Abstract
Ninety-six clinical isolates of Staphylococcus aureus from Nigeria were characterized phenotypically and genetically. Twelve multidrug-resistant methicillin (meticillin)-resistant S. aureus (MRSA) isolates carrying a new staphylococcal cassette chromosome mec element and a high proportion of Panton-Valentine leukocidin (PVL)-positive methicillin-susceptible S. aureus (MSSA) isolates were observed. The cooccurrence of multidrug-resistant MRSA and PVL-positive MSSA isolates entails the risk of emergence of a multidrug-resistant PVL-positive MRSA clone.
Staphylococcus aureus is a major cause of both hospital- and community-acquired infections. In particular, methicillin (meticillin)-resistant S. aureus (MRSA) strains have been detected worldwide (15), and the prevalence of MRSA varies among countries and health institutions (2, 4, 27). The emergence of MRSA strains resistant to glycopeptides, as well as the increasing prevalence in the community (7), highlights the need for worldwide epidemiological studies of this pathogen. However, data about the epidemiology and prevalence of staphylococcal infections in Africa are scarce compared to information about such infections in the rest of the world. Studies have indicated low prevalences of MRSA in Nigeria, Somalia, and Tanzania (1), but high prevalences in South Africa, Zimbabwe, Kenya, Ethiopia, Egypt, Senegal, and the Ivory Coast have been reported (2, 9, 18). In addition, a recent study of the genetic diversity of S. aureus strains in a carriage population from Mali showed a high frequency of a Panton-Valentine leukocidin (PVL)-positive clone (25). The mechanisms for the emergence and spread of S. aureus clones in Africa are largely unknown; hence, the characterization of isolates may provide baseline information needed in establishing effective infection control measures in Nigeria.
In this study, a total of 96 S. aureus isolates obtained between January and December 2007 from clinical specimens in six tertiary-care hospitals located in northeastern Nigeria were characterized. The isolates were identified based on standard bacteriological procedures (i.e., Gram staining and catalase, tube coagulase, and DNase testing), and susceptibilities to 12 antibiotics (Table 1) were determined by the disk diffusion method according to the CLSI guidelines. All the isolates were susceptible to vancomycin, fusidic acid, and mupirocin, and 12 (12.5%) were resistant to methicillin (i.e., oxacillin and cefoxitin resistant) (Table 1). The MRSA isolates were multidrug resistant (i.e., resistant to beta-lactams, along with at least three other classes of antibiotics), a finding similar to previously reported findings in other African countries like Morocco, Kenya, Cameroon, and South Africa (17). MRSA resistance to non-beta-lactams may further increase the medical expenses and the complexity of patient management, as well as morbidity and mortality rates since alternative antibiotics may not be affordable in many African countries.
TABLE 1.
Frequency of resistance of S. aureus (MSSA and MRSA) isolates to antibiotics
| Antibiotic | % of resistant isolates among:
|
||
|---|---|---|---|
| MSSA isolates (n = 84) | MRSA isolates (n = 12) | All isolates (n = 96) | |
| Penicillin | 91.6 | 100 | 92.7 |
| Oxacillin | 0 | 100 | 12.5 |
| Cefoxitin | 0 | 100 | 12.5 |
| Gentamicin | 2.4 | 100 | 14.6 |
| Erythromycin | 3.6 | 100 | 15.6 |
| Clindamycin | 0 | 75 | 9.4 |
| Co-trimoxazole | 8.3 | 100 | 19.8 |
| Ciprofloxacin | 3.6 | 100 | 15.6 |
| Rifampin | 2.4 | 0 | 2.1 |
