PBS washes, fresh minimum essential medium was added before a further 1 h of incubation. Removal and addition of fresh medium were repeated another four times before adherent bacteria were detached using TrypLE Express (Gibco). The detached bacteria were then diluted 1:100 in TS broth with catalase, and assays were performed as described above for the biofilm assay. Experiments were repeated four times, each time in sextuplicate using epithelial cells without bacteria as a negative control. When inhibitors were used, they were present during epithelial cell interaction and in microtiter trays for biofilm formation. Inhibition with N-acetylneuraminic acid (NANA) was performed using 0.2% (wt/vol) NANA (Sigma) (56). Images of crystal violet-stained microtiter wells were taken with a standard digital camera. Fluorescence microscopy was performed using a Zeiss Observer Z1 inverted fluorescence microscope with ApoTome (Zeiss) for optical sectioning and AxioVision software (version 4.6.2.0; Zeiss). Microtiter wells were stained using the BacLight live/dead stain from Invitrogen (Carlsbad, CA).
Phylogenetic analysis.
Our sampling strategy was aimed at maximizing phylogenetic breadth in order to understand the overall pattern of evolution in the neuraminidase-sialidase gene family. We began with a list of well-known neuraminidases, including those from Vibrio cholerae, Salmonella enterica serovar Typhimurium, Clostridium perfringens, S. pneumoniae, Trypanosoma cruzi, and Pseudomonas aeruginosa. For each sequence we performed a standard BLAST search and collected one sequence from each genus in the list of hits that had an e value of 1 × 10−5 or less. Duplicates were deleted. We also included sequences that have been included in previous studies of the evolution of sialidases (43). The GenBank (http://www.ncbi.nlm.nih.gov/Entrez/) accession numbers of the sequences utilized for the phylogenetic analysis are as follows: Verrucomicrobium spinosum, gi 164421336:353068-354225; Blastopirellula marina, gi 87311313:89394-90503; Lentisphaera araneosa, gi 149198907:89577-90743; Propionibacterium acnes, gi 50841496:752060-754375; Ruminococcus lactaris, gi 197302028:32228-35857; Erysipelothrix rhusiopathiae, gi 13516389:295-3807; Pasteurella multocida, gi 15601865:1176085-1179327; Actinomyces odontolyticus, gi 145845834:308755-310992; Mannheimia haemolytica, gi 125433996:1-2376; Haemophilus parasuis, gi 167854877:54475-56886; Bacteroides fragilis, gi 53711291:4836372-4838006; Akkermansia muciniphila, gi 187734516:2229943-2231967; Capnocytophaga canimorsus, gi 194454827:2197-3765; Parabacteroides distasonis, gi 150006674:3525685-3527310; Shewanella pealeana, gi 157959830:1838982-1841831; Flavobacteriales bacterium, gi 88710837:680637-681797; Rhodopirellula baltica, gi 32470666:1724290-1725519; Opitutaceae bacterium, gi 153892517:3249-4847; Sassharopolyspora erythraea, gi 134096620:5769332-5771182; Pseudoalteromonas haloplanktis, gi 77361923:196316-197458; Chthoniobacter flavus, gi 196231426:66519-67730; Janibacter sp., gi 84494251:767782-770736; Monosiga brevicollis, gi 167534964:1-984; Strongylocentrotus purpuratus, gi 115616575:1-719; Planctomyces maris, gi 149177549:10030-11205; Acinetobacter baumannii, gi 169632029:647539-649371; Opitutaceae bacterium, gi 153890920:5481-6641; Danio rerio, gi 148539964:69-1220; Corynebacterium diphtheriae, gi 38232642:512872-515037; Gemmata obscuriglobus, gi 163804184:63331-64515; Streptomyces coelicolor, gi 32141095:7255596-7257542; Takifugu rubripes, gi 148372013:1-87; V. cholerae, gi 12057212:1933231-1935654; C. perfringens nanI, gi 18308982:900997-903081; C. perfringens nanH, gi 18308982:904693-905499; P. aeruginosa, gi 110227054:3150886-3152202; Clostridium septicum, gi 40662; Clostridium sordellii, gi 1710442; Actinomyces viscosus, gi 39254; Trypanosoma rangeli, gi 2894809; T. cruzi, gi 162265; T. cruzi SAPA (shed acute-phase antigen), gi 10943; Micromonosporta viridifaciens, gi 216782:816-2759; Arthrobacter ureafaciens, gi 60544840; influenza virus A H5N1, gi 108671038; Macrobdella decora, gi 1353880; S. enterica serovar Typhimurium, gi 16763390:1002088-1003326; S. pneumoniae, gi 116515308:1522475-1525468; Arcanobacterium pyogenes, gi 18146340:1026-6239; Xenopus laevis, gi 148228846:180-1376; Trichomonas vaginalis, gi 123473002:1-1050; Rattus norvegicus, gi 71896601:59-1288; Bos taurus, gi 149676185:61-219, 650-842, 1541-1803, 2490-2672, 2849-3071, and 3185-3411; and Monodelphis domestica, gi 126309689:1-1404. Sequences were aligned using the ClustalW algorithm as implemented in the program BioEdit using default settings. Amino acid sequences were aligned and then transposed to obtain the original nucleotide sequences, maintaining the gaps determined by the initial alignment (a total of 5,394 characters, 4,124 parsimony-informative characters with gaps as a fifth state, and 3,766 parsimony-informative characters with gaps treated as missing).
