Abstract
In the spring of 2009, a novel influenza A (H1N1) virus (swine origin influenza virus [S-OIV]) emerged and began causing a large outbreak of illness in Milwaukee, WI. Our group at the Midwest Respiratory Virus Program laboratory developed a semiautomated real-time multiplex reverse transcription-PCR assay (Seasonal), employing the NucliSENS easyMAG system (bioMérieux, Durham, NC) and a Raider thermocycler (HandyLab Inc., Ann Arbor, MI), that typed influenza A virus, influenza B virus, and respiratory syncytial virus (RSV) and subtyped influenza A virus into the currently circulating H1 and H3 subtypes, as well as a similar assay that identified H1 of S-OIV. The Seasonal and H1 S-OIV assays demonstrated analytical limits of detection of <50 50% tissue culture infective doses/ml and 3 to 30 input copies, respectively. Testing of the analytical specificities revealed no cross-reactivity with 41 and 26 different common organisms and demonstrated outstanding reproducibility of results. Clinical testing showed 95% sensitivity for influenza A virus and influenza B virus and 95 and 97% specificity compared to tissue culture. Comparisons of results from other molecular tests showed levels of positive agreement with the Seasonal and H1 S-OIV assay results of 99 and 100% and levels of negative agreement of 98 and 100%. This study has demonstrated the use of a semiautomated system for sensitive, specific, and rapid detection of influenza A virus, influenza B virus, and RSV and subtyping of influenza A virus into human H1 and H3 and S-OIV strains. This assay/system performed well in clinical testing of regular seasonal influenza virus subtypes and was outstanding during the 2009 Milwaukee S-OIV infection outbreak. This recent outbreak of infection with a novel influenza A (H1N1) virus also demonstrates the importance of quickly distributing information on new agents and of having rapid influenza virus subtyping assays widely available for clinical and public health decisions.
In 2005 an adolescent in Wisconsin developed a brief respiratory illness later determined to be caused by a novel influenza A virus that was a mixture of swine, avian, and human virus gene segments. This infection was thought to be zoonotic, originating from pigs that the individual had helped butcher a few days before. No other cases could be linked to this particular virus or individual. In the early spring of 2009, this scenario appeared to occur again in Mexico. An outbreak of respiratory illness caused by a novel influenza A (H1N1) virus (swine origin influenza virus [S-OIV]) shown also to be a triple reassortant began, with significant numbers of individuals being infected (2-4, 20, 21). Soon a widespread outbreak occurred in Milwaukee, WI (13). Our group at the Midwest Respiratory Virus Program laboratory was able to molecularly confirm the first case of infection in Wisconsin on 29 April 2009 and worked closely with the state and city public health officials to provide rapid influenza virus subtyping for a large number of samples.
We have recently developed a number of rapid assays to type and subtype influenza virus. During the 2 weeks prior to 27 April 2009, very little respiratory virus activity was detected in our community or within the entire state of Wisconsin. Because of the concern over S-OIV, we began to perform confirmatory influenza virus typing and subtyping on influenza A virus-positive specimens from the Children's Hospital of Wisconsin (CHW; E. T. Beck, L. A. Jurgens, S. C. Kehl, M. E. Bose, T. Patitucci, E. LaGue, P. Darga, K. Wilkinson, L. M. Witt, J. Fan, J. He, S. Kumar, and K. J. Henrickson, unpublished data) and Dynacare Laboratories (DL) (6, 17) by using multiplex real-time reverse transcription-PCR (rRT-PCR) assays for influenza A virus, influenza B virus, and respiratory syncytial virus (RSV). The presence of these influenza virus subtyping assays in our laboratory provided the necessary tools for us to quickly respond to the emergence of a novel influenza virus subtype within our community.
The Seasonal assay is a semiautomated multiplex rRT-PCR assay that types influenza A virus, influenza B virus, and RSV and subtypes influenza virus by targeting the H1 (human) and H3 (human) hemagglutinin (HA) genes with a noncompetitive RNA internal control (MS2 RNA phage). The FluPlex is a large multiplex RT-PCR enzyme hybridization assay that types influenza A virus and influenza B virus and identifies H1 (human), H2, H3, H5, H7, H9, N1 (human), N1 (animal), N2, and N7 subtypes. We initially were able to use the Seasonal assay to identify influenza A virus samples that did not type as H1 or H3. We then performed the FluPlex assay, which confirmed the samples to be positive for influenza A virus and subtyped them as negative for human HA and neuraminidase subtypes and positive for animal (swine) N1 virus (7). Three days after subtyping with the FluPlex began, we had developed an S-OIV-specific semiautomated assay (the H1 S-OIV assay) with extraction by the NucliSENS easyMAG system (bioMérieux, Durham, NC) and amplification by the Raider thermocycler (HandyLab Inc., Ann Arbor, MI) and the same protocols and formats used for our Seasonal assay. With these tools, we were able to rapidly subtype the influenza A viruses sent to us from two large clinical laboratories. This paper reports the use of the semiautomated multiplex real-time typing and subtyping assays during this outbreak.
MATERIALS AND METHODS
Primer and probe design.
The Seasonal assay has primers and probes designed to correspond to highly conserved regions of the influenza A virus matrix (M) gene, the influenza B virus M gene, the RSV polymerase (L) gene, the HA genes of the H1 and H3 subtypes of human influenza A virus, and the bacteriophage MS2 (internal control) (Table 1). Influenza virus primers were designed using the Influenza Primer Design Resource (http://www.ipdr.mcw.edu), and RSV primers were designed by aligning the 16 RSV L gene sequences found in GenBank (1). Primers and probes utilize proprietary superbases and a 5′ minor groove binder (Pleiades probes; Nanogen, Inc., Bothell, WA) (18). The H1 S-OIV multiplex rRT-PCR assay (H1 S-OIV assay) primer/probe set was designed to detect the HA gene segment from the currently circulating S-OIV. In silico coverage by the primer/probe sets was determined using an in-house program. A sequence was considered to be hit by the primers if there were no mutations within 5 bases from the 3′ end, one or no mutation within 10 bases from the 3′ end, and two or fewer mutations in the whole region corresponding to the oligonucleotide or to be hit by the probes if there were two or fewer mutations in the whole region corresponding to the oligonucleotide. The number of gaps was determined by looking at an alignment of the sequences for which coverage was being determined and counting the number of sequences in the alignment that did not have a full sequence in the target region for the primers and probes. To calculate the percent coverage, the number of sequences hit was divided by the total number of sequences with the number of gaps subtracted, and the quotient was then multiplied by 100 [hits ÷ (total − gaps) × 100]. In silico coverage rates for the primer/probe sets in the Seasonal assay were greater than 95% for all of the targets in the assay. For the H1 S-OIV assay, the coverage was 99.1% for all sequences available as of 11 July 2009 (Table 1). One of the sequences not covered in silico was from one of our own isolates that we had detected with the H1 S-OIV assay and then sequenced.