| Vancomycin | 0 | 0 | 0 |
| Fusidic acid | 0 | 0 | 0 |
| Mupirocin | 0 | 0 | 0 |
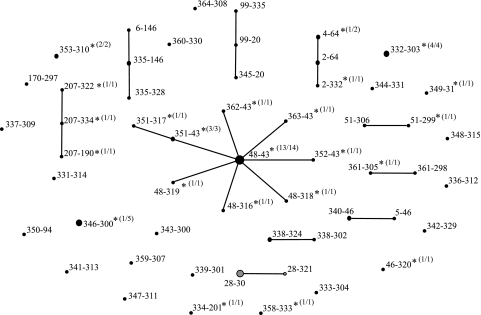
The genetic diversity of the S. aureus population was assessed by the highly discriminatory double-locus sequence typing (DLST) method as described previously (20). This method is based on the analysis of partial sequences (about 500 bp) of the variable clfB and spa genes. A total of 41 clfB and 46 spa alleles were observed among the 96 S. aureus isolates evaluated by DLST, and these alleles represented 53 different DLST types. The eBURST software was used to cluster DLST types with identical sequences of at least one allele. Cluster analysis showed a low level of diversity among the 12 MRSA isolates, which belonged to a single cluster, while a high level of diversity among the methicillin-susceptible S. aureus (MSSA) isolates (i.e., 10 single-locus variant clusters and 23 singletons) was observed (Fig. 1). However, one cluster (DLST type 48-43) was predominant among the MSSA isolates. To confirm the relationship between S. aureus genotypes from Nigeria and worldwide clonal complexes (CCs), multilocus sequence typing (MLST) of at least one representative strain from each of the main DLST clusters (Table 2) was performed as described earlier (10). A total of 12 sequence types (STs) were observed among the 16 isolates analyzed by MLST. The MRSA cluster belonged to ST 241, while the predominant MSSA cluster was grouped into ST 152. With the exception of the genetically divergent ST 152, all the STs belonged to one of eight internationally recognized S. aureus CCs: CC1, CC5, CC8, CC9, CC15, CC30, CC80, and CC121.
FIG. 1.
DLST single-locus variant clustering of 96 S. aureus isolates from northeastern Nigeria by using eBURST. Each circle represents one DLST type, and the diameter of the circle reflects the frequency (i.e., the number of isolates) of that type. Linked DLST types differ at one of the two loci (clfB or spa). DLST types represented by only MSSA or MRSA isolates are indicated in black or gray, respectively. DLST types including PVL-positive isolates are indicated by asterisks (values in parentheses indicate the number of PVL-positive isolates/total number of isolates of that DLST type).
TABLE 2.
Multilocus STs of representative isolates of the major S. aureus DLST clones observed in hospitals in northern Nigeria
| Strain no. | Identification | PVL statusa | DLST type | MLST profile | ST | CCb |
|---|---|---|---|---|---|---|
| H18134 | MSSA | Pos | 361-305 | 10-1-1-1-1-1-1 | New | 1 |
| H18192 | MSSA | Pos | 340-46 | 1-new-1-1-1-1-1 | New | 1 |
| H18109 | MSSA | Pos | 2-64 | 10-8-1-4-12-1-10 | 5 | 5 |
| H18105 | MRSA | Neg | 28-30 | 2-3-1-1-4-4-30 | 241 | 8 |
| H18113 | MRSA | Neg | 28-321 | 2-3-1-1-4-4-30 | 241 | 8 |
| H18132 | MSSA | Pos | 346-300 | 3-3-1-1-4-4-3 | 8 | 8 |
| H18196 | MSSA | Pos | 99-20 | 3-3-1-1-4-4-3 | 8 | 8 |
| H18166 | MSSA | Pos | 338-324 | 3-3-1-1-1-1-10 | 9 | 9 |
| H18127 | MSSA | Pos | 335-146 | New-13-1-1-12-11-13 | New | 15 |
| H18118 | MSSA | Pos | 353-310 | 2-2-2-7-6-3-2 | 30 | 30 |
| H18165 | MSSA | Pos | 46-320 | 1-3-1-14-11-51-10 | 80 | 80 |
| H18129 | MSSA | Pos | 332-303 | 6-5-6-new-7-14-5 | New | 121 |
| H18101 | MSSA | Pos | 207-334 | 6-5-6-2-7-14-5 | 121 | 121 |
| H18100 | MSSA | Pos | 48-43 | 46-75-49-44-13-68-60 | 152 | NA |
| H18106 | MSSA | Pos | 48-316 | 46-75-49-44-13-68-60 | 152 | NA |
| H18172 | MSSA | Pos | 51-299 | 46-75-49-44-13-68-60 | 152 | NA |
Pos, positive; neg, negative.
NA, not applicable.
