Abstract
Since 2000, our geographical region in France systematically surveys bloodstream infections (BSI) due to Staphylococcus aureus. This survey involves 39 health care institutions (HCIs) encompassing 6,888 short-stay beds and was performed during two 3-month periods during 2007 and 2008. The study periods of this survey identified 292 S. aureus isolates causing BSI. Extensive molecular characterization, including genotyping as well as toxin, agr, and staphylococcal cassette chromosome content determinations, allowed us to describe epidemiological evolution in comparison to that discussed in our previous study. Our main epidemiological observation shows that the incidence of BSI remained constant but that methicillin (meticillin)-resistant S. aureus strains with a wider variety of genetic backgrounds now harbor pyl, as has already been reported in different European countries. We noticed stable numbers of BSI episodes involving community-acquired methicillin-sensitive S. aureus (MSSA), whereas a drastic increase in the number of strains harboring the tst gene was recorded. The increase in the number of tst gene-harboring strains is related to known hospital-acquired MSSA isolates and appears related to epidemic episodes in specific HCIs. Monitoring the increase in prevalence of specific strains helps us understand where the standard precautions are not satisfactorily applied or do not efficiently prevent the spread of epidemic MSSA strains in these HCIs. The recent increases in incidence of these strains call for particular vigilance to avoid the spread of potentially virulent MSSA strains harboring the tst gene and for continuance of this strategy of BSI surveillance.
Reporting of methicillin-resistant Staphylococcus aureus (MRSA) bloodstream infections (BSI) is often mandatory, and reduction of BSI rates is a performance target (2, 15, 16, 19, 22, 23). Since 2000, a prospective, longitudinal survey of BSI has been under way at a regional level in France. Results obtained during the first 7 years of surveillance have been reported (26, 27). After sustained and decreasing incidence rates of S. aureus BSI attributed to successful infection control efforts in participating health care institutions (HCIs), we reported in 2006 a sudden increase in incidence that involved two populations of S. aureus strains: one of methicillin-sensitive S. aureus (MSSA) strains and one of MRSA isolates.
First, an increasing incidence of BSI was observed due to MSSA strains, including (i) strains associated with epidemic phenomena in HCIs and (ii) a genetically homogeneous population of tst-positive MSSA isolates, mostly associated with community-acquired (CA) BSI, suggesting clonal spread at a regional level.
Second, we observed the emergence of BSI associated with genetically diverse nonmultiresistant staphylococcal cassette chromosome mec type IV (SCCmec IV) MRSA strains (named NORSA) that could not be related to any local outbreak in the participating HCIs.
We report here data collected in 2007 and 2008 using exactly the same study design. Again, a large S. aureus population genetic study, including MSSA and MRSA isolates in parallel, was conducted. Strain characterization included antimicrobial susceptibility profiles; luk, tst, eta, and etb gene detection; and SCCmec and pulsed-field gel electrophoresis (PFGE) typing. In addition, and to better understand how populations evolved and interacted, characterization of the S. aureus strains was completed by accessory global regulator (agr) typing and multiple-locus variable-number tandem-repeat analysis (MLVA) and multilocus sequence typing (MLST) analysis.
MATERIALS AND METHODS
BSI epidemiological survey method.
A BSI surveillance program and a microbiological study of S. aureus strains isolated from BSI cases have been conducted since 2000 in the central region of France (2.5 million inhabitants) (Fig. 1) (26, 27). In 2007 and 2008, 39 HCIs, comprising a total of 6,888 short-stay beds, participated in this program, which involves an annual 3-month survey of all cases of BSI. The survey covered 1,518,475 patient days (PD). The methods, study design, and data for the years 2000 to 2006 have been reported previously (26, 27). Briefly, BSI is defined as a positive (not contaminated) blood culture from a patient with clinical or laboratory evidence of infection. A blood culture was considered to be contaminated when only one blood culture was positive for a bacterium that is frequently found as a contaminant (i.e., coagulase-negative staphylococci, Corynebacterium spp., or Propionibacterium spp.) and when the clinician did not prescribe any antibiotic treatment against the bacterium. The variables studied included patient age and sex, portal of entry (skin [primitive cutaneous form or superinfection of a skin breach], surgical site, lungs, urine, intravascular device, or digestion), community-associated or hospital-acquired (HA) BSI, death within 7 days of BSI diagnosis, and duration of hospital stay. The data were analyzed with Epi Info version 6 software. Ethical approval of the BSI surveillance program was obtained at the national level from the Réseau Alerte Investigation Surveillance des Infections Nosocomiales (RAISIN).
