Abstract
Tuberculosis is one of the important public health problems in Egypt. However, limited information on the Mycobacterium tuberculosis genotypes circulating in Egypt is available. A total of 151 M. tuberculosis strains were characterized by spoligotyping. The results revealed that 74.8% of M. tuberculosis isolates grouped into 13 different clusters, while 25.2% had unique spoligotype patterns. Comparison with an international spoligotyping database (the SITVIT2 database) showed that types SIT53 (T1 variant) and SIT54 (Manu2 variant) were the most common types between cluster groups. In addition, new shared types SIT2977, SIT2978, and SIT2979 were observed. The results identified for the first time an unusually high proportion of ancestral Manu strains of M. tuberculosis from patients in Egypt. The percentage of the Manu clade in this study (27.15%) was significantly higher than its overall representation of 0.4% in the SITVIT2 database. We show that in Egypt tuberculosis is caused by a predominant M. tuberculosis genotype belonging to the ancestral Manu lineage which could be a missing link in the split between ancestral and modern tubercle bacilli during the evolution of M. tuberculosis.
Despite the availability of antituberculosis (anti-TB) drugs, the disease burden of human TB remains a very serious and widespread public health problem. Many of the 22 countries most affected by human TB identified by the World Health Organization (WHO) are developing countries (11). Although Egypt is not on the WHO list of 22 high-TB-burden countries, it is considered one of the high-burden countries in WHO's Eastern Mediterranean region, where TB constitutes the second most important public health problem after schistosomiasis (21). Every year, the National Tuberculosis Control Program of the Ministry of Health and Population (MOHP) registers over 12,000 new TB patients; more than 50% of the cases are sputum smear-positive pulmonary TB. On the basis of an annual risk of infection of 0.32%, it is estimated that about 8,000 people receive a diagnosis of TB at facilities other than those of MOHP (25).
In the context described above, it is important to have a precise picture of the predominant and emerging Mycobacterium tuberculosis clones in Egypt, in order to have a first snapshot of the epidemiology of TB. The advent of molecular typing methods has given a new boost to epidemiological studies of TB in recent years (3); the most commonly used methods are IS6110-based restriction fragment length polymorphism (RFLP) (IS6110-RFLP) and spoligotyping (12, 22). As opposed to IS6110-RFLP, which was considered the “gold standard” for the genotyping of M. tuberculosis in the early 1990s (22) and which is time-consuming and labor-intensive and requires large amounts of highly purified DNA, PCR-based spoligotyping avoids the problems associated with the slow growth of these bacteria, thereby offering the possibility of the rapid recognition of outbreaks (2, 4, 12). The clinical usefulness of spoligotyping is determined by its rapidity both in detecting the causative bacteria and in providing epidemiological information on strain identities (12). Spoligotyping, which allows determination of the phylogeographical specificity of circulating clades of tubercle bacilli, is also useful for population-based studies (4, 10). Recent data from international spoligotyping studies have identified a growing number of important clades or genogroups, further facilitating the study of the spread of the disease due to human migratory movements (4, 8, 9, 17). To date, there is very limited genotyping information on the M. tuberculosis strains circulating in Egypt. The SITVIT2 database, which is an updated version of the previously released SpolDB4 database (4; available online at http://www.pasteur-guadeloupe.fr:8081/SITVITDemo), showed very little information on the isolates circulating in Egypt, with only 79 of 69,000 clinical isolates, i.e., 0.1% of the total recruitment. The aim of this study was to identify the predominant international spoligotypes responsible for the transmission and prevalence of TB in Egypt. The spoligotypes obtained were further compared with those in an updated version of the international database named SITVIT2.
MATERIALS AND METHODS
Patients and specimens.