TABLE 1.
In silico coverage by the primers and probes used in the Seasonal assay and the H1 S-OIV assay
| Organism | Primer name | Primer sequencea | Target | Total no. of sequences | No. of hits | No. of gaps | Coverage (%) |
|---|---|---|---|---|---|---|---|
| RSV | RSV-L3 | AATAAATCATAAGTCA*GTAGTA*GACCATGT | L gene | 16 | 16 | 0 | 100.0 |
| RSV-E4 | AATAAATCATAATAAGCT*GGTA*TTGA*TGCA | L gene | 16 | 16 | 0 | 100.0 | |
| RSV-FAM12 | MGB-Fam-TTGAT*GCA*GG*GA*ATTCA*CA-EDQ | L gene | 16 | 16 | 0 | 100.0 | |
| Total for RSV | 16 | 16 | 0 | 100.0 | |||
| Influenza A virus | INFA-L30 | AATAAATCATAAGTCAGAGGTGACAGGATTGG | M gene | 3,767 | 3,723 | 6 | 99.0 |
| INFA-E7 | AATAAATCATAACTCA*TGGA*ATGGCTAAAG | M gene | 3,767 | 3,705 | 9 | 98.6 | |
| INFA-E8 | AATAAATCATAACTCA*TGGA*GTGGCTAAAG | M gene | 3,767 | 3,496 | 9 | 93.0 | |
| INFA-FAM42 | MGB-Fam-AAG*ACA*A*GA*CCZ*A*T*-EDQ | M gene | 3,767 | 3,760 | 7 | 100.0 | |
| Total for influenza A virus | 3,767 | 3,677 | 9 | 97.8 | |||
| Influenza B virus | DIF430-L30 | AATAAATCATAAGCCTTCTCCA*TCTTCTG | M gene | 364 | 282 | 82 | 100.0 |
| DIF430-E24 | AATAAATCATAAGTCGCTGTTTGGA*GACAC | M gene | 364 | 314 | 42 | 97.5 | |
| DIF430-FAM37 | MGB-Fam-AGCAGGTA*GGCA*ATTGT-EDQ | M gene | 364 | 291 | 73 | 100.0 | |
| Total for influenza B virus | 364 | 274 | 82 | 97.2 | |||
| H1 (human) virus | H1-L17 | AATAAATCATAAGTA*GTGTCTTCACA*TTATAGCA | HA gene | 1,463 | 1,436 | 27 | 100.0 |
| H1-E19 | AATAAATCATAATGATCTCTCA*CCTTGGGTCTT | HA gene | 1,463 | 1,379 | 27 | 96.0 | |
| H1-E20 | AATAAATCATAATGA*TCTCTTA*CTTTGGGTCTT | HA gene | 1,463 | 1,411 | 27 | 98.3 | |
| H1-AP593-16 | MGB-AP-593-CAGAAATAGCCAAAAGAC-EDQ | HA gene | 1,463 | 1,436 | 27 | 100.0 | |
| Total for H1 (human) virus | 1,463 | 1,411 | 27 | 98.3 | |||
| H3 virus | H3-L4 | AATAAATCATAACCCT*GT*GCTGT*T*A*ATCA | HA gene | 4,016 | 3,906 | 6 | 97.4 |
| H3-E9 | AATAAATCATAAGA*ATAAGCA*TCTA*TTGGAC | HA gene | 4,016 | 4,004 | 6 | 99.9 | |
| H3-AP593-2 | MGB-AP-593-G*G*TTTTA*CTATTGTCCAA-EDQ | HA gene | 4,016 | 4,005 | 6 | 99.9 | |
| Total for H3 virus | 4,016 | 3,900 | 6 | 97.3 | |||
| H1 virus (S-OIV) | H1Sw_For652 + 22 | AATAAATCATAAGTGGGGTCATCAAGATACAGCA | HA gene | 555 | 551 | 1 | 99.5b |
| H1Sw_Rev719-21 | AATAAATCATAATGATCCCTCACTTTGGGTCTT | HA gene | 555 | 552 | 1 | 99.6 | |
| H1Sw_Probe684 + 20FAM | 6-Fam-GCCGGAAATAGCAATAAGAC-BHQ1 | HA gene | 555 | 554 | 1 | 100.0 | |
| Total for H1 virus (S-OIV) | 555 | 549 | 1 | 99.1 | |||
| MS2 phage | MS2-L23 | AATAAATCATAAGGTCGGTA*CTAACA*TCAAG | NAd | NA | NA | NA | |
| MS2-E7 | AATAAATCATAAGCA*CGTTGT*CTGGAAGTT | NA | NA | NA | NA | ||
| MS2-AP593-7c | MGB-AP-593-CG*TATCCA*G*CTG*CA*AA*CT-EDQ | NA | NA | NA | NA | ||
| MS2-AP593-6c | MGB-AP-593-CGTATCCA*GCTGCA*AA*CT-EDQ | NA | NA | NA | NA |
* indicates that the previous base is a proprietary superbase (Nanogen, Inc., Bothell, WA). MGB is a proprietary minor groove binder (Nanogen, Inc., Bothell, WA). Fam is a proprietary fluorophore (Nanogen, Inc., Bothell, WA) similar to 6-Fam (Applied Biosystems Inc., Foster City, CA). AP-593 is a proprietary fluorophore (Nanogen, Inc., Bothell, WA) similar to CalRed. EDQ is Eclipse dark quencher (Glen Research Corp., Sterling, VA). BHQ1 is Black Hole Quencher 1 (Biosearch Technologies Inc., Novato, CA).