We performed a rigorous phylogenetic analysis using the maximum parsimony algorithm implemented in PAUP* (50). We used 1,000 replicates for random addition, followed by the tree branch reconnection algorithm using the “multrees” option to save more than one optimal tree if more than one optimal tree was discovered in the search. All characters and state transformations were given equal weight. Terminal gaps were scored as missing data in all analyses. We performed two analyses, designating internal gaps as a fifth character state in one analysis and as missing data in the other analysis. Although the trees had many nodes in agreement, there were major differences between the structures of the two trees. Since gaps can be informative characters (14, 42), we favor the analysis in which internal gaps are counted as character states. We performed nonparametric bootstrap analyses with 100 iterations consisting of 100 random addition replicates, followed by tree branch reconnection to gauge the robustness of the tree.
Statistics.
The significance of data was determined using a Student t test and, for multivariant data, analysis of variance followed by a Dunnetti posttest using GraphPad Prism software.
RESULTS
Biochemical properties of NanA.
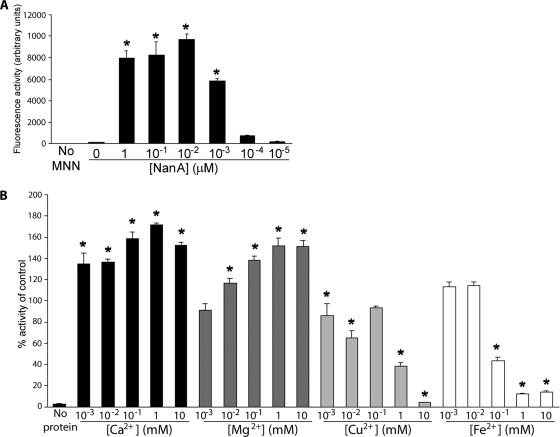
To better understand the biochemical properties of NanA and to facilitate screening of potential inhibitory compounds, we established an assay to measure its neuraminidase activity. The biochemical activity of NanA was assayed using the fluorogenic sialic acid derivative 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid. NanA cleaved the fluorogenic substrate significantly at low nanomolar and even picomolar concentrations (Fig. 1A). The Km of NanA for this substrate is about 1.4 mM (data not shown), which is generally comparable to the Km values reported for other neuraminidases. The neuraminidase from V. cholerae requires divalent cations, specifically calcium, to be active (10, 17). We investigated the effect of adding ions to the reaction mixture and observed that calcium was not essential for NanA activity but did increase the activity by 70% at a concentration of 1 mM (Fig. 1B), that there was a 50% increase in the activity in the presence of magnesium ions (Fig. 1B), and that there was decreased activity in the presence of iron and copper ions, presumably due to the higher molecular masses of these ions (Fig. 1B). The presence of either copper or ferric ions decreased the activity by 90% or more at millimolar levels.
FIG. 1.
Activity of S. pneumoniae neuraminidase. (A) Titration of activity using different concentrations of purified NanA. (B) Effect of divalent cations on the activity of purified NanA compared to the activity of the wild-type enzyme (control). The assay was performed using 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MNN). *, P < 0.05.
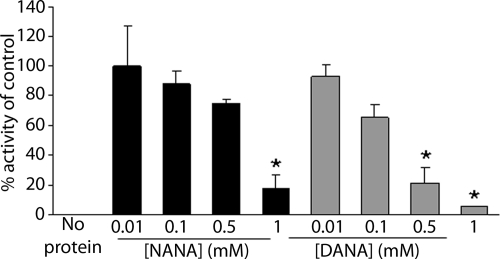
The ability of sialic acids to competitively inhibit NanA activity was also tested. The two sialic acids used, NANA and 2,3-dehydro-2-deoxy-N-acetylneuraminic acid (DANA), were both utilized to obtain cocrystals of NanA (19). NANA caused 50% inhibition at a concentration of 600 μM (Fig. 2). We observed greater inhibition with the transition state analog DANA than with NANA. DANA reduced the activity by 50% at a concentration of 200 μM.
FIG. 2.
Inhibition of NanA neuraminidase activity by the sialic acid compounds NANA and DANA. The activity is expressed as a percentage of the activity of NanA without an inhibitor (control). The assay was performed using 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid. *, P < 0.05.
Phylogeny of the neuraminidases.
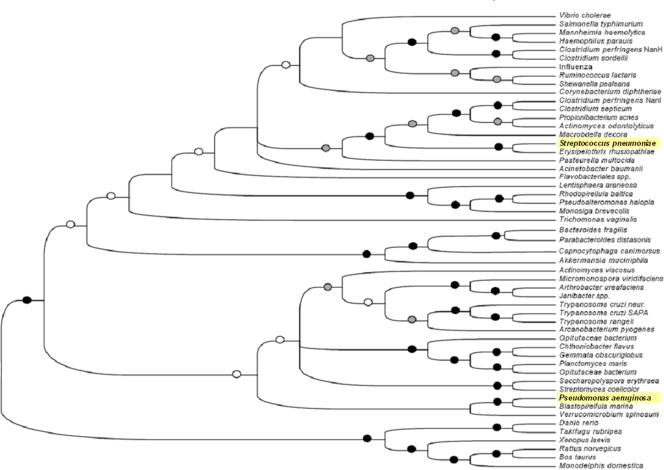
The neuraminidase superfamily is known to be highly divergent (43, 51). We postulated that neuraminidases from organisms that infect the same site may have similar functions and hence may be evolutionarily related, which might be useful in the identification of common inhibitors. To better ascertain the evolutionary relatedness of NanA, a phylogenetic analysis of a number of bacterial, eukaryotic, and viral neuraminidases was conducted (Fig. 3). NanA clustered closely with the large neuraminidase of C. perfringens (38) and was more closely related to well-characterized bacterial neuraminidases. NanA did not cluster with the neuraminidase from P. aeruginosa, which is also involved in respiratory tract colonization.