The clonality of MRSA strains was further confirmed by the typing of the staphylococcal cassette chromosome mec (SCCmec) elements observed in these isolates. Using the multiplex PCRs described by Kondo et al. (19) and Milheiriço et al. (23), we found that all the Nigerian isolates carried ccr type 5 and mec class A, as well as a J2 region similar to SCCmec type III. So far, the combination of these elements had been observed only in strains simultaneously carrying two SCCmec elements: SCCmec type III and SCCmercury (6, 19). However, we did not detect the presence of the mercury operon, suggesting that the Nigerian cassette does not carry SCCmercury and that it is a new SCCmec element. Recombination between different SCCmec types and/or local acquisitions may explain the emergence of new resistance elements (5, 12, 13). Recent data indicated that the local acquisition of SCCmec elements is a frequent phenomenon (24), highlighting the need to compare the molecular epidemiologies of MSSA and MRSA. However, we were not able to establish a link between these two categories since the genetic background of MRSA was clearly distinct from that of MSSA. SCCmec elements are often associated with resistance to multiple classes of antibiotics (8). However, resistance determinants may also be carried on other mobile elements, such as plasmids, transposons, and phages (22), and further investigations are needed to characterize this new cassette and unambiguously link the multidrug resistance pattern with this element.
PVL is a toxin responsible for skin and soft-tissue infections and is often associated with community-acquired MRSA infections. All isolates were tested for the presence of PVL genes as described elsewhere (21). Among the 96 isolates, 41 (42.7%) were PVL positive, but the MRSA isolates were PVL negative (Fig. 1). The prevalence of PVL-positive S. aureus isolates in this study was high compared with the data in recent reports indicating prevalences of less than 10% in several European countries (16, 26). The Nigerian PVL-positive MSSA isolates were well distributed among the hospitals, and more (39%) were recovered from wound specimens than from any other source. Interestingly, the PVL genes were noted to be present in almost all the MSSA isolates in the predominant group (DLST cluster 48-43). This observation supports the finding of a high prevalence of PVL-positive S. aureus isolates (ST 152) in a carriage population from Mali (25). Furthermore, a PVL-positive community-acquired MRSA clone (ST 152) has been observed in the Balkans and Central Europe (3, 11, 14). The presence of PVL-positive MSSA isolates (ST 152) in Nigeria and Mali supports the hypothesis that the MRSA clone originated in Africa, migrated throughout central Europe, and acquired methicillin resistance (25).
In conclusion, our analysis of isolates from northeastern Nigeria indicated a high number of PVL-positive MSSA isolates, along with a multidrug-resistant MRSA clone carrying a novel SCCmec element. The cooccurrence of multidrug-resistant MRSA and PVL-positive MSSA highlights the risk for the emergence of a multidrug-resistant PVL-positive MRSA clone. This point further underlines the need for surveillance studies in Africa and the enforcement of antibiotic stewardship and infection control to prevent further dissemination of epidemic clones.
Acknowledgments
We are grateful to Caroline Choulat for technical assistance in Switzerland and to Ahmadu Danhajja, Victoria Bitrus, and Saleh Harun for their help in the collection of Nigerian S. aureus isolates. We thank Valérie Vogel and Giorgio Zanetti for insightful comments on the manuscript.