One of the sequences missed in silico was actually detected experimentally.
The MS2-AP593-6 probe is used in the S-OIV assay, and the MS2-AP593-7 probe is used in the Seasonal subtyping assay.
NA, not applicable.
Sample preparation.
A sample of 400 μl was combined with 10 μl of MS2 bacteriophage (5 × 105 PFU/ml) and 1 ml of lysis buffer and incubated at room temperature for 10 min. After lysis, the samples were loaded onto the NucliSENS easyMAG system (bioMérieux, Durham, NC). Total nucleic acid extractions proceeded according to the manufacturer's protocol. Samples were eluted in 25 μl of elution buffer.
rRT-PCR and melt analysis.
Following elution, 3.4 μl of RNA was mixed with 4.6 μl of supermix containing primers and probes, Platinum Tfi, and SuperScript III (Invitrogen, Carlsbad, CA) to yield a one-step RT-PCR mixture with an 8-μl final volume. The reaction mixture was manually loaded into a microfluidic Raider cartridge and placed into the Raider high-speed thermocycler (HandyLab Inc., Ann Arbor, MI). The Raider thermocycler utilizes a proprietary microfluidic cartridge that is approximately 1.5 mm thick and utilizes 4.2-μl reaction wells allowing for rapid heating and cooling. Each cartridge can run up to 12 reactions. The cycling parameters used are as follows: 15 min at 50°C; 2 min at 95°C; 25 cycles of 1 s at 95°C, 15 s at 61°C, and 10 s at 76°C; 20 cycles of 1 s at 95°C, 15 s at 56°C, and 10 s at 76°C; a 60-s light-emitting diode warm-up; and a subsequent melt analysis at 45 to 85°C with a melting rate of 0.3°C/s.
Analysis of results.
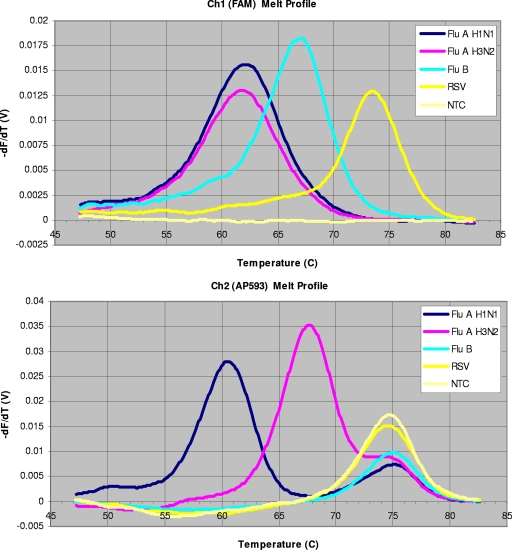
Assay results were analyzed using an in-house-developed Excel workbook (computational algorithm) that converts raw data into the final output format. Data were analyzed and the tests were scored based on the amplification and melting profiles of the sample. The melting profile was created as the change in fluorescence versus temperature (Fig. 1) in two different channels. Influenza A virus, influenza B virus, and RSV were labeled with Fam (a proprietary fluorophore created at Nanogen, Inc.). The H1 S-OIV probe was labeled with 6-carboxyfluorescein (6-Fam; a dye similar to Fam) from Applied Biosystems, Foster City, CA. In this article, Fam, 6-Fam, and FAM refer to the fluorophore from Nanogen, Inc., the fluorophore from Applied Biosystems, and the fluorescence channel on the Raider thermocycler that detects fluorescence of the Fam and 6-Fam fluorophores. The H1 (human) virus and the H3 virus and both internal control MS2 probes were labeled with AP-593 (a proprietary fluorophore from Nanogen, Inc.). Samples were considered positive if the threshold cycle (CT) value was ≤40.0, the amplification curve shape was appropriate, and the melting profiles yielded melting temperatures (Tm) within 2°C of those expected for RSV (74°C), influenza B virus (66°C), influenza A virus (60°C), H1 (human) virus (60°C), H3 (human) virus (68°C), or H1 S-OIV (60°C). Sample results were considered indeterminate if CT values were >40.0, with an appropriate Tm and amplification curve shape. Samples were considered negative if the internal control (MS2) Tm was appropriate (73 ± 2°C in the AP-593 channel), the AP-593 CT was <40.0 with the appropriate amplification curve shape, the FAM CT was >40.0 with an incorrect or nonexistent Tm or an abnormal amplification curve, and no melt profiles in the AP-593 channel for H1 or H3 were obtained.
FIG. 1.
Melting profile from the Seasonal subtyping assay. The melting curves for influenza A and influenza B viruses and RSV are visible on the FAM channel, and the H1, H3, and MS2 melting curves are visible on the AP-593 channel. NTC, no-template (negative) control.
Analytical sensitivity (LODs).
Serial 10-fold dilutions from 104 to 10−2 TCID50/ml of different subtypes of influenza virus were prepared in M4 viral transport medium (Remel, Lenexa, KS) and tested in the Seasonal and H1 S-OIV assays (Table 2). The limits of detection (LODs) for the Seasonal assay were determined using a probit analysis based on 10 separate experiments to calculate the LODs. Several of the S-OIV-positive samples were cultured and quantitated using an in-house quantitative rRT-PCR assay that targets the matrix gene of influenza A virus. Serial 10-fold dilutions of these virus isolates were prepared in M4 and tested in the H1 S-OIV assay to determine the lowest concentrations that could successfully be detected (Table 2). LODs for this assay were determined with only three replicates because time was of the essence during the beginning of the pandemic.
TABLE 2.