FIG. 1.
Map of France and the central region.
Microbiological methods. (i) Bacteriology.
BSI-associated S. aureus strains were collected during the survey period. The strains were sent to the laboratory of the Réseau des Hygiénistes du Centre. The isolates were identified as S. aureus on the basis of their characteristic growth pattern, colony morphology, Gram staining result, catalase test result, coagulase production (rabbit plasma; Difco Laboratories, Elancourt, France), slide test result for clumping factor (Pastorex Plus-Staph; Bio-Rad, Ivry-sur-Seine, France), and results obtained with ID 32 Staph cards (VITEK2 system; bioMérieux, Lyon, France).
(ii) Antimicrobial susceptibility testing.
We used the disk diffusion method (Bio-Rad, France) to test the antibiotic susceptibilities of S. aureus strains. According to the French National Committee recommendations (25), the antibiotics tested were penicillin G, cefoxitin, erythromycin, lincomycin, pristinamycin, tetracycline, kanamycin, tobramycin, gentamicin, rifampin (rifampicin), fusidic acid, fosfomycin, pefloxacin, cotrimoxazole, vancomycin, and teicoplanin. The cefinase test (bioMérieux, Lyon, France) was used to detect β-lactamase production. The MRSA-Screen latex agglutination assay was performed in accordance with the manufacturer's protocol (bioMérieux, Lyon, France). Agglutination was assessed visually within 3 min (18). The mecA gene was detected by PCR according to the previously described procedure (27). MRSA strains displaying typical resistance to multiple antibiotics were referred to as CMRSA (classic multiresistant MRSA) strains, whereas MRSA strains resistant to no more than three antimicrobial agents (excluding penicillin and methicillin) were referred to as NORSA.
(iii) Multiplex PCR assay for typing of SCCmec elements.
Multiplex PCR was used to type the SCCmec element in MRSA strains as described by Oliveira and de Lencastre (21).
(iv) VNTR analysis.
All S. aureus strains were analyzed by VNTR analysis according to François et al. (11). Multiplex PCR amplifications with eight primer pairs that target gene regions with variable numbers of tandem repeats were resolved by microcapillary electrophoresis and automatically assessed by cluster analysis.
(v) DNA macrorestriction and PFGE.
PFGE was used as a typing technique according to the previously described procedure (27). PFGE patterns (pulsotypes) were analyzed with the Taxotron package (Taxolab, Institut Pasteur, Paris, France). The images were transferred to a microcomputer, and the distances migrated by each band in each lane were determined in pixel units by RestrictoScan. The molecular size of each fragment was calculated from the distance migrated using cubic Schaffer and Sederoff method algorithms with RestrictoTyper. The relationships between pulsotypes were determined by the unweighted-pair group method using arithmetic means and the Adanson pulsogrouping program (dissimilarity). A dendrogram was drawn with Dendrograf.
(vi) agr typing.
The agr gene is a crucial regulatory component of Staphylococcus aureus involved in the control of bacterial virulence factor expression. The agr types 1 to 4 were determined for S. aureus strains using a real-time multiplex quantitative PCR assay (13).
(vii) MLST.
MLST is a highly discriminatory method of characterizing bacterial isolates on the basis of the sequences of ∼450-bp internal fragments of seven housekeeping genes. For each gene fragment, the different sequences are assigned as distinct alleles, and each isolate is defined by the alleles at each of the seven housekeeping loci (the allelic profile or sequence type [ST]). S. aureus strains (n = 120) representative of the genetic diversity of the panel of studied strains were analyzed by MLST according to the procedure described by Enright et al. (9).