FIG. 3.
Phylogenetic analysis of neuraminidases: unrooted phylogenetic tree based on our broad survey of neuraminidase phylogeny. The tree is the strict consensus of three most parsimonious trees (45,885steps; consistency index, 0.326; retention index, 0.385; rescaled consistency, 0.125). Black circles indicate branches with bootstrap values greater than 80%. Gray circles indicate branches with bootstrap values between 50 and 80%. Open circles indicate nodes with bootstrap values less than 50% for agreement with the consensus bootstrap tree. Branches without an indication of the bootstrap value are found in the maximum parsimony tree but not in the bootstrap tree.
Biological activity of pneumococcal NanA.
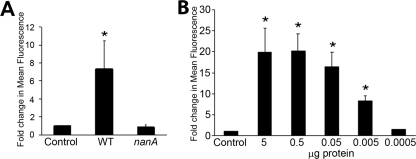
Many lung pathogens, including S. pneumoniae and P. aeruginosa, bind to asialylated ganglioside receptor GM1 (asialo-GM1) (Galβ1-3GalNAcβ1-4Galβ1-4Glcβ1-1Cer) (26). Either purified NanA or concentrated supernatant from wild-type S. pneumoniae strain D39, but not concentrated supernatant from an isogenic nanA mutant of this strain, exposed asialo-GM1 on the surface of 16HBE14o− epithelial cells (Fig. 4). However, no effect on bacterial adherence was observed, nor was there a growth advantage in the presence of airway epithelial cells (data not shown).
FIG. 4.
Release of sialic acid from the surface of airway epithelial cells by NanA. (A) Exposure of asialo-GM1 to concentrated supernatant from wild-type (WT) and nanA strains. (B) Exposure of asialo-GM1 to purified NanA. Cells were stained with antibody to asialo-GM1 and quantified by flow cytometry, and the results are expressed as the changes compared to a medium-only control. *, P < 0.05.
NanA is involved in biofilm formation.
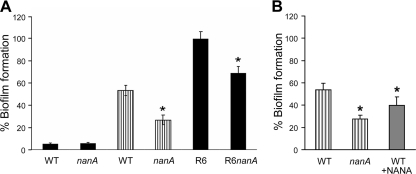
As S. pneumoniae nanA expression is upregulated in lung tissue and in biofilm-grown cells (39), the contribution of nanA to the formation of biofilms was examined (Fig. 5). The increase in expression was exploited by initially growing the bacteria on airway epithelial cells over a day (see Materials and Methods). Adherent bacterial cells were then recovered before the standard microtiter biofilm assay was performed. Exposure of S. pneumoniae to airway epithelial cells before the biofilm assay was performed not only resulted in a significant increase in biofilm formation but also showed that the nanA mutant had a significantly reduced capacity to form biofilms. When S. pneumoniae was not exposed previously to airway epithelial cells, no difference in biofilm formation was observed between the wild-type and nanA strains, nor was much biofilm formation observed. In an S. pneumoniae R6, unencapsulated background, significantly more biofilm was produced, and the ability of the nanA strain to form biofilms was also significantly reduced (Fig. 5A). Consistent with the hypothesis that NANA acts as an inhibitor in the neuraminidase assay (Fig. 2), adding exogenous NANA to the biofilm assay mixture also resulted in decreased biofilm formation by the wild-type strain (Fig. 5B).
FIG. 5.
S. pneumoniae biofilm formation. (A) Encapsulated (D39 background) strains were grown in microtiter trays without (filled bars) or with (striped bars) previous exposure to epithelial cells. Unencapsulated R6 strains were grown in microtiter trays without exposure to epithelial cells. (B) Incubation with NANA results in reduced biofilm formation by wild-type strain D39 (WT). Biofilms were measured by using crystal violet staining. Biofilm formation was normalized to growth and expressed as a percentage compared to the R6 wild-type strain. *, P < 0.05.