Analytical sensitivities of the Seasonal subtyping assay and the H1 S-OIV assay
| Assay | Virus | Analyte | LODa |
|---|---|---|---|
| Seasonal subtyping | A/New Caledonia/20/1999 (H1N1) | Influenza A virus | 16 |
| assay | A/New Caledonia/20/1999 (H1N1) | H1 virus | 1 |
| A/Hawaii/15/2001 (H1N1) | Influenza A virus | 1 | |
| A/Hawaii/15/2001 (H1N1) | H1 virus | 2 | |
| A/New York/55/2004 (H3N2) | Influenza A virus | 34 | |
| A/New York/55/2004 (H3N2) | H3 virus | 53 | |
| A/Wisconsin/67/2005 (H3N2) | Influenza A virus | 8 | |
| A/Wisconsin/67/2005 (H3N2) | H3 virus | <0.3 | |
| B/Ohio/01/2005 (Victoria/2/87-like) | Influenza B virus | <0.1 | |
| B/Florida/07/2004 (Yamagata/16/88-like) | Influenza B virus | 12 | |
| RSV-A/WI/629-5/0708 | RSV | <0.0003 | |
| H1 S-OIV assay | A/WI/629-S128/09 (H1N1 S-OIV) | H1 virus (S-OIV) | 3-30 |
| A/WI/629-S129/09 (H1N1 S-OIV) | H1 virus (S-OIV) | 3-30 |
The LODs for the Seasonal subtyping assay are expressed as the number of TCID50 per reaction, and those for the H1 S-OIV assay are expressed as the number of copies per reaction.
Influenza A virus subtype specificity.
Both assays were tested against influenza A virus strains representing H1 to H15 and N1 to N9 at >105 TCID50/ml to determine cross-reactivity with other influenza A virus subtypes.
Analytical specificity against other common respiratory organisms.
M4 viral transport medium was spiked with high concentrations (>104 TCID50, PFU, or CFU/ml) of common respiratory pathogens and commensal organisms and tested in both of the assays (Table 3) .
TABLE 3.
List of common respiratory organisms tested to evaluate specificity
| Organism(s) |
|---|
| Organisms tested in both assays |
| Adenovirus type 3 |
| Coronavirus OC43 |
| Herpes simplex virus type 1 |
| Human metapneumovirus (A1, A2, B1, and B2) |
| Human parainfluenza viruses 1 to 4 |
| Human rhinovirus 1B |
| Chlamydophila pneumoniae |
| Eikenella corrodens |
| Enterococcus faecalis |
| Escherichia coli |
| Haemophilus influenzae |
| Moraxella catarrhalis |
| Neisseria sicca |
| Pseudomonas aeruginosa |
| Staphylococcus aureus |
| Staphylococcus epidermidis |
| Streptococcus agalactiae |
| Streptococcus mitis |
| Streptococcus pyogenes |
| Streptococcus sanguinis |
| Organisms tested in the Seasonal assay only |
| Adenovirus types 1, 5, 7, 10, and 18 |
| Coxsackievirus E9 and B5 |
| Cytomegalovirus |
| Echovirus 2 |
| Human rhinovirus 2, 14, and 16 |
| Measles virus |
| Mumps virus |
| Rubella virus |
| Varicella-zoster virus |
| Acinetobacter calcoaceticus |
| Bacteroides fragilis |
| Bordetella pertussis |
| Candida albicans |
| Corynebacterium diphtheriae |
| Gardnerella vaginalis |
| Klebsiella pneumoniae |
| Lactobacillus casei |
| Lactobacillus plantarum |
| Legionella pneumophila |
| Listeria monocytogenes |
| Mycobacterium avium |
| Mycoplasma orale |
| Mycoplasma pneumoniae |
| Neisseria gonorrhoeae |
| Neisseria meningitidis |
| Neisseria subflava |
| Proteus vulgaris |
| Streptococcus mutans |
| Streptococcus pneumoniae |
| Streptococcus salivarius |
| Streptococcus groups B, C, F, and G |
Reproducibility.
Interrun reproducibility of the Seasonal assay results was determined by calculating the standard deviations of the CT values and Tm of the positive controls in seven runs with the same sets of samples representing each of the targets in the assay. The positive controls consisted of quantitated virus diluted in M4 at the following concentrations: 100 and 10−3 TCID50/ml (RSV-A) and 103 and 102 TCID50/ml (influenza A [H1N1 and H3N2] virus and influenza B virus) (see Table 5). Intrarun reproducibility was determined by calculating the standard deviations of the CT values and Tm for an influenza A H1N1 virus at 106 TCID50/ml and negative samples. The H1N1 virus samples and negative samples were prepared in M4 and then divided into 400-μl aliquots. Five runs with six H1N1 virus samples and six negative samples per run were performed. Intrarun variability for each run was then assessed.
TABLE 5.
Reproducibility of Seasonal assay CT and Tm results in seven runs on the same day during a throughput study
| Virus | Subtype | Concn (TCID50/ml) | FAM channel
|
AP-593 channel
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean CT | CT SD | n | Mean Tm (°C) | Tm SD | n | Mean CTa | CT SD | n | Mean Tm (°C) | Tm SD | n | |||
| A/New Caledonia/20/1999 | H1N1 | 100 | 36 | 1 | 6 | 60.2 | 0.2 | 5 | 35.8 | 1.7 | 6 | 60.2 | 0.1 | 6 |
| A/New Caledonia/20/1999 | H1N1 | 1,000 | 32.8 | 0.5 | 7 | 60.4 | 0.3 | 7 | 31.4 | 0.3 | 7 | 60.1 | 0.1 | 7 |
| A/Hawaii/15/2001 | H1N1 | 100 | 34.8 | 0.8 | 7 | 60.6 | 0.1 | 7 | 33.8 | 0.6 | 7 | 60.5 | 0.1 | 7 |
| A/Hawaii/15/2001 | H1N1 | 1,000 | 32.9 | 0.4 | 7 | 60.7 | 0.4 | 7 | 31.2 | 0.4 | 7 | 60.4 | 0.2 | 7 |
| A/Wisconsin/67/2005 | H3N2 | 100 | 33.2 | 0.7 | 7 | 61.3 | 0.2 | 7 | 30.5 | 0.8 | 7 | 68.4 | 0.1 | 7 |
| A/Wisconsin/67/2005 | H3N2 | 1,000 | 30.6 | 0.6 | 7 | 61.6 | 0.2 | 7 | 27.1 | 0.5 | 7 | 68.3 | 0.2 | 7 |
| A/New York/55/2004 | H3N2 | 100 | 36.3 | 0.8 | 7 | 60.9 | 0.3 | 7 | 35.7 | 0.8 | 7 | 68.3 | 0.1 | 7 |
| A/New York/55/2004 | H3N2 | 1,000 | 32.4 | 0.5 | 7 | 61 | 0.2 | 7 | 31 | 0.4 | 7 | 68.2 | 0.1 | 7 |
| RSV-A/WI/625-5/0708 | A | 0.001 | 38.4 | 0.6 | 7 | 74.1 | 0.1 | 7 | NAc | NA | NA | NA | NA | NA |
| RSV-A/WI/625-5/0708 | A | 1 | 25.3 | 0.8 | 6 | 74.5 | 0.2 | 6 | NA | NA | NA | NA | NA | NA |
| B/Florida | Yamagata | 100 | 35.1 | 0.7 | 7 | 66.8 | 0.3 | 7 | NA | NA | NA | NA | NA | NA |
| B/Florida | Yamagata | 1,000 | 29.2 | 0.9 | 7 | 66.2 | 0.1 | 7 | NA | NA | NA | NA | NA | NA |
| MS2 bacteriophageb | NA | NA | NA | NA | NA | NA | 37.3 | 1.5 | 111 | 75 | 0.5 | 150 | ||
AP-593 CT values for H1N1 and H3N2 viruses reflect simultaneous amplification of the HA subtype and the MS2 internal control amplicons.