The stay of K.O.O. in the Lausanne laboratory was supported by Swiss National Science Foundation grant no. IZKOBO-121266.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Adesida, S., H. Boelens, B. Babajide, A. Kehinde, S. Snijders, W. van Leeuwen, A. Coker, H. Verbrugh, and A. van Belkum. 2005. Major epidemic clones of Staphylococcus aureus in Nigeria. Microb. Drug Resist. 11115-121. [DOI] [PubMed] [Google Scholar]
- 2.Bell, J. M., and J. D. Turnidge. 2002. High prevalence of oxacillin-resistant Staphylococcus aureus isolates from hospitalized patients in Asia-Pacific and South Africa: results from SENTRY antimicrobial surveillance program, 1998-1999. Antimicrob. Agents Chemother. 46879-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc, D. S., C. Petignat, A. Wenger, G. Kuhn, Y. Vallet, D. Fracheboud, S. Trachsel, A. Reymond, N. Troillet, H. H. Siegrist, S. Oeuvray, M. Bes, J. Etienne, J. Bille, P. Francioli, and G. Zanetti. 2007. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small geographic area over an eight-year period. J. Clin. Microbiol. 453729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchillon, S. K., B. M. Johnson, D. J. Hoban, J. L. Johnson, M. J. Dowzicky, D. H. Wu, M. A. Visalli, and P. A. Bradford. 2004. Determining incidence of extended spectrum beta-lactamase producing Enterobacteriaceae, vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus in 38 centres from 17 countries: the PEARLS study 2001-2002. Int. J. Antimicrob. Agents 24119-124. [DOI] [PubMed] [Google Scholar]
- 5.Branger, C., C. Gardye, J. O. Galdbart, C. Deschamps, and N. Lambert. 2003. Genetic relationship between methicillin-sensitive and methicillin-resistant Staphylococcus aureus strains from France and from international sources: delineation of genomic groups. J. Clin. Microbiol. 412946-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 501001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deresinski, S. 2005. Methicillin-resistant Staphylococcus aureus: an evolutionary, epidemiologic, and therapeutic odyssey. Clin. Infect. Dis. 40562-573. [DOI] [PubMed] [Google Scholar]
- 8.Deurenberg, R. H., and E. E. Stobberingh. 2008. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8747-763. [DOI] [PubMed] [Google Scholar]
- 9.El Kholy, A., H. Baseem, G. S. Hall, G. W. Procop, and D. L. Longworth. 2003. Antimicrobial resistance in Cairo, Egypt 1999-2000: a survey of five hospitals. J. Antimicrob. Chemother. 51625-630. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faria, N. A., D. C. Oliveira, H. Westh, D. L. Monnet, A. R. Larsen, R. Skov, and H. de Lencastre. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 431836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 1853307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 988821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harbarth, S., P. Francois, J. Schrenzel, C. Fankhauser-Rodriguez, S. Hugonnet, T. Koessler, A. Huyghe, and D. Pittet. 2005. Community-associated methicillin-resistant Staphylococcus aureus, Switzerland. Emerg. Infect. Dis. 11962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9486-493. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, A., M. Ganner, S. McGuane, T. L. Pitt, B. D. Cookson, and A. M. Kearns. 2005. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 432384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kesah, C., S. Ben Redjeb, T. O. Odugbemi, C. S. B. Boye, M. Dosso, J. O. N. Achola, S. Koulla-Shiro, M. Benbachir, K. Rahal, and M. Borg. 2003. Prevalence of methicillin-resistant Staphylococcus aureus in eight African hospitals and Malta. Clin. Microbiol. Infect. 9153-156. [DOI] [PubMed] [Google Scholar]
- 18.Klugman, K. P. 1998. Emerging infectious diseases—South Africa. Emerg. Infect. Dis. 4517-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn, G., P. Francioli, and D. S. Blanc. 2007. Double-locus sequence typing using clfB and spa, a fast and simple method for epidemiological typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 4554-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 291128-1132. [DOI] [PubMed] [Google Scholar]
- 22.Lindsay, J. A., and M. T. Holden. 2006. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics 6186-201. [DOI] [PubMed] [Google Scholar]
- 23.Milheiriço, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 514537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nübel, U., P. Roumagnac, M. Feldkamp, J. H. Song, K. S. Ko, Y. C. Huang, G. Coombs, M. Ip, H. Westh, R. Skov, M. J. Struelens, R. V. Goering, B. Strommenger, A. Weller, W. Witte, and M. Achtman. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 10514130-14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruimy, R., A. Maiga, L. Armand-Lefevre, I. Maiga, A. Diallo, A. K. Koumare, K. Ouattara, S. Soumare, K. Gaillard, J. C. Lucet, A. Andremont, and E. J. Feil. 2008. The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J. Bacteriol. 1903962-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wannet, W. J., E. Spalburg, M. E. Heck, G. N. Pluister, E. Tiemersma, R. J. Willems, X. W. Huijsdens, A. J. de Neeling, and J. Etienne. 2005. Emergence of virulent methicillin-resistant Staphylococcus aureus strains carrying Panton-Valentine leucocidin genes in The Netherlands. J. Clin. Microbiol. 433341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinn, C. S., H. Westh, and V. T. Rosdahl. 2004. An international multicenter study of antimicrobial resistance and typing of hospital Staphylococcus aureus isolates from 21 laboratories in 19 countries or states. Microb. Drug Resist. 10160-168. [DOI] [PubMed] [Google Scholar]