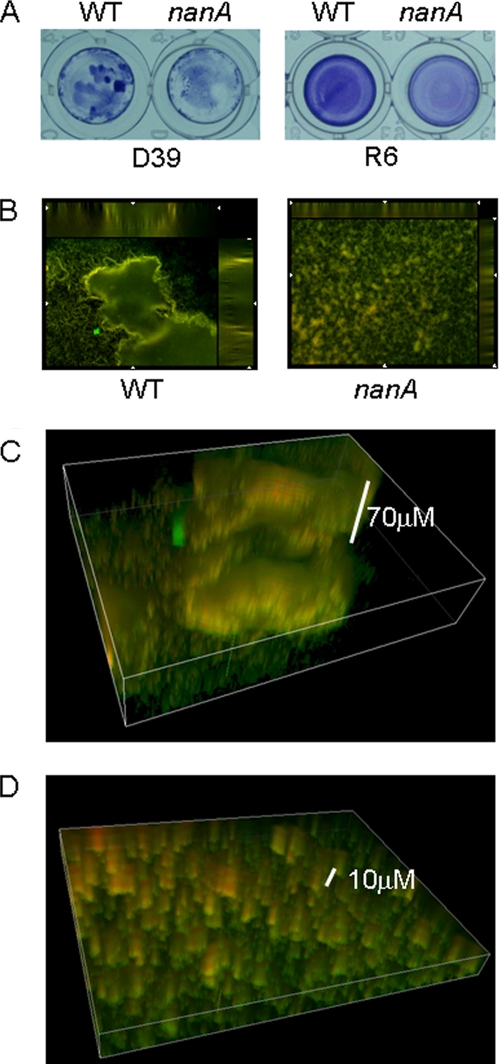
The biofilms produced by R6 were even layers of cells on microtiter plates, and the reduction in biofilm formation observed with the nanA mutant was indicated by reduced intensity of crystal violet staining (Fig. 6A). In the D39 background, after exposure to epithelial cells, the wild-type strain had more intense crystal violet staining overall than the nanA strain (Fig. 6A). Macroscopically, the wild-type strain had regions of intense crystal violet staining in clumps. When the D39 biofilms were examined under the microscope (Fig. 6B), a lattice-like arrangement of cells was observed for both the wild-type and nanA strains. Regions of concentrated cells were observed for the nanA strain, but no macrostructures were observed. However, for the wild-type background, structures that had significant height were observed (Fig. 6B and C). These structures were approximately 70 μm high, as shown by the three-dimensional reconstruction (Fig. 6C). For the nanA background only cells in the initial sections were attached to the microtiter wells (Fig. 6B and D).
FIG. 6.
Imaging of S. pneumoniae biofilms. (A) Images of crystal violet-stained biofilms in microtiter wells for wild-type (WT) and nanA strains with D39 (after exposure to epithelial cells) and R6 backgrounds. (B) Fluorescence microscopy of wild-type strain D39 and nanA biofilms grown in microtiter trays after exposure to epithelial cells and stained with BacLight live/dead stain. Magnification, ×200. (C) Three-dimensional reconstruction of the biofilm structure for the wild-type strain shown in panel B. (D) Three-dimensional reconstruction of cells of the nanA strain shown in panel B.
Identification of small-molecule inhibitors of the neuraminidases.
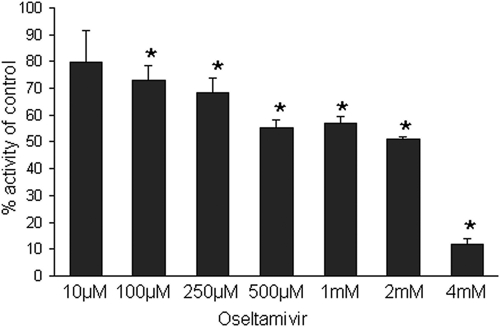
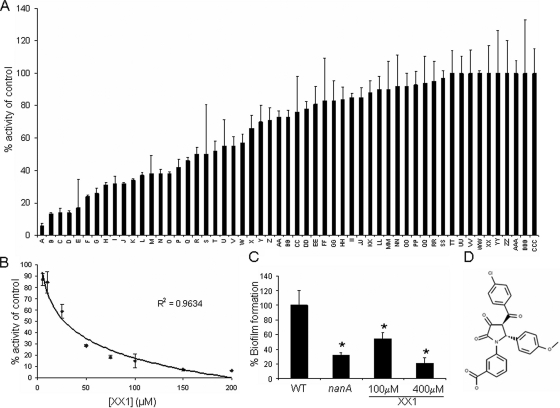
Virtual library screening was performed (Schrödinger LLC, Portland, OR) (25, 44) that identified small molecules that were predicted to interact with the active site of the enzyme. As a control we tested the ability of the influenza virus neuraminidase inhibitor oseltamivir to inhibit NanA (Fig. 7). Oseltamivir had a 50% inhibitory concentration (IC50) of 2 mM. A number of compounds identified in this screen showed high degrees of inhibition in vitro (Fig. 8A). A lead compound designated XX1 (Fig. 8D) with a pyrrolidine-2,3-dione chemical scaffold was found to inhibit NanA over a range of concentrations in a dose-dependent manner (Fig. 8B). An IC50 of 28 μM was determined. The inhibition of NanA by XX1 was more than 7, 20, and 70 times more effective than the inhibition by DANA, NANA, and oseltamivir, respectively. At a XX1 concentration of 100 μM we observed a significant reduction in biofilm formation, and at a concentration of 400 μM the levels of biofilm formation were comparable to those of the nanA strain (Fig. 8C). XX1 did not affect the growth of planktonic cells (data not shown).
FIG. 7.
Inhibition of NanA by oseltamivir. The activity is expressed as a percentage of the activity without inhibitor. The assay was performed using 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid. *, P < 0.05.