Means and standard deviations for MS2 were calculated using negative, influenza B virus, and RSV samples in which the only target in the AP-593 channel was MS2.
NA, not applicable.
Clinical sensitivity and specificity.
Three hundred and fifteen deidentified nasopharyngeal swabs were collected at TriCore Laboratories (Albuquerque, NM). Following collection, Dacron swabs were placed into M5 viral transport medium (Remel, Lenexa, KS) and shipped to TriCore Laboratories. Upon arrival, samples were inoculated into shell vials containing R-Mix cells. Positive samples were analyzed at TriCore by a fluorescent-antibody assay for the presence of influenza A virus or influenza B virus. Influenza virus-positive samples were then subtyped using type-specific serum. Samples were then frozen at −80°C until testing with the Seasonal assay could be performed (as described previously).
Subtyping of influenza A virus-positive samples.
A total of 2,517 nasopharyngeal, nasal, and/or throat specimens submitted to the CHW or DL between 27 April and 11 May 2009 were tested for influenza A virus by using multiplex rRT-PCR assays. Samples from the CHW were tested using a fully automated multiplex RT-PCR on a Jaguar extractor/thermocycler (HandyLab, Inc., Ann Arbor, MI), while samples from DL were tested using a semiautomated multiplex RT-PCR consisting of extraction on an easyMAG system and real-time amplification on a smart cycler (Cepheid, Sunnyvale, CA) using Cepheid's assay-specific reagents. Both assays are capable of simultaneously detecting influenza A virus, influenza B virus, and RSV (6, 17; Beck et al., unpublished). Three hundred and five influenza A virus-positive specimens, 2 influenza B virus-positive specimens, and 22 negative specimens were sent to the Midwest Respiratory Virus Program lab for influenza virus subtyping. Raw specimens (from the CHW) subjected to extraction as described above or total nucleic acid previously extracted from 255 μl of sample material on the easyMAG system with elution in 55 μl (from DL) was used in the assay. Influenza A virus-positive samples were typed and subtyped with the Seasonal assay, the H1 S-OIV assay, and the FluPlex (7). The FluPlex targets different genetic regions from those targeted by the Seasonal or H1 S-OIV assay. The first 127 influenza A virus-positive and 22 negative clinical samples were tested by the Seasonal, FluPlex, and H1 S-OIV assays. Thereafter, all influenza A virus-positive samples were subtyped with the Seasonal and H1 S-OIV assays. All samples with discrepant results were tested by the FluPlex. A segment of the H1 gene was sequenced using 13 random clinical S-OIV-positive samples for subtype confirmation. For sequencing, 3 μl of nucleic acid was reverse transcribed in a 20-μl reaction mixture with murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA). Ten microliters of this reaction mixture was used for PCR with the following primers: H1sw_For403 + 21SQ (TGTAAAACGACGGCCAGTCCCAAGACAAGTTCATGGCCC) and H1sw_Rev906-21SQ (AGGAAACAGCTATGACCATAGCACCCTTGGGTGTTTGACA) (underlining indicates M13 primer binding sequences used during subsequent reactions with M13 primers). Following amplification, PCR products were purified with the QiaQuick gel extraction kit and sent to Retrogen, Inc. (San Diego, CA), along with the primers for sequence analysis. In addition to being analyzed in our laboratory, 23 clinical samples (collected early in the course of the outbreak) were sent to the Wisconsin State Laboratory of Hygiene for confirmation of results by the CDC Laboratory Response Network influenza virus typing and subtyping assays and the CDC H1N1 S-OIV-specific assay.
RESULTS
Analytical sensitivity and subtype specificity.
The analytical sensitivities, or LODs, of the Seasonal assay and the H1 S-OIV assay are shown in Table 2. The Seasonal assay was able to detect ∼50 TCID50/ml or fewer with all of the targets in the assay. The H1 S-OIV assay had a sensitivity of 102 to 103 copies/ml, or 3 to 30 copies/reaction. Testing of the two assays with viruses representing 15 different HA types and 9 different neuraminidase types showed no cross-reactivity with other subtypes and no cross-reactivity between human H1 virus and H1 S-OIV (Table 4) .
TABLE 4.