A total of 151 Mycobacterium tuberculosis strains from as many patients collected over a period of 1 year (2005) at three different chest hospitals in Egypt were included in this study. The isolates were recovered from sputum specimens from patients diagnosed with pulmonary TB on the basis of clinical symptoms, chest X ray, and bacteriological examination. Despite an attempt to ensure that the sample is an accurate representation of the pulmonary TB patients attending the three chest hospitals over a 1-year period, it instead represents a coincidental group (convenience sample) of patients originating from 14 different governorates across Egypt. Demographic and epidemiological data were obtained from the medical records of each patient. Three sputum specimens were collected in the early morning on consecutive days and transported to the Tuberculosis Center Laboratory, Faculty of Medicine, Cairo University, Cairo, Egypt. Samples were decontaminated with N-acetyl-l-cysteine and sodium hydroxide, as described previously (13). The decontaminated specimens were concentrated by centrifugation at 3,000 × g for 20 min. All handling and manipulations of the clinical specimens were done in a certified biosafety level II cabinet. The processed specimens were subjected to Ziehl-Neelsen staining, followed by culture onto Lowenstein-Jensen medium. The cultures were identified as M. tuberculosis complex on the basis of the growth rate, colony morphology, and cultural and biochemical properties by standard microbiological procedures. The phenotypic identification of the M. tuberculosis complex was further confirmed by the amplification of a 540-bp DNA fragment specific for the IS6110 gene (14). For this purpose, bacterial DNA was extracted by using a Qiagen DNA isolation kit, according to the manufacturer's protocol.
Spoligotyping.
Spoligotyping was carried out as described previously (12). Briefly, the direct repeat (DR) region of the TB genome was amplified with primers DRa and DRb, and the amplified biotinylated products were hybridized to a set of 43 oligonucleotides covalently bound to a membrane (Isogen Life Science B.V., Utrecht, The Netherlands). The hybridized PCR products were then incubated with streptavidin-peroxidase conjugate, and the membrane was exposed to chemoluminescence (Amersham, Little Chalfont, England) and X-ray film (Hyperfilm ECL; Amersham), according to the manufacturer's instructions. The X-ray film was developed by standard photochemical procedures after overnight exposure. To avoid any possibility that nonspecific hybridization spots would appear on the commercial membranes, appropriate controls that included DNA from the known strain M. tuberculosis H37Rv and deionized autoclaved water were used in each experiment. The reproducibility of spoligotyping was confirmed by repeating the test. Spoligotyping in a binary format was converted to an octal code (7; the conversion tool is available at http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/outilsConversion.jsp) for comparison with the codes in the SITVIT2 proprietary database of the Pasteur Institute of Guadeloupe.
Comparison of spoligotyping results with database types.
Spoligotypes in binary format were entered into the SITVIT2 database (Pasteur Institute of Guadeloupe), which is an updated version of the previously released SpolDB4 database (4; available at http://www.pasteur-guadeloupe.fr:8081/SITVITDemo). At the time of the present study, SITVIT2 contained genotyping information on 69,000 M. tuberculosis clinical isolates from 160 countries of origin. In this database, the Spoligotype International Type (SIT) designates spoligotypes shared by two or more patient isolates, whereas “orphan” designates the patterns reported for a single isolate. Major phylogenetic clades were assigned according to the signatures provided in the SpolDB4 database, which defined 62 genetic lineages and sublineages (4). These include specific signatures for various M. tuberculosis complex members, as well as rules defining the major lineages and sublineages for M. tuberculosis sensu stricto. These include specific signatures for various M. tuberculosis complex species, such as M. bovis, M. microti, M. caprae, M. pinipedii, and M. africanum, as well as rules defining the major lineages and sublineages of M. tuberculosis sensu stricto. These include the Beijing clade, the Central Asian (CAS) clade and 2 sublineages, the East African-Indian (EAI) clade and 9 sublineages, the Haarlem clade and 3 sublineages, the Latin American-Mediterranean (LAM) clade and 12 sublineages, the Manu family and 3 sublineages, the S clade, the IS6110-low-banding X clade and 4 sublineages, and an ill-defined T clade with 5 sublineages.
Phylogenetic analysis.