FIG. 8.
Inhibitory activity of potential NanA inhibitors. (A) Screening of potential inhibitors was performed with NanA and inhibitors at a concentration of 100 μM in the neuraminidase assay. Results are percentages of the level for the control without any inhibitor. (B) Dose-response curve for NanA with lead compound XX1. Data were fitted with a logarithmically based trend line. The data are the percentages of activity compared to the control with only the vehicle (dimethyl sulfoxide). (C) Biofilm formation by wild-type strain D39 (WT) grown in the presence of XX1 during exposure to epithelial cells and growth in microtiter trays. The nanA strain was used as a reference. Biofilm formation was normalized to growth and was expressed as a percentage compared to the results for the wild-type control. (D) Chemical structure of XX1. *, P < 0.05.
DISCUSSION
For S. pneumoniae, the biofilm phenotype is most evident in vivo (15) or, as we show, under in vitro conditions that favor neuraminidase expression. We were able to demonstrate a difference in biofilm production after growth on human airway cells, which has been shown to induce nanA expression (27, 39, 46) and reduce capsule production (16). By selecting for organisms on human airway cells, we apparently mimicked a likely in vivo process in which encapsulated organisms that are able to avoid mucus entrapment and clearance gradually produce less capsule to facilitate epithelial attachment (20). This association between capsule expression and biofilm formation was evident with the R6 strain. R6 does not produce capsule, and we observed that it produced significant biofilms and that a nanA mutant of R6 displayed a reduced propensity for biofilm formation. The reduced capsule is important in initial airway colonization both for facilitating the initial adherence and for promoting cell-cell interactions for biofilm formation (20). This conclusion is also consistent with other studies that showed that cells induced to form a biofilm had a greater propensity to cause pneumonia than planktonic cells (39).
It is likely that the role of the neuraminidase is somewhere between the cluster formation and biofilm maturation processes (1). This conclusion is based on the fact that the wild-type strain biofilms observed resembled the mature biofilms grown under flow conditions (2), and while the nanA strain still produced regions of clustered cells, it had no large cellular structures. It should be noted that some differences are also expected when static and continuous-flow systems are compared. Even though the development and composition of biofilms are complex and numerous genes and proteins are differentially expressed (1, 34, 39), the high level of expression of nanA in biofilms and other data presented here indicate that the neuraminidase has a crucial role in biofilm production. A recent study by Trappetti et al. (56) demonstrated the role of sialic acid in pneumococcal biofilm formation. Addition of sialic acid, but not addition of other sugars, resulted in increased biofilm formation, as well as increases in the number of organisms present in the nasopharynx in a murine model of colonization (56). We found that NANA at high concentrations inhibits biofilm formation by S. pneumoniae by occupying the binding site of NanA. Based on this study and our other work, the liberation of sialic acid residues from the airway epithelium by the action of NanA appears to contribute to biofilm formation by S. pneumoniae and hence is a desirable target for therapeutic intervention.
Even though nanA is the most highly expressed neuraminidase gene, the other neuraminidases of S. pneumoniae may contribute to biofilm formation. In the serotype 4 background (strain TIGR4) nanB is involved to a small extent in biofilm formation (36). It should be noted that there is a mutation in nanA in this strain that affects its cell wall attachment. These studies highlighted the important role that biofilms play in pathogenesis, establishing a link between biofilm formation and colonization of the murine nasopharynx. Our data are consistent with these observations, suggesting that a reduced capacity to form biofilms is behind the reduced pathogenesis of the neuraminidase (nanA) mutant in animal models (31, 40). Based on this work and other work, a model for the establishment of infection in the respiratory tract by S. pneumoniae can be established. Encapsulated cells reach the mucus layer, where the capsule is required to avoid mucous entrapment. NanA is also able to utilize the mucin as a carbon source (61). When the respiratory epithelium is reached, capsule expression is downregulated to facilitate intimate attachment. It is at this point that neuraminidase expression is increased and cells begin to establish a biofilm.
Another respiratory pathogen, P. aeruginosa, also produces a neuraminidase (NanPs) that is important for biofilm formation and pathogenesis in animal models (47), indicating that the role in pathogenesis of the two neuraminidases is conserved. Desialylated glycolipids provide receptors for many of the common bacterial pulmonary pathogens, including both S. pneumoniae and P. aeruginosa (26), which bind to the exposed GalNAcβ1-4Gal residues when terminal sialic acid is cleaved. We demonstrated that NanA, like P. aeruginosa neuraminidase (47), was capable of exposing this receptor on human airway cells. The neuraminidase activity associated with intact organisms, either S. pneumoniae or P. aeruginosa (47), was not associated with increased bacterial attachment, which is an interesting observation given that the desialylation of airway mucosal cells by the influenza virus neuraminidase increases susceptibility to secondary infections often caused by S. pneumoniae (33).
The crystal structures of both NanA and NanPs were recently solved (19, 60). Structural analysis indicated that while these two enzymes had similar overall structures, their active sites were remarkably different, indicating likely differences in substrate specificity and biochemical function (9, 47). We also observed that the two enzymes are phylogenetically distinct, with NanA clustering closer to other well-characterized canonical neuraminidases and NanPs located in a phylogenetically diverse branch of the tree. Yet despite their differences in structure and likely different substrates, these two neuraminidases have similar functions in pathogenesis.