Results from evaluation of the Seasonal subtyping assay and the H1 S-OIV assay against other influenza virus subtypes to determine cross-reactivity
| Strain name | Concna (TCID50/ml or copies/ml) | Seasonal assay result | S-OIV assay resultb |
|---|---|---|---|
| A/New Caledonia/20/1999 (H1N1) | 105 | A, H1 | N |
| A/Mallard/NY/6750/78 (H2N2) | 107 | A | N |
| A/WI/67/2005 (H3N2) | 105 | A, H3 | N |
| A/Mallard/OH/330/86 (H4N8) | 107 | A | N |
| Anhui/02/2005/PR8-IBCDC-RG5 (H5N1) | 105.05 | A | N |
| A/Chicken/CA/431/00 (H6N2) | 107 | A | N |
| A/Chicken/NJ/15086-3/94 (H7N3) | 106 | A | N |
| A/Blue-Winged Teal/LA/B194/86 (H8N4) | 106 | A | N |
| A/Chicken/NJ/12220/97 (H9N2) | 106 | A | N |
| A/GWT/LA/169GW/88 (H10N7) | 106 | A | N |
| A/Ch/MJ/15902-9/96 (H11N9) | 106 | A | N |
| A/Duck/LA/188D/87 (H12N5) | 106 | A | N |
| A/Gull/MD/704/77 (H13N6) | 105.8 | A | N |
| A/Mallard/GurjevRussia/262/82 (H14N5) | 106 | A | N |
| A/Shearwater/Australia/2576/79 (H15N9) | 106 | A | N |
| B/Ohio/01/2005 (Victoria/2/87-like) | 105 | B | N |
| B/Florida/07/2004 (Yamagata/16/88-like) | 105 | B | N |
| S-OIV A/WI/629-S128/2009 (H1N1) | 106 | A | H1 S-OIV |
The S-OIV concentration is given in copies per milliliter; all other concentrations are in TCID50 per milliliter.
N indicates a negative result.
Analytical specificity with other common respiratory organisms.
Testing of the analytical specificities of both assays against 26 common respiratory organisms revealed no cross-reactivity between the assay mixtures and the nonspecific targets. Additional testing of the Seasonal assay against 41 more respiratory organisms also showed no cross-reactivity.
Reproducibility.
The interrun variability of the Seasonal assay showed a standard deviation of the CT of less than 1 cycle and of the Tm of less than 0.5°C for all of the targets except the internal control, which had a standard deviation of 1.5 cycles and 0.5°C (Table 5). The intrarun variability was similar with an average standard deviation of 0.6 cycles for the influenza A CT, 0.8 cycles for the MS2 Ct, 0.5°C for the influenza A Tm, 0.3°C for the H1 Tm, and 0.4°C for the MS2 Tm.
Clinical sensitivity and specificity.
Of the 315 nasopharyngeal swabs tested, 20% (65) were positive for influenza A virus, 18% (57) were positive for influenza B virus, and 65% (205) were negative for both viruses by tissue culture. Testing with the Seasonal assay showed 95% sensitivity for both of these viruses, with 95% specificity for influenza A virus and 97% specificity for influenza B virus (Table 6). While these specificity numbers are excellent, it is probable that the real specificity is higher since RT-PCR is known to be more sensitive than tissue culture. In addition, of the 60 samples called influenza A virus by tissue culture and the Seasonal assay, all 60 gave a subtype result (50 H1 samples and 10 H3 samples).
TABLE 6.
Performance characteristics of the Seasonal assay compared to tissue culturea
| Virus | % Clinical sensitivity (95% CI) | % Clinical specificity (95% CI) | PPV | NPV |
|---|---|---|---|---|
| Influenza A virus | 95 (87-99) | 95 (92-98) | 0.83 | 0.99 |
| Influenza B virus | 95 (85-99) | 97 (94-99) | 0.89 | 0.99 |
95% CI, 95% confidence interval; PPV, positive predictive value; NPV, negative predictive value.
Clinical testing of samples during the S-OIV infection outbreak.
The results of the influenza virus typing and subtyping can be seen in Table 7 . The Seasonal and H1 S-OIV assay results had outstanding agreement and correlation with each other and with the results of the other molecular assays tested (7; Beck et al., unpublished). In addition, 23 of 23 of the clinical samples were confirmed to be positive for influenza A H1N1 virus (19 for S-OIV and 4 for human virus) by Wisconsin State Laboratory of Hygiene. Thirteen clinical isolates were also confirmed by sequencing to be influenza A H1N1 S-OIV. One influenza A virus sample (328) could not be subtyped in any assay, nor could it be sequenced, while all other influenza A virus isolates could be sequenced. This may indicate a sample with a low virus titer or an incomplete RNA genome.
TABLE 7.
Results of influenza virus subtyping during S-OIV infection outbreak in Milwaukee, WI, in 2009
| Sample group (no. of samples) | Result for target (no. of samples) or no. of samples designated:
|
Agreement (95% CI) (comparison)g:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Influenza A virus positive | Influenza A virus negative | Not H1/H3 human virus | H1 human virus | N1 human virus | H3 human virus | N2 human virus | H1 (S-OIV)f | N1 (S-OIV) | Influenza B virus positive | Untyped | Negative | Positive | Negative | |
| Samples with resolved results | 303 | 2 | 10 | 2 | 288 | 110 | 2 | 1 | 22 | |||||
| Samples subjected to: | ||||||||||||||
| Influenza A/B virus-RSV screeninga (329) | 305 (4c) | NAd | NA | NA | NA | NA | NA | NA | 2 | NA | 22 | 99 (98-100) | 100 (92-100) | |
| Testing in Seasonal assay (subtyping) (329) | 301 | 4c | 291e | 8e | NA | 2 | NA | NA | NA | 2 | 0 | 22 | 99 (98-100) | 98 (87-100) |
| Testing in H1 S-OIV assay (329) | NA | Negative (3c) | Negative (10) | NA | Negative (2) | NA | 288 | NA | Negative (2) | 0 | 22 | 100 (97-100) | 100 (91-100) | |
| Testing in FluPlex assayb (149) | 123 (1c) | 2c | 10 | 10 | 2 | 2 | NA | 110 | 2 | 1 | 22 | |||
Testing was performed at the CHW (Beck et al., unpublished) and DL.
Multiplex RT-PCR-enzyme hybridization influenza virus subtyping assay (4).
Samples with discrepant results (no. 266, 301, 307, and 328): no. 301, influenza A virus (H1N1 S-OIV) positive in H1 S-OIV assay and FluPlex assay, with CT value just over the cutoff limit in the Seasonal assay; no. 328, influenza A (untyped) virus positive in FluPlex assay (only M gene detected); no. 266 and 307, subjected to repeated testing and found to be negative by the screening assay and negative by the Seasonal and FluPlex assays, with previous results all determined to be false positives.