The evolutionary relationships among all the spoligotype patterns in our sample were studied by drawing a minimum-spanning tree (MST) with Bionumerics software (version 3.5, Applied Maths, Sint-Marteen-Latem, Belgium). MST is an undirected network in which all the samples are linked together with the smallest possible linkages between nearest neighbors. By this approach, one considers that all intermediate stages are present within the sample analyzed, by including first the individual that shows the most possible linkages to other individuals in the population studied.
RESULTS AND DISCUSSION
TB remains a great public health concern in Egypt; however, little information on the prevailing genotypes of the tubercle bacilli and their spread is available (1, 6). In a country where TB is endemic, it is critical to identify the genotypes of the predominant strains in order to study the transmission patterns and the epidemiological features of the disease. The present study is a first attempt to provide insight into the population structure of M. tuberculosis in Egypt. We selected spoligotyping to create this snapshot, as it has been shown to be a valuable method for determination of the clonal and phylogenetic relationships of M. tuberculosis strains, providing in parallel useful information about TB in a given population, in a given country, among countries, and throughout the world (4, 5, 9, 10, 12, 16, 17, 19).
This study was performed over a period of 1 year (2005) with a total of 151 M. tuberculosis complex strains from as many patients from Egypt. The demographic information for the patients (Table 1) underlined that within our sample the disease predominated in male patients (male-to-female sex ratios, 3.52 for the cluster group and 3.75 for the unclustered group) and individuals aged 15 to 45 years (70.7% of the cases grouped in clusters and 68.4% of the unclustered group). Of all cases, 118/151 (78.1%) patients had newly diagnosed TB, whereas 33/151 (21.9%) patients were previously treated. Lastly, 135/151 (90%) patients were native and 15/151 (10%) were immigrants (the origin was unreported for 1 patient). However, none of the parameters shown in Table 1 varied significantly among any of the groups (by Fisher's exact test with an online statistical calculator [http://www.graphpad.com/quickcalcs/contingency1.cfm]).
TABLE 1.
Clinical and epidemiological characteristics of patients harboring clustered versus nonclustered strains
| Parameter (n = 151) | Cluster groups (n = 113)
|
Noncluster group (n = 38) | |||
|---|---|---|---|---|---|
| All clusters (n = 113) | SIT53 (n = 51) | SIT54 (n = 33) | Other cluster groups (n = 29) | ||
| No. (%) of patients ages (yr): | |||||
| 15-45 | 80 (70.7) | 39 (76.6) | 22 (66.6) | 19 (65.5) | 26 (68.4) |
| ≥46 | 33 (29.3) | 12 (23.3) | 11 (33.4) | 10 (34.5) | 12 (31.6) |
| No. (%) of patients of the following sex: | |||||
| Male | 88 (77.8) | 39 (76.6) | 28 (84.8) | 21 (72.4) | 30 (78.9) |
| Female | 25 (22.2) | 12 (23.3) | 5 (15.2) | 8 (27.6) | 8 (21.1) |
| Sex ratioa | 3.52 | 3.25 | 5.6 | 2.6 | 3.75 |
| No. (%) of patients with history of TB | |||||
| No previous therapy | 86 (76) | 42 (82.3) | 21 (63.6) | 23 (79.3) | 32 (84.2) |
| Previously treated | 27 (24) | 9 (17.7) | 12 (36.4) | 6 (20.7) | 6 (15.8) |
| No. (%) of patients with the following country of origin: | |||||
| Egyptian | 101 (89.3) | 46 (90.1) | 27 (81.8) | 28 (96.5) | 34 (89.4) |
| Foreign born | 12 (10.7) | 5 (9.9) | 6 (18.2) | 1 (3.5) | 3 (10.6) |
The sex ratio is the number of males/number of females.