(viii) Virulence factors.
Given the diffusion of Panton-Valentine leukocidin- and toxic shock syndrome toxin-1-producing MRSA strains in France since 2002, we sought sequences corresponding to lukS-PV and lukF-PV, encoding Panton-Valentine leukocidin, to tst, encoding toxic shock syndrome toxin-1, and to the eta and etb genes, encoding exfoliatins A and B, respectively. Genomic DNA was extracted from staphylococcal cultures and used as the template for PCR amplification using a procedure previously described by Jarraud et al. (16).
(ix) Statistical data.
Chi-square tests and Fisher's exact test (two-tailed) were used to test associations, and a P value of 0.050 was considered significant.
RESULTS
S. aureus BSI incidence.
During the study period, 292 cases of S. aureus BSI were identified in 188 males and 104 females (median age, 68 years). The mean incidence of S. aureus BSI was 0.192 per 1,000 PD (Table 1).
TABLE 1.
BSI incidence per 1,000 PD according to the annual 3-month survey, with epidemiological and strain characteristics
| Yr | No. of PD | No. of patient admissions | BSI incidence per 1,000 PD according to the annual 3-month surveya
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus | MRSA | CMRSA | HA CMRSA | CA CMRSA | NORSA | HA NORSA | CA NORSA | MSSA | EMSSA | HA MSSA | CA MSSA | tst gene + MRSA | tst gene + MSSA | luk gene + MRSA | luk gene + MSSA | eta gene + MSSA | |||
| 2002 | 604,850 | 89,959 | 0.202 | 0.055 | 0.046 | 0.008 | 0.147 | 0.015 | NK | NK | NK | ||||||||
| 2003 | 662,555 | 86,530 | 0.178 | 0.047 | 0.047 | 0.006 | 0.125 | 0.023 | NK | NK | NK | ||||||||
| 2004 | 729,013 | 81,867 | 0.167 | 0.059 | 0.045 | 0.014 | 0.108 | 0.008 | 0.012 | NK | |||||||||
| 2005 | 607,300 | 71,579 | 0.13 | 0.038 | 0.03 | 0.023 | 0.006 | 0.008 | 0.007 | 0.001 | 0.092 | 0.005 | 0.048 | 0.042 | 0.008 | NK | |||
| 2006 | 671,368 | 80,499 | 0.234 | 0.068 | 0.04 | 0.03 | 0.009 | 0.028 | 0.022 | 0.005 | 0.165 | 0.016 | 0.068 | 0.078 | 0.004 | 0.030 | 0.001 | NK | |
| 2007 | 759,214 | 86,938 | 0.175 | 0.053 | 0.038 | 0.024 | 0.008 | 0.014 | 0.013 | 0.001 | 0.122 | 0.01 | 0.074 | 0.037 | 0.001 | 0.018 | 0.001 | 0.001 | |
| 2008 | 759,261 | 86,876 | 0.209 | 0.058 | 0.029 | 0.018 | 0.008 | 0.029 | 0.018 | 0.008 | 0.151 | 0.01 | 0.086 | 0.056 | 0.005 | 0.024 | 0.001* | 0.001 | 0.001 |
All S. aureus strains were submitted to a cefoxitin disk diffusion test and MRSA-Screen latex agglutination assay. According to the French National Committee recommendations (25), an S. aureus strain was defined as a MRSA strain when the cefoxitin disk diffusion test was positive (diameter < 25 mm) or when the MRSA-Screen latex agglutination assay was positive. Also, strains negative by the cefoxitin disk diffusion test and MRSA-Screen latex agglutination assay but resistant to one antibiotic were furthermore studied by PCR for mecA gene detection. Among these 292 S. aureus strains, 208 (71%) were MSSA and 84 (29%) were MRSA strains (Table 2). The mean incidence of MSSA BSI was 0.137 per 1,000 PD. Thirteen of the 208 MSSA strains (6.3%) displayed fluoroquinolone (FQ) resistance. The mean incidence of MRSA BSI was 0.055 per 1,000 PD. The mean incidence of NORSA BSI was 0.022 per 1,000 PD (Table 1). Twenty-five of the 33 NORSA strains (76%) were resistant only to FQs and methicillin (designated NORSA FQ strains) (Table 2).