The neuraminidase of V. cholerae requires divalent cations, specifically calcium, to be active (10, 17), and while we observed an increase in activity in the presence of calcium ions, such ions were not an absolute requirement for activity. This is an observation analogous to the observation for the neuraminidase of P. aeruginosa (9). The inhibition that we did observe with iron and copper ions is consistent with inhibition of such enzymes by metal ions (17). The increase in activity with calcium might suggest the presence of a calcium binding site, like that in V. cholerae (10) or C. perfringens (38), although this is not a categorical feature of neuraminidases nor was such a site identified from the crystal structure (9, 11, 19, 48, 60). However, the presence of a calcium binding site would not be incongruous given the close phylogenetic relationship between NanA and the other well-characterized bacterial neuraminidases.
There is a need for new therapeutic strategies against S. pneumoniae as the prevalence of serotypes not covered by available vaccines is increasing due to genetic recombination and such serotypes are increasingly associated with invasive disease (7, 20, 45, 59); in addition, antibiotic resistance of S. pneumoniae strains is a growing problem (32). Using in silico docking studies, we were able to identify a number of compounds that were significantly more inhibitory than the sialic acids NANA and DANA and the influenza virus inhibitor oseltamivir. Many neuraminidases have been crystallized in the presence of the sialic acid DANA (11, 19, 35, 38), and we observed that this compound inhibits the neuraminidase activity of NanA better than it inhibits the activity of NANA. Although the inhibition by DANA observed was not as great as the inhibition of some neuraminidases tested to date, it is within the observed range of inhibition (12, 48). Our lead compound XX1 was found to inhibit the neuraminidase activity of NanA at concentrations in the low-micromolar range and also to inhibit the ability of organisms to form biofilms. Some work examining inhibition of other bacterial neuraminidases has been done, but most of the work to date has been done with the trypanosome trans-sialidases (37, 48, 49). Trypanosome trans-sialidase studies have identified inhibitors with IC50s in the range from 100 to 300 μM, indicating that the studies presented here are progressing well. We are actively involved in synthesizing XX1 in an attempt to examine its ability to prevent infection in vivo.
Based on structural data, it is likely that inhibitors of the pneumococcal neuraminidase will be organism specific. The NanPs neuraminidase of P. aeruginosa has an active site that is significantly different. The binding pocket of NanA is tight, while NanPs has an open conformation that likely requires different inhibitor structures for effective inhibition (19). We envision that delivery of a neuraminidase inhibitor to the lung could be used as prophylaxis to circumvent bacterial pneumonia after influenza virus infection and also in at-risk populations.
Despite biochemical, structural, and phylogenetic differences, we demonstrate a common role for the neuraminidases of S. pneumoniae and P. aeruginosa in biofilm formation and the pathogenesis of respiratory tract infection. Due to the importance of biofilms in S. pneumoniae colonization, we have begun to identify inhibitors targeting the pneumococcal neuraminidase to prevent infection. This study represents a starting point in the development of a potentially novel drug against pneumococcal pneumonia.
Acknowledgments
We thank Jeffrey Weiser for providing S. pneumoniae strains.
D.P. was the recipient of an NHRMC Biomedical Overseas Fellowship. This work was supported by the Thrasher Research Fund (D.P.) and by a Deans pilot grant from Columbia University (A.P.).
Editor: J. N. Weiser
Footnotes
Published ahead of print on 29 June 2009.
REFERENCES
- 1.Allegrucci, M., F. Z. Hu, K. Shen, J. Hayes, G. D. Ehrlich, J. C. Post, and K. Sauer. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 1882325-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allegrucci, M., and K. Sauer. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J. Bacteriol. 1892030-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, B., J. Dahmen, T. Frejd, H. Leffler, G. Magnusson, G. Noori, and C. S. Eden. 1983. Identification of an active disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells. J. Exp. Med. 158559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry, A. M., R. A. Lock, and J. C. Paton. 1996. Cloning and characterization of nanB, a second Streptococcus pneumoniae neuraminidase gene, and purification of the NanB enzyme from recombinant Escherichia coli. J. Bacteriol. 1784854-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry, A. M., and J. C. Paton. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect. Immun. 68133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brueggemann, A. B., R. Pai, D. W. Crook, and B. Beall. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnaugh, A. M., L. J. Frantz, and S. J. King. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cacalano, G., M. Kays, L. Saiman, and A. Prince. 1992. Production of the Pseudomonas aeruginosa neuraminidase is increased under hyperosmolar conditions and is regulated by genes involved in alginate expression. J. Clin. Investig. 891866-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crennell, S., E. Garman, G. Laver, E. Vimr, and G. Taylor. 1994. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure 2535-544. [DOI] [PubMed] [Google Scholar]