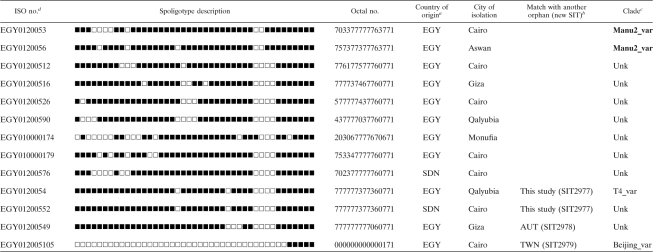
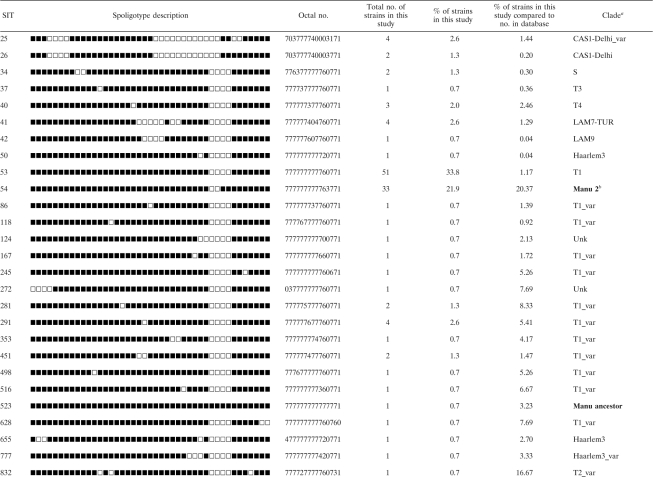
Detailed spoligotyping results are summarized in Tables 2 and 3, while the evolutionary relationships of the strains are illustrated in an MST shown in Fig. 1. Starting with our sample of 151 clinical isolates, spoligotyping produced a total of 51 different patterns. A total of 12 patterns corresponding to 13 strains were not present in the database and could be termed orphans (Table 2); nonetheless, 2 strains (1 from a patient born in Egypt and another from a patient born in Sudan) matched each other, thereby creating a new shared type, SIT2977. Two more orphans matched one strain each from Austria and Taiwan, thereby creating SIT2978 and SIT2979, respectively. As illustrated in Table 3, 138/151 strains from this study belonged to a total of 39 SITs (not including the 3 newly created SITs shown in Table 2). The binary and octal descriptions, the total number of strains, and the percentage of strains in the present study compared to the same data in the SITVIT2 database at the time of comparison and the corresponding genetic lineages and sublineages are summarized for each of the shared types shown.
TABLE 2.
Description of the orphan strains and newly created shared types in case of a match with another orphan from the database
aEGY, Egypt; SDN, Sudan.
bConcerns only new SITs created after a match with another orphan strain in the SITVIT2 database. AUT, Austria; TWN, Taiwan.
cClade designations according to SITVIT2 by use of revised SpolDB4 rules. Unk, unknown patterns within any of the major clades described in SITVIT2. The Manu lineage strains are indicated in boldface.
dISO, International Organization for Standardization.
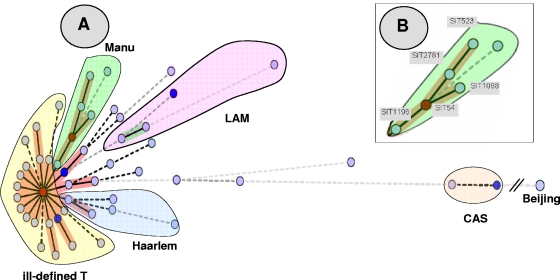
FIG. 1.
MST of potential evolutionary relationships of M. tuberculosis spoligotypes from Egypt built with Bionumerics software. Solid lines, single spacer change; dotted lines, two spacer changes (black) or more spacer changes (gray). The color of the circles is proportional to the number of clinical isolates in our study, illustrating unique isolates (sky blue) versus clustered isolates (deep blue, two to five strains; red, six strains and more). (A). As opposed to the clades belonging to the principal genetic group 1 (such as Beijing and CAS) in the MST, the Manu strains are the ones situated closest to the principal genetic group 2 or 3 lineages, such as Haarlem, LAM, and T. The central node of this unrooted tree is represented by SIT53, which is the prototype of the T1 lineage. (B). A zoom of the Manu lineage representing SIT523 (Manu ancestor), SIT2781 (Manu ancestor var_Δ33), SIT1088 (Manu2_var), SIT54 (Manu2), and SIT1196 (Manu2_var). In this scenario, the evolution is supposed to happen starting from SIT523 (Manu ancestor), which is the most conserved spoligotype in the SITVIT2 database (with positive hybridization for all 43 spacers of the DR locus) to SIT1196 (Manu2_var, with absence of consecutive spacers 33 to 35).