NA, not applicable.
Two influenza A H1N1 (human) virus samples (no. 39 and 107) were not subtyped by the Seasonal assay.
Of the 110 H1 S-OIV assay-positive samples tested in the FluPlex, all 110 were positive for N1 S-OIV, showing a 100% correlation between the results of the two assays.
Percent agreement is given for the following comparisons, from top to bottom: initial screening results versus Seasonal assay results for identification of influenza virus; Seasonal assay results versus H1 S-OIV results for designation as negative for H1/H3 versus positive for S-OIV; and H1 S-OIV assay results versus FluPlex assay results. 95% CI, 95% confidence interval.
DISCUSSION
We report the development and use of semiautomated rRT-PCR assays to type and subtype influenza viruses and the ease and efficiency with which this technology was adapted to detect a novel influenza virus subtype very early in an outbreak. Ultimately, the ability to perform influenza virus subtyping on large numbers of clinical samples each day led to improved patient care and greatly facilitated timely and informed public health decisions throughout the epidemic.
A goal of our laboratory has been to develop rapid, sensitive, and specific semiautomated and automated multiplex assays for the detection of common community-acquired respiratory viruses (5, 9, 11, 14). Tissue culture had been the “gold standard” for respiratory virus detection until approximately the late 1990s, when large multiplex RT-PCR assays first became available clinically and commercially (5). RT-PCR was quickly shown to be more sensitive than tissue culture and highly specific for the detection of influenza virus and RSV (and most other respiratory viruses) (5, 8-11, 14).
Just prior to the S-OIV infection outbreak, we had developed a rapid, sensitive, multiplex rRT-PCR assay (the Seasonal assay) capable of detecting and differentiating influenza A virus, influenza B virus, and RSV and identifying the H1 and H3 subtypes of influenza A virus. We had also developed a reflex (nonseasonal) assay for potential pandemic situations to further subtype influenza A virus-positive samples as H5, H7, or H9 if they were not subtyped by the Seasonal assay. These avian subtypes had been identified as possible causes of the next influenza pandemic, and many laboratories around the world have been focusing their efforts on being able to detect these subtypes, especially H5. The emergence of S-OIV and its ability to efficiently spread from human to human has effectively demonstrated that widely available influenza virus subtyping assays that include broader subtyping ability may be critical in the next pandemic.
The LODs for the Seasonal assay are less then 102 TCID50/ml for RSV-A and RSV-B, influenza B virus, and H1N1 and H3N2 viruses, and those for the H1 S-OIV assay are 103 copies/ml or less. These results compare well to those reported for the FDA-approved ProFlu+ (Prodesse Inc., Waukesha, WI) and xTAG respiratory virus panel (Luminex Corp., Austin, TX) assays (12, 15, 16). Our assays, however, can be completed much faster than either of these two assays, with a time from sample collection to result of just under 2 h, with the ProFlu+ assay taking 3.5 h and the xTAG respiratory virus panel assay taking up to 8 h (12, 16, 17, 19). Throughput studies with the Seasonal assay demonstrated that as many as 144 samples can be processed and tested in an 8-h shift by using one easyMAG extractor and two Raider thermocyclers. In addition to the impressive sensitivity, we demonstrated that our assay has a high level of specificity, showing no cross-reactivity with a panel of common respiratory organisms. We also tested subtypes H1 to H15 of influenza A virus with both the assays. All viruses were typed as influenza A virus, and only H1 and H3 subtypes were positive in the Seasonal assay and only H1 S-OIV was positive in the H1 S-OIV assay.
Testing of the Seasonal and H1 S-OIV assays demonstrated outstanding clinical sensitivities, specificities, and agreement of results with those of other molecular assays (7; Beck et al., unpublished). One advantage to real-time melt analysis for influenza virus subtyping is that mutations in the probe region can be readily seen by the shift of the melting curve. Two of the 288 S-OIV-positive clinical samples demonstrated a significant melting-curve shift, suggesting mutations in the HA gene of S-OIV. Upon HA gene sequence analysis for these two samples, different single mutations in the probe region were discovered, explaining the observed shift in the Tm.
The easyMAG/Raider system developed for our Seasonal assay demonstrated significant flexibility, allowing us to quickly respond to the emergence of S-OIV infection in Milwaukee. We were able to have a validated assay for the H1 gene of S-OIV up and clinically available within 4 days of the first S-OIV sequence being available. The H1 S-OIV assay not only uses the same internal control (MS2) but also runs on the same cycling parameters and can be run at the same time in the same microfluidic cartridges as the Seasonal assay. The only currently available FDA-approved influenza virus subtyping assays that are comparable in speed to our assay are the CDC singleplex assays for H1, H3, and H5 and now their S-OIV assay. These are only available through the Laboratory Response Network and have not yet been distributed to other laboratories in our area. The ability to rapidly test 329 samples and provide specific influenza virus subtyping information in as little as 3 h during the first 2 weeks of the S-OIV infection outbreak allowed for timely and effective clinical and public health decision making by health officials. On any one day, we reported the presence of human H3N2 virus, H1N1 virus, S-OIV, influenza B virus, and many other community-acquired respiratory viruses. The fact that the influenza viruses have different antiviral susceptibilities makes rapid subtype reporting critical to clinical management. Working closely with the state and city public health officials, we provided rapid subtyping of S-OIV which helped them recognize the extent of the Milwaukee outbreak earlier than would normally have been possible. This recent outbreak of infection with a novel influenza A (H1N1) virus demonstrates the importance of quickly distributing information on new agents and of having rapid influenza virus subtyping assays widely available.
Acknowledgments
We thank Jacob Metallo for his help with bioinformatics, Cecilia Rebuffo-Scheer for help with assay development, and Jessica Trost and Rose Chen for growing and quantitating the virus isolates used in this study.
This research was supported in part by grants UO1-AI77988, U01-AI070428, and U01-AI066584 from the NIAID and by the Centers for Disease Control and Prevention contract 200-2008-25466, which is a cooperative agreement with Nanogen, Inc. (San Diego, CA) and HandyLab, Inc. (Ann Arbor, MI).