- 11.Crennell, S. J., E. F. Garman, W. G. Laver, E. R. Vimr, and G. L. Taylor. 1993. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc. Natl. Acad. Sci. USA 909852-9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crennell, S. J., E. F. Garman, C. Philippon, A. Vasella, W. G. Laver, E. R. Vimr, and G. L. Taylor. 1996. The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution. J. Mol. Biol. 259264-280. [DOI] [PubMed] [Google Scholar]
- 13.Gingles, N. A., J. E. Alexander, A. Kadioglu, P. W. Andrew, A. Kerr, T. J. Mitchell, E. Hopes, P. Denny, S. Brown, H. B. Jones, S. Little, G. C. Booth, and W. L. McPheat. 2001. Role of genetic resistance in invasive pneumococcal infection: identification and study of susceptibility and resistance in inbred mouse strains. Infect. Immun. 69426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giribet, G., and W. C. Wheeler. 1999. On gaps. Mol. Phylogenet. Evol. 13132-143. [DOI] [PubMed] [Google Scholar]
- 15.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammerschmidt, S., S. Wolff, A. Hocke, S. Rosseau, E. Muller, and M. Rohde. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 734653-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmquist, L. 1975. Activation of Vibrio cholerae neuraminidase by divalent cations. FEBS Lett. 50269-271. [DOI] [PubMed] [Google Scholar]
- 18.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiao, Y. S., D. Parker, A. J. Ratner, A. Prince, and L. Tong. 2009. Crystal structures of respiratory pathogen neuraminidases. Biochem. Biophys. Res. Commun. 380467-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadioglu, A., J. N. Weiser, J. C. Paton, and P. W. Andrew. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6288-301. [DOI] [PubMed] [Google Scholar]
- 21.Kelly, R. T., S. Farmer, and D. Greiff. 1967. Neuraminidase activities of clinical isolates of Diplococcus pneumoniae. J. Bacteriol. 94272-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King, S. J., K. R. Hippe, J. M. Gould, D. Bae, S. Peterson, R. T. Cline, C. Fasching, E. N. Janoff, and J. N. Weiser. 2004. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol. Microbiol. 54159-171. [DOI] [PubMed] [Google Scholar]
- 23.King, S. J., K. R. Hippe, and J. N. Weiser. 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59961-974. [DOI] [PubMed] [Google Scholar]
- 24.King, S. J., A. M. Whatmore, and C. G. Dowson. 2005. NanA, a neuraminidase from Streptococcus pneumoniae, shows high levels of sequence diversity, at least in part through recombination with Streptococcus oralis. J. Bacteriol. 1875376-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontoyianni, M., L. M. McClellan, and G. S. Sokol. 2004. Evaluation of docking performance: comparative data on docking algorithms. J. Med. Chem. 47558-565. [DOI] [PubMed] [Google Scholar]
- 26.Krivan, H. C., D. D. Roberts, and V. Ginsburg. 1988. Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence GalNAc beta 1-4Gal found in some glycolipids. Proc. Natl. Acad. Sci. USA 856157-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeMessurier, K. S., A. D. Ogunniyi, and J. C. Paton. 2006. Differential expression of key pneumococcal virulence genes in vivo. Microbiology 152305-311. [DOI] [PubMed] [Google Scholar]
- 28.Linder, T. E., R. L. Daniels, D. J. Lim, and T. F. DeMaria. 1994. Effect of intranasal inoculation of Streptococcus pneumoniae on the structure of the surface carbohydrates of the chinchilla eustachian tube and middle ear mucosa. Microb. Pathog. 16435-441. [DOI] [PubMed] [Google Scholar]
- 29.Lock, R. A., J. C. Paton, and D. Hansman. 1988. Comparative efficacy of pneumococcal neuraminidase and pneumolysin as immunogens protective against Streptococcus pneumoniae. Microb. Pathog. 5461-467. [DOI] [PubMed] [Google Scholar]
- 30.Long, J. P., H. H. Tong, and T. F. DeMaria. 2004. Immunization with native or recombinant Streptococcus pneumoniae neuraminidase affords protection in the chinchilla otitis media model. Infect. Immun. 724309-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manco, S., F. Hernon, H. Yesilkaya, J. C. Paton, P. W. Andrew, and A. Kadioglu. 2006. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect. Immun. 744014-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCormick, A. W., C. G. Whitney, M. M. Farley, R. Lynfield, L. H. Harrison, N. M. Bennett, W. Schaffner, A. Reingold, J. Hadler, P. Cieslak, M. H. Samore, and M. Lipsitch. 2003. Geographic diversity and temporal trends of antimicrobial resistance in Streptococcus pneumoniae in the United States. Nat. Med. 9424-430. [DOI] [PubMed] [Google Scholar]
- 33.McCullers, J. A. 2006. Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. 19571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moscoso, M., E. Garcia, and R. Lopez. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 1887785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moustafa, I., H. Connaris, M. Taylor, V. Zaitsev, J. C. Wilson, M. J. Kiefel, M. von Itzstein, and G. Taylor. 2004. Sialic acid recognition by Vibrio cholerae neuraminidase. J. Biol. Chem. 27940819-40826. [DOI] [PubMed] [Google Scholar]