TABLE 3.
Description of 39 shared types representing 138 isolates that matched a preexisting shared type in the SITVIT2 database
aClade designations are according to the SITVIT2 database by use of revised SpolDB4 rules. Unk, unknown patterns within any of the major clades described in the SITVIT2 database. All Manu lineage strains are indicated in boldface.
Irrespective of the database comparison, a total of 113/151 (74.8%) clinical isolates were clustered in the present study (13 clusters containing 2 to 51 isolates per cluster). Twelve clusters are shown in Table 3, and 1 additional cluster of two strains is represented by the newly created shared type (SIT2977; Table 2). Thus, 38/151 (25.2%) of the strains in this study were unclustered. SIT53, which belongs to the ill-defined T clade (the T family was defined by default [4]), predominated in our setting and contained 51/151 (33.8%) of the isolates. That was followed by SIT54, which belongs to the Manu2 clade and which contained 33/151 (21.8%) of strains, followed by SIT25, a CAS1-Delhi variant (4/151 [2.7%]); SIT41 of the LAM7-TUR lineage (4/151 [2.7%]); and SIT291 of the ill-defined T1 variant (4/151 [2.7%]). Overall, 51 strains were identified in SIT53, making up 33.8% of the strains studied. These were from different location in Egypt: 36 from Cairo; 7 from Qalyubia; 4 from Giza; and 1 each from Monufia, Sohag, BeniSuef, and Asyut. The second largest cluster, SIT54 (21.8%), contained 21 strains from Cairo; 4 from Giza; 3 from Qalyubia; and 1 from each Sohag, Helwan, Monufia, Sinai, and Kafr-elSheikh.
The overall classification of the strains according to the major M. tuberculosis genotypic lineages (and not the shared types stricto sensu) showed that the most predominant group was the ill-defined T superfamily (81/151 [53.6%]). However, the T genotype does not represent a clade in a strict evolutionary sense, since it was defined by default to include strains that cannot be classified in one of the established genotypic lineages, such as the Haarlem, LAM, CAS, and EAI lineages, with well-established phylogeographical specificity (4). In this sense, the most predominant clade in our setting was the ancestral Manu lineage, which comprised not only the 33 isolates of SIT54 (Manu2_var) but also a group of other Manu strains (Tables 2 and 3): 2 Manu2_var orphans; 1 SIT523 strain classified as a Manu ancestor; 3 Manu2_var strains belonging to SIT1088, SIT1196, and SIT1690, respectively; a single SIT1378 (Manu3) strain; and 1 SIT2781 strain classified as Manu ancestor var_Δ33 (lacking a single spacer at position 33). Thus, by taking all the Manu isolates together (shown in boldface in Tables 2 and 3), they represented 41 of 151 (27.15%) of the strains in the present study. This is one of the highest distribution patterns ever observed for the ancestral Manu lineage in any single setting. The predominance of the Manu lineage was followed by the LAM (7/151 [4.6%]), CAS1-Delhi (6/151 [4%]), Haarlem (3/151 [2%]), and S (2/151 [1.3%]) lineages.
Before the entry of our data, spoligotypes from 79 strains were recorded in the SITVIT2 database. These strains were isolated from patients either in Egypt or from elsewhere in the world from a patient of Egyptian origin (56 strains classified within 30 different SITs and 23 orphans). Some previously found SITs belong to predominant lineages in Egypt, particularly SIT53 (T1 prototype), SIT54 (Manu2 prototype), SIT25 (CAS1-Delhi_var), and SIT41 (LAM7-TUR), while others (SIT34, SIT37, SIT42) are found in smaller numbers (Table 2). On the other hand, many of the SITs found in patients of Egyptian origin but isolated in distant countries were not found in the present study performed in Egypt. Such examples include SIT35 (Haarlem3), SIT47 (Haarlem1), SIT52 (T2), SIT390 (Haarlem3), SIT460 (EAI5), SIT717 (T1), SIT732 (T1), SIT798 (T), and SIT878 (X1).