TABLE 2.
Distribution of S. aureus strains associated with BSI according to the annual 3-month survey and the antibiogroup
| Antibiogroup | No. (%) of strains, according to annual 3-month survey period, ina:
|
||||||
|---|---|---|---|---|---|---|---|
| 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | |
| MRSA | 33 (27) | 35 (30) | 43 (35) | 23 (29) | 46 (29) | 40 (30) | 44 (28) |
| CMRSA | 28 (85) | 31 (89) | 33 (77) | 18 (78) | 27 (59) | 29 (72) | 22 (50) |
| NORSA | 5 (15) | 4 (11) | 10 (23) | 5 (22) | 19 (41) | 11 (28) | 22 (50) |
| NORSA FQb | 1 | 4 | 7 | 4 | 16 | 9 | 16 |
| Other NORSA | 4 | 3 | 1 | 3 | 2 | 6 | |
| FQ resistance | 31 (94) | 35 (100) | 43 (100) | 23 (100) | 46 (100) | 38 (97) | 38 (89) |
| MSSA | 89 (73) | 83 (70) | 79 (65) | 56 (71) | 111 (71) | 93 (70) | 115 (72) |
| EMSSAc | 9 | 15 | 6 | 3 | 11 | 8 | 8 |
| EMSSA FQd | 4 | 7 | 6 | ||||
| FQ resistance | 6 (7) | 14 (17) | 8 (8) | 4 (7) | 9 (8) | 7 (8) | 5 (4) |
| Total | 122 | 118 | 122 | 79 | 157 | 133 | 159 |
Results obtained before 2007 were described previously (26, 27) and are reported to show the changes through time affecting the different groups of strains. Percentages indicate the proportions of S. aureus strains that are of the indicated antibiogroup.
NORSA strains resistant only to FQs.
EMSSA, MSSA strains with resistance to FQs or at least two antibiotics.
EMSSA FQ, MSSA strains with resistance to FQs only.
CA and HA S. aureus BSI.
According to the study design, BSI cases were defined as either CA or HA. The mean incidences of CA BSI did not vary significantly according to the year of survey and whether the strains were MSSA or MRSA (Table 1). For HA BSI, the mean incidences varied distinctly according to the strains, as incidence decreased with MRSA and increased with MSSA.
SCCmec typing.
Sixty-four of the 78 MRSA strains were of SCCmec type IV (82%). Four strains were of SCCmec type I, and no SCCmec fragment could be amplified by PCR from the remaining 10 strains (13%), which were therefore designated nontypeable.
luk, tst, eta, and etb genes.
Thirty-seven BSI cases, identified in 24 males and 13 females (median age, 70 years), were associated with strains scoring positive for the tst gene. The strains that scored positive for the tst gene included 32 MSSA strains (32/200 [16%]) and five MRSA strains (5/78 [6.4%]). One MSSA strain and two MRSA strains, the first ever since 2000, scored positive for luk genes. Two MSSA strains were positive for the eta gene.
VNTR typing.