- 36.Munoz-Elias, E. J., J. Marcano, and A. Camilli. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect. Immun. 765049-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neres, J., M. L. Brewer, L. Ratier, H. Botti, A. Buschiazzo, P. N. Edwards, P. N. Mortenson, M. H. Charlton, P. M. Alzari, A. C. Frasch, R. A. Bryce, and K. T. Douglas. 2009. Discovery of novel inhibitors of Trypanosoma cruzi trans-sialidase from in silico screening. Bioorg. Med. Chem. Lett. 19589-596. [DOI] [PubMed] [Google Scholar]
- 38.Newstead, S. L., J. A. Potter, J. C. Wilson, G. Xu, C. H. Chien, A. G. Watts, S. G. Withers, and G. L. Taylor. 2008. The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J. Biol. Chem. 2839080-9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oggioni, M. R., C. Trappetti, A. Kadioglu, M. Cassone, F. Iannelli, S. Ricci, P. W. Andrew, and G. Pozzi. 2006. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol. Microbiol. 611196-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orihuela, C. J., G. Gao, K. P. Francis, J. Yu, and E. I. Tuomanen. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 1901661-1669. [DOI] [PubMed] [Google Scholar]
- 41.Pettigrew, M. M., K. P. Fennie, M. P. York, J. Daniels, and F. Ghaffar. 2006. Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect. Immun. 743360-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips, A., D. Janies, and W. Wheeler. 2000. Multiple sequence alignment in phylogenetic analysis. Mol. Phylogenet. Evol. 16317-330. [DOI] [PubMed] [Google Scholar]
- 43.Roggentin, P., R. Schauer, L. L. Hoyer, and E. R. Vimr. 1993. The sialidase superfamily and its spread by horizontal gene transfer. Mol. Microbiol. 9915-921. [DOI] [PubMed] [Google Scholar]
- 44.Sherman, W., T. Day, M. P. Jacobson, R. A. Friesner, and R. Farid. 2006. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 49534-553. [DOI] [PubMed] [Google Scholar]
- 45.Singleton, R. J., T. W. Hennessy, L. R. Bulkow, L. L. Hammitt, T. Zulz, D. A. Hurlburt, J. C. Butler, K. Rudolph, and A. Parkinson. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 2971784-1792. [DOI] [PubMed] [Google Scholar]
- 46.Song, X. M., W. Connor, K. Hokamp, L. A. Babiuk, and A. A. Potter. 2008. Streptococcus pneumoniae early response genes to human lung epithelial cells. BMC Res. Notes 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soong, G., A. Muir, M. I. Gomez, J. Waks, B. Reddy, P. Planet, P. K. Singh, Y. Kaneko, M. C. Wolfgang, Y. S. Hsiao, L. Tong, and A. Prince. 2006. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Investig. 1162297-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Streicher, H. 2004. Inhibition of microbial sialidases—what has happened beyond the influenza virus? Curr. Med. Chem. 3149-161. [Google Scholar]
- 49.Streicher, H., and H. Busse. 2006. Building a successful structural motif into sialylmimetics—cyclohexenephosphonate monoesters as pseudo-sialosides with promising inhibitory properties. Bioorg. Med. Chem. 141047-1057. [DOI] [PubMed] [Google Scholar]
- 50.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0 beta ed. Sinauer, Sunderland, MA.
- 51.Taylor, G. 1996. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 6830-837. [DOI] [PubMed] [Google Scholar]
- 52.Tong, H. H., L. E. Blue, M. A. James, and T. F. DeMaria. 2000. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 68921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong, H. H., D. Li, S. Chen, J. P. Long, and T. F. DeMaria. 2005. Immunization with recombinant Streptococcus pneumoniae neuraminidase NanA protects chinchillas against nasopharyngeal colonization. Infect. Immun. 737775-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong, H. H., X. Liu, Y. Chen, M. James, and T. Demaria. 2002. Effect of neuraminidase on receptor-mediated adherence of Streptococcus pneumoniae to chinchilla tracheal epithelium. Acta Otolaryngol. 122413-419. [DOI] [PubMed] [Google Scholar]
- 55.Tong, H. H., M. A. McIver, L. M. Fisher, and T. F. DeMaria. 1999. Effect of lacto-N-neotetraose, asialoganglioside-GM1 and neuraminidase on adherence of otitis media-associated serotypes of Streptococcus pneumoniae to chinchilla tracheal epithelium. Microb. Pathog. 26111-119. [DOI] [PubMed] [Google Scholar]
- 56.Trappetti, C., A. Kadioglu, C. Carter, J. Hayre, F. Iannelli, G. Pozzi, P. W. Andrew, and M. R. Oggioni. 2009. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J. Infect. Dis. 1991497-1505. [DOI] [PubMed] [Google Scholar]
- 57.von Itzstein, M. 2007. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 6967-974. [DOI] [PubMed] [Google Scholar]
- 58.Williamson, Y. M., R. Gowrisankar, D. L. Longo, R. Facklam, I. K. Gipson, E. P. Ades, G. M. Carlone, and J. S. Sampson. 2008. Adherence of nontypeable Streptococcus pneumoniae to human conjunctival epithelial cells. Microb. Pathog. 44175-185. [DOI] [PubMed] [Google Scholar]
- 59.World Health Organization. 2007. Pneumococcal conjugate vaccine for childhood immunization. W.H.O. position paper. Weekly epidemiological record. World Health Organization, Geneva, Switzerland.
- 60.Xu, G., X. Li, P. W. Andrew, and G. L. Taylor. 2008. Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA. Acta Crystallogr. F 64772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yesilkaya, H., S. Manco, A. Kadioglu, V. S. Terra, and P. W. Andrew. 2008. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett. 278231-235. [DOI] [PubMed] [Google Scholar]