The unusually high proportion of strains belonging to the Manu clade (27.15%) in the present study is really exceptional. This percentage is significantly higher than its overall representation (0.4%) in the SITVIT2 database (n = 261). The Manu lineage was initially described as a new family from India in 2004 (16), and later, similar strains in small numbers were reported in a study from Madagascar (8), Soon afterwards, it was tentatively subdivided into the Manu1 (deletion of spacer 34), Manu2 (deletion of spacers 33 and 34), and Manu3 (deletion of spacers 34 to 36) sublineages; and it was suggested that this lineage could represent an ancestral clone of principal genetic group 1 strains (4). More recently, Manu lineage strains were reported from Saudi Arabia (2) and Tunisia (15).
Interestingly, the Manu2 strains (prototype SIT54) have been reported only scarcely in the SITVIT2 database (n = 130, not counting this study). In the updated database, SIT54 strains are present in the highest numbers in India (n = 21), followed by Saudi Arabia (n = 20), the United States (n = 20), South Africa (n = 17), Russia (n = 9), Brazil (n = 6), Great Britain (n = 3), Madagascar (n = 3), and Poland (n = 3). Smaller numbers of SIT54 strains (two or less) have been reported from Armenia, Austria, Bangladesh, China, Cuba, Italy, France, Georgia, Greece, Guinea-Bissau, Iran, Latvia, Panama, Turkey, Senegal, Sweden, and Sudan. Although the origin of the patient was not always indicated in the SITVIT2 database, many of these patients were foreign born; e.g., the two patients from Saudi Arabia were born in Chad, while one patient was from Indonesia. The single case from Australia was from Eritrea, and the two cases from South Africa were from Egypt and Zambia, respectively. One patient each from Sweden and Turkey were from Azerbaijan.
The evolutionary relationships among the strains with all the different spoligotype patterns observed in our sample were further studied by use of an MST (Fig. 1). This tree summarizes the phylogenetic links between two spoligotypes differing by changes observed in direct variable repeats (23); the length of the branches indicates the level of change induced by the loss or the gain of spoligotype spacers in the 43-oligonucleotide format to induce a shift from one allele to another. In Fig. 1, the solid lines show a single spacer change, while dotted lines show two spacer changes (black) or more spacer changed (gray). The color of the circles in Fig. 1 is proportional to the number of clinical isolates in our study, illustrating unique isolates (sky blue) versus clustered isolates (deep blue, two to five strains; red, six strains and more). As opposed to other clades belonging to the principal genetic group 1, such as Beijing and CAS (18), the Manu strains in this MST are the ones situated the closest to the principal genetic group 2 or 3 lineages, such as Haarlem, LAM, and T. Interestingly, the T1 strain represented by its prototype SIT53 constitutes the central node of this unrooted tree (Fig. 1A) and has the highest number of isolates from our study (51/151 [33.8%]). Its prevalence is immediately followed by that of the Manu2 prototype SIT54 (33/151 [21.9%]), suggesting that these two lineages might be phylogenetically linked. As shown in Fig. 1B, the evolution of the Manu lineage was supposed to have happened starting from SIT523 (Manu ancestor), which is the most conserved spoligotype in the SITVIT2 database and which has positive hybridization for all the spacers in the standard 43-spacer format. The potential evolution of modern T1 (SIT53) strains from the Manu lineage via Manu2 (SIT54) and Manu2_var (SIT1196) will now be investigated in detail in a parallel investigation by using 24-locus mycobacterial interspersed repetitive units (20) and other single-nucleotide polymorphisms, such as katG-gyrA polymorphisms (18).