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Bose, M. E., J. C. Littrell, A. D. Patzer, A. J. Kraft, J. A. Metallo, J. Fan, and K. J. Henrickson. 2008. The Influenza Primer Design Resource: a new tool for translating influenza sequence data into effective diagnostics. Influenza Other Respir. Viruses 223-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. 2009. Swine influenza A (H1N1) infection in two children—Southern California, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58400-402. [PubMed] [Google Scholar]
- 3.CDC. 2009. Swine-origin influenza A (H1N1) virus infections in a school—New York City, April 2009. MMWR Morb. Mortal. Wkly. Rep. 58470-472. [PubMed] [Google Scholar]
- 4.CDC. 2009. Update: infections with a swine-origin influenza A (H1N1) virus—United States and other countries, April 28, 2009. MMWR Morb. Mortal. Wkly. Rep. 58431-433. [PubMed] [Google Scholar]
- 5.Fan, J., K. J. Henrickson, and L. L. Savatski. 1998. Rapid simultaneous diagnosis of RSV A, B, influenza A, B, human parainfluenza virus type 1, 2, and 3 infection by multiplex quantitative RT-PCR enzyme hybridization (Hexaplex) assay. Clin. Infect. Dis. 261397-1402. [DOI] [PubMed] [Google Scholar]
- 6.Goodrich, J. S., and M. B. Miller. 2007. Comparison of Cepheid's analyte-specific reagents with BD Directigen for detection of respiratory syncytial virus. J. Clin. Microbiol. 45604-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He, J., M. E. Bose, E. T. Beck, J. Fan, S. Tiwari, J. Metallo, L. A. Jurgens, S. C. Kehl, N. Ledeboer, S. Kumar, W. Weisberg, and K. J. Henrickson. 2009. Rapid multiplex reverse transcription-PCR typing of influenza A and B virus, and subtyping of influenza A virus into H1, 2, 3, 5, 7, 9, N1 (human), N1 (animal), N2, and N7, including typing of novel swine origin influenza A (H1N1) virus, during the 2009 outbreak in Milwaukee, Wisconsin. J. Clin. Microbiol. 472772-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrickson, K. J. 2004. Advances in the laboratory diagnosis of viral respiratory disease. Pediatr. Infect. Dis. J. 23(1 Suppl.)S6-10. [DOI] [PubMed] [Google Scholar]
- 9.Henrickson, K. J. 2005. Cost-effective use of rapid diagnostic techniques in the treatment and prevention of viral respiratory infections. Pediatr. Ann. 3424-31. [DOI] [PubMed] [Google Scholar]
- 10.Henrickson, K. J., S. Hoover, K. S. Kehl, and W. Hua. 2004. National disease burden of respiratory viruses detected in children by polymerase chain reaction. Pediatr. Infect. Dis. J. 23(1 Suppl.)S11-S18. [DOI] [PubMed] [Google Scholar]
- 11.Huang, Y., H. Tang, S. Duffy, Y. Hong, S. Norman, M. Ghosh, J. He, M. Bose, K. J. Henrickson, J. Fan, A. J. Kraft, W. G. Weisburg, and E. L. Mather. 2009. Multiplex assay for simultaneously typing and subtyping influenza viruses by use of an electronic microarray. J. Clin. Microbiol. 47390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krunic, N., T. D. Yager, D. Himsworth, F. Merante, S. Yaghoubian, and R. Janeczko. 2007. xTAG RVP assay: analytical and clinical performance. J. Clin. Virol. 40(Suppl. 1)S39-S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar, S., M. J. Chusid, R. E. Willoughby, P. L. Havens, S. C. Kehl, N. A. Ledeboer, S. Li, and K. J. Henrickson. 2009. Introduction of a novel swine-origin influenza A (H1N1) virus into Milwaukee, Wisconsin in 2009. Viruses 173-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar, S., L. Wang, J. Fan, A. Kraft, M. E. Bose, S. Tiwari, M. Van Dyke, R. Haigis, T. Luo, M. Ghosh, H. Tand, M. Haghnia, E. L. Mather, W. G. Weisburg, and K. J. Henrickson. 2008. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J. Clin. Microbiol. 463063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, W. M., K. Grindle, T. Pappas, D. J. Marshall, M. J. Moser, E. L. Beaty, P. A. Shult, J. R. Prudent, and J. E. Gern. 2007. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J. Clin. Microbiol. 452626-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeGoff, J., R. Kara, F. Moulin, A. Si-Mohamed, A. Krivine, L. Bélec, and P. Lebon. 2008. Evaluation of the one-step multiplex real-time reverse transcription-PCR ProFlu-1 assay for detection of influenza A and influenza B viruses and respiratory syncytial viruses in children. J. Clin. Microbiol. 46789-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao, R. S., L. L. Tomolty, A. Majury, and D. E. Zoutman. 2009. Comparison of viral isolation and multiplex real-time reverse transcription-PCR for confirmation of respiratory syncytial virus and influenza virus detection by antigen immunoassays. J. Clin. Microbiol. 47527-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukhtanov, E. A., S. G. Lokhov, V. V. Gorn, M. A. Podyminogin, and W. Mahoney. 2007. Novel DNA probes with low background and high hybridization-triggered fluorescence. Nucleic Acids Res. 35e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahony, J., S. Chong, F. Merante, S. Yaghoubian, T. Sinha, C. Lisle, and R. Janeczko. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 452965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. 7 May 2009, posting date. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. doi:10.1056/NEJMoa0903810. [DOI] [PubMed]
- 21.Shinde, V., C. B. Bridges, T. M. Uyeki, B. Shu, A. Balish, X. Xu, S. Lindstrom, L. V. Gubareva, V. Deyde, R. J. Garten, M. Harris, S. Gerber, S. Vagasky, F. Smith, N. Pascoe, K. Martin, D. Dufficy, K. Ritger, C. Conover, P. Quinlisk, A. Klimov, J. S. Bresee, and L. Finelli. 7 May 2009, posting date. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N. Engl. J. Med. doi:10.1056/NEJMoa0903810. [DOI] [PubMed]