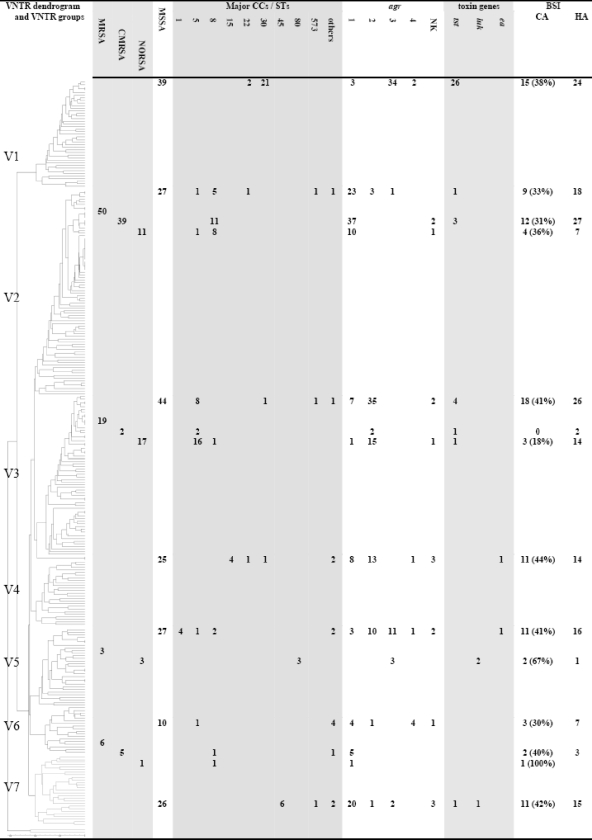
VNRT typing was conducted with 275 of the 292 S. aureus BSI strains. VNTR typing revealed seven major divisions (designated V1 to V7) (Fig. 2). Divisions V1, V4, and V7 were composed exclusively of MSSA strains, and MSSA strains were predominant in the divisions V3, V5, and V6. MRSA strains were predominant only in division V2. The CMRSA strains predominated in divisions V2 (39 CMRSA and 11 NORSA strains) and V6 (5 CMRSA strains and 1 NORSA strain). In contrast, NORSA strains were predominant in divisions V3 (17 NORSA and 2 CMRSA strains) and V5, where the 3 MRSA strains were all NORSA strains. The NORSA FQ strains distributed among divisions V2, V3, and V6. The two luk-positive MRSA strains were observed in V5, whereas the luk-positive MSSA strains clustered in V7. Division V1 contained the most tst-positive strains (26/32 [81%]). The five tst-positive MRSA strains were in V2 (n = 3) and V3 (n = 2).
FIG. 2.
Characteristics of strains and BSI according to VNTR groups. The dendrogram was calculated using the MLVA data. On the basis of their MLVA fingerprints, the isolates segregated into different clusters, showing a distance limit of 0.1 for clonality and a distance limit of 0.2 for related isolates, as previously described (10). Results of other molecular tests, including agr and SCCmec typing as well as determination of toxin content, are also indicated. Based on previous determinations, the origins of isolates composing each cluster (either HA or CA strains) appear in the figure. NK, not known.
SmaI macrorestriction.
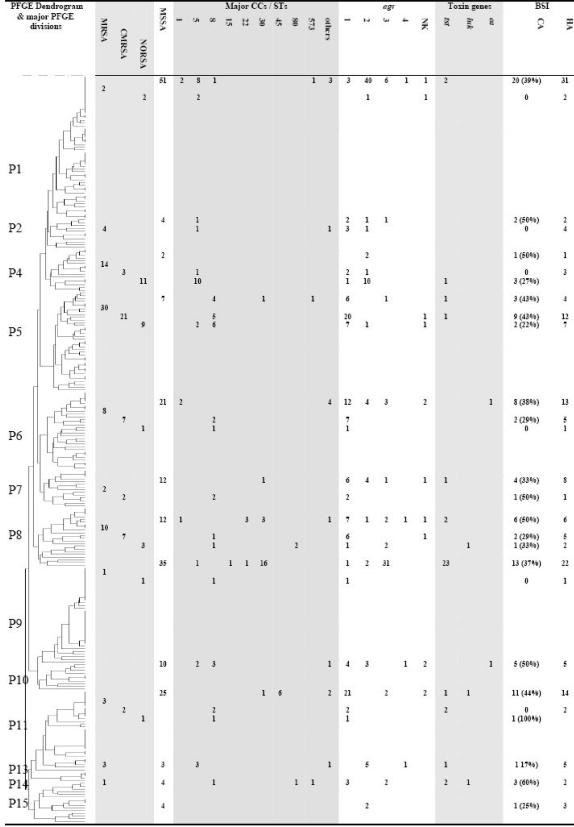
PFGE has been conducted with 267 of the 292 S. aureus BSI strains. PFGE revealed 184 different pulsotypes across the studied strains. After performance of an analysis by the unweighted-pair group method using average linkages, the strains were distributed into 15 major divisions (P1 to P15) (Fig. 3). Divisions P3, P10, P12, and P15 were exclusively composed of MSSA strains, and MSSA strains were also predominant in divisions P1, P6, P7, P9, P11, and P14. MRSA strains were distributed across 11 divisions. CMRSA strains were predominant in divisions P5 and P6; in contrast, NORSA strains were predominant in P4. The two luk-positive MRSA strains were in P8, and the luk-positive MSSA strain was in P11. Division P9 contained most of the tst-positive strains (23/32 [72%]), and the five tst-positive MRSA strains were in P4 (n = 1), P5 (n = 1), P11 (n = 2), and P13 (n = 1). A significant link was observed between VNTR divisions and the major PFGE divisions (Table 3): V1 was associated with P9 (P < 0.001), V2 with P5 (P < 0.001), V3 with P1 (P < 0.001), V5 with P1 (P = 0.002), and V7 with P11 (P < 0.001). To evaluate the likelihood of epidemic phenomena, we studied the occurrence, in the same HCI, of at least two BSI cases with strains presenting similar pulsotypes. Here, 80 of the 200 MSSA strains (40%) and 14 of the 78 MRSA strains (18%) (P < 0.001) were involved in such clusters of strains.
FIG. 3.
Characteristics of strains and BSI according to PFGE groups. The dendrogram was calculated using the relationships between pulsotypes determined by the unweighted-pair group method using arithmetic means and the Adanson pulsogrouping program (dissimilarity). Results of other molecular tests, including agr and SCCmec typing as well as determination of toxin content, are also indicated. Based on previous determinations, the origins of isolates composing each cluster (either HA or CA strains) appear in the figure. NK, not known.
TABLE 3.
Distribution of strains among major PFGE divisions according to the VNTR groups
| VNTR group | No. of strains in indicated PFGE division
|
Total | |||||
|---|---|---|---|---|---|---|---|
| P1 | P5 | P6 | P9 | P11 | Other PFGE divisions | ||
| 1 | 1 | 30 | 1 | 7 | 39 | ||
| 2 | 3 | 31 | 14 | 2 | 3 | 24 | 77 |
| 3 | 33 | 1 | 2 | 1 | 1 | 25 | 63 |
| 4 | 2 | 4 | 1 | 3 | 15 | 25 | |
| 5 | 12 | 1 | 5 | 1 | 1 | 10 | 30 |
| 6 | 1 | 3 | 2 | 9 | 15 | ||
| 7 | 2 | 1 | 20 | 2 | 25 | ||
| Singletons | 1 | 4 | 5 | ||||
| Total | 53 | 37 | 29 | 35 | 29 | 96 | 279 |
MLST.
MLST was performed with 120 strains representative of the genetic diversity of the studied strains according to PFGE results and included 44 MRSA and 76 MSSA strains (Fig. 2 and 3). The 44 MRSA strains were distributed into four clonal complexes (CCs) or STs: 18 strains were of CC5, 22 of CC8, 3 of CC80, and 1 of ST247, the gentamicin-resistant Iberian clone that was the most frequent clone in France in the 1990s. Among the 29 NORSA strains studied, 16 were of CC5, 10 were of CC8, and 3 were of CC80. The five tst-positive MRSA strains were distributed into CC8 (n = 3) and CC5 (n = 2). Both of the two luk-positive MRSA strains were found in CC80.
The 76 MSSA strains studied were distributed into 17 CCs or STs, and at least 4 MSSA strains were clustered in 7 CCs or STs: 23 strains were clustered in CC30, 11 in CC5, 8 in CC8, 6 in CC45, 5 in CC1, 4 in CC22, and 4 in ST15. The 25 tst-positive MSSA strains were distributed among five CCs or STs, but most (20/25 [80%]) were observed in CC30. The luk-positive MSSA strain was found in ST45. The two eta-positive MSSA strains were in ST1 and ST6.