Our findings are of the utmost importance in relation to the findings of a recently published study showing that the most common ancestor of the M. tuberculosis complex emerged some 40,000 years ago from its progenitor in East Africa and was followed by the dissemination of two major basal lineages that spread out of Mesopotamia 10,000 to 20,000 years later (24). In this evolutionary scenario of the spread of TB out of Mesopotamia, the later human migration (8,000 to 5,000 years ago) to Africa, Asia, and Europe is supposed to have played a dynamic role leading to locally adapted strains of tubercle bacilli and further diversifications (24). Nonetheless, this evolutionary scenario does not take into account any of the Manu lineage strains, simply because of the paucity of this genotype in reported TB cases worldwide. Our report shows for the first time an unusually high proportion of Manu strains from Egypt, which should be taken into account for the study of the origin and the evolution of the M. tuberculosis complex. In this context, a retrospective analysis of the results from a study dealing with ancient DNA from Egyptian mummies (26) showed that 2 of 12 strains (strains TT196-44 and TT196-M5) that were thought to have patterns close to those for M. africanum (the absence of spacers 7 to 9 and spacer 39) did share major characteristics of the Manu1 variants, i.e., the presence of spacer 33 and the absence of spacer 34. One may hypothesize that these ancient strains could represent an intermediate clone between an M. africanum lineage and the Manu lineage. In conclusion, our study shows that in Egypt TB is caused by a predominant genotype that belongs to the ancestral Manu lineage and that could be a missing link of the split between ancestral and modern tubercle bacilli during the evolution of M. tuberculosis. We believe that detailed investigations of the Manu lineage strains by 24-locus mycobacterial interspersed repetitive unit analysis, extended spoligotyping, analysis of regions of deletions, and single-nucleotide polymorphism analysis would shed light on the central role of isolates from Egypt and Mesopotamia in the origin, evolution, and global spread of TB.
Acknowledgments
The work done at the Pasteur Institute of Guadeloupe benefited through research grants awarded to N.R. by the European Regional Development Fund, the European Commission (grant ERDF/FEDER, A34-05), and the Regional Council of Guadeloupe (grant CR/08-1612, Biodiversité et Risque Infectieux dans les Modèles Insulaires). T.Z. was awarded a Ph.D. fellowship by the European Social Funds through the Regional Council of Guadeloupe.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Abbadi, S., H. G. Rashed, G. P. Morlock, C. L. Woodley, O. El Shanawy, and R. C. Cooksey. 2001. Characterization of IS6110 restriction fragment length polymorphism patterns and mechanisms of antimicrobial resistant isolates Mycobacterium tuberculosis from a major reference hospital in Assiut, Egypt. J. Clin. Microbiol. 392330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Hajoj, S. A., T. Zozio, F. Al-Rabiah, V. Mohammad, M. Al-Nasser, C. Sola, and N. Rastogi. 2007. First insight into the population structure of Mycobacterium tuberculosis in Saudi Arabia. J. Clin. Microbiol. 452467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, P. F., and M. D. Cave. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 3491149-1156. [DOI] [PubMed] [Google Scholar]
- 4.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Gutierrez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, H. M. Ly, C. Martin, I. Mokrousov, O. Narvskaya, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, M. Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. Shemyakin, U. B. Singh, A. Somoskov, R. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. T. Vincent, T. C. Victor, R. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudey, K., M. C. Gutierrez, V. Vincent, L. M. Parsons, M. Salfinger, N. Rastogi, and C. Sola. 2004. Mycobacterium africanum genotyping using novel spacer oligonucleotides in the direct repeat locus. J. Clin. Microbiol. 425053-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooksey, R. C., S. H. Abdadi, C. L. Woodley, D. Sikes, M. Wasfy, J. T. Crawford, and F. Mahoney. 2002. Characterization of Mycobacterium tuberculosis complex isolates from the cerebrospinal fluid of meningitis patients at six fever hospitals in Egypt. J. Clin. Microbiol. 401651-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins, J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. Quitugua, N. Rastogi, R. A. Skuce, C. Sola, D. Van Soolingen, and V. Vincent. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis. 5216-219. [PubMed] [Google Scholar]
- 8.Ferdinand, S., C. Sola, S. Chanteau, H. Ramarokoto, T. Rasolonavalona, V. Rasolofo-Razanamparany, and N. Rastogi. 2005. A study of spoligotyping-defined Mycobacterium tuberculosis clades in relation to the origin of peopling and the demographic history in Madagascar. Infect. Genet. Evol. 5340-348. [DOI] [PubMed] [Google Scholar]
- 9.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valétudie, D. A. Dang, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, M. L. Ho, A. Makristathis, C. Mammina, G. Martin, P. Moström, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2003. A snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 411963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbón, M. Bobadilla del Valle, J. Fyfe, L. García-García, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. L. León, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendón, J. Sifuentes-Osornio, A. Ponce de León, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Floyd, K., L. Blanc, M. Raviglione, and J. W. Lee. 2002. Resources required for global tuberculosis control. Science 2952040-2041. [DOI] [PubMed] [Google Scholar]
- 12.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent, B. S., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Publication 86-8230. U.S. Department of Health and Human Services, Washington, DC.
- 14.Kox, L. F. F., D. Rhienthong, A. Miranda, N. Udomasantisuk, K. Ellis, J. van Leeuwen, S. vanHeusden, S. Kuijper, and A. H. J. Kolk. 1994. A more reliable PCR for detection of Mycobacterium tuberculosis in clinical samples. J. Clin. Microbiol. 32672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Namouchi, A., A. Karboul, B. Mhenni, N. Khabouchi, R. Haltiti, R. Ben Hassine, B. Louzir, A. Chabbou, and H. Mardassi. 2008. Genetic profiling of Mycobacterium tuberculosis in Tunisia: predominance and evidence for the establishment of a few genotypes. J. Med. Microbiol. 57864-872. [DOI] [PubMed] [Google Scholar]
- 16.Singh, U. B., N. Suresh, N. V. Bhanu, J. Arora, H. Pant, S. Sinha, R. C. Aggarwal, S. Singh, J. N. Pande, C. Sola, N. Rastogi, and P. Seth. 2004. Predominant tuberculosis spoligotypes, Delhi, India. Emerg. Infect. Dis. 101138-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53680-689. [DOI] [PubMed] [Google Scholar]
- 18.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 949869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Streicher, E. M., R. M. Warren, C. Kewley, J. Simpson, N. Rastogi, C. Sola, G. D. van der Spuy, P. D. van Helden, and T. C. Victor. 2004. Genotypic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from rural districts of Western Cape Province of South Africa. J. Clin. Microbiol. 42891-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rüsch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 444498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.USAID. 2004. Infectious diseases. USAID, Washington, DC. http://www.usaid.gov/ourwork/globalhealth/id/tuberculosis/countries/ane/Egypt.
- 22.Van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendation for a standardized methodology. J. Clin. Microbiol. 31406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Embden, J. D., T. Van Gorkom, K. Kremer, R. Jansen, B. A. Van Der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 1822393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wirth, T., F. Hildebrand, C. Allix-Béguec, F. Wölbeling, T. Kubica, K. Kremer, D. van Soolingen, S. Rüsch-Gerdes, C. Locht, S. Brisse, A. Meyer, P. Supply, and S. Niemann. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4e1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaher, H. A., M. A. Tag El Din, and P. J. M. van Maaren. 2008. Tuberculosis control and intersectoral collaboration: the experience of Egypt. National Tuberculosis Control Programme, Cairo, Egypt. http://www.emro.who.int/stb/egypt/Collaboration.htm#section2.
- 26.Zink, A. R., C. Sola, U. Reischl, W. Grabner, N. Rastogi, H. Wolf, and A. G. Nerlich. 2003. Characterization of Mycobacterium tuberculosis complex DNAs from Egyptian mummies by spoligotyping. J. Clin. Microbiol. 41359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]