Again, a significant link was observed between the major VNTR divisions and the major CCs and STs (Fig. 1): V1 was associated with CC30 (P < 0.001), V2 with CC8 (P < 0.001), V3 with CC5 (P < 0.001), V7 with CC45 (P < 0.001), and V5 with CC80 (P < 0.001).
Determination of the agr groups.
The agr groups was determined for 266 strains (Fig. 2 and 3). The MSSA strains were rather equally distributed among agr group 1 (68/189 [36%]), agr group 2 (63/189 [33%]), and agr group 3 (49/189 [26%]). In contrast, the MRSA strains were mostly of agr group 1 (56/77 [73%]), while agr groups 2 and 3 were found, respectively, in 17 (22%) and 3 (3.9%) MRSA strains. The six agr group 4 strains were all MSSA strains. The distributions of strains across the various agr groups differed significantly between MSSA and MRSA strains (P = 0.010). agr genes of group 3 were mostly recovered among tst gene-positive MSSA isolates (25/31 [81%]). In contrast, tst-positive MRSA strains were of agr groups 1 (n = 3) and 2 (n = 2). The two luk-positive MRSA strains were of agr group 3, and the luk-positive MSSA strain was placed into agr groups 1 and 3. A significant link was found between the major VNTR divisions and the various agr types (Table 3): type V1 was associated with agr group 3 (P < 0.001), V2 with agr group 1 (P < 0.001), V3 with agr group 2 (P < 0.001), and V6 with agr group 4 (P = 0.035).
DISCUSSION
In our reporting of S. aureus BSI since 2000 (26, 27), we observed a regular decrease in incidence, followed in 2006 by a sudden increase. The data obtained during the years 2007 and 2008 according to the same study design (26) confirmed the changes observed in 2006. The surge in S. aureus BSI incidence was first associated with the diffusion of epidemic MSSA strains responsible for outbreaks in participating HCIs (27). The new data indicate (i) a high level of MSSA BSI incidence, (ii) a stable CA-MSSA BSI incidence, (iii) an increase in HA-MSSA BSI incidence, and (iv) the persistence of MSSA epidemic phenomena at the HCI level. This indicates that the standard precautions may not be satisfactorily applied or that they are not sufficient to prevent the diffusion of the MSSA strains that are spreading today. These data must prompt us to have particular vigilance against the spread of these strains and especially against the tst-positive MSSA strains. Second, the surge of the S. aureus BSI incidence was also linked to NORSA strains. The new data indicate (i) a stable level of MRSA BSI incidence, (ii) stable CA- and HA-MRSA BSI incidences, (iii) a progressive replacement of the CMRSA by NORSA strains, (iv) the preponderance of MRSA strains resistant only to FQs, and (v) limited MRSA epidemic phenomena. Despite infection control efforts conducted in the HCIs to prevent transmission of MRSA strains, the MRSA BSI incidence did not decrease significantly. This suggests that measures to prevent the diffusion of MRSA strains either are not satisfactorily applied or are not efficient at preventing the diffusion of these strains. As most MRSA strains harbor type IV SCCmec, the small SCCmec element that may enable more-efficient horizontal gene transfer and permit potentially higher growth rates (8), we cannot exclude the possibility that these factors may be related to the spread of such strains and thus concur with the limited impact of the measures conducted.
MLST allows us to better characterize S. aureus strains spreading in our region. The MSSA and MRSA clones belonging to CC5, CC8, CC22, CC30, and CC45 are pandemic lineages (3, 4, 9, 12, 17, 19, 20, 24). MSSA strains were highly diverse and rather equally distributed across such lineages. In contrast, MRSA isolates were much more homogenous and mostly distributed with the pandemic Lyon clone (ST8-IV-agr1) and the pediatric clone (ST5-IV-agr2) (6, 7).
Nevertheless, while CMRSA strains were homogenous and mostly of the CC8 background, NORSA strains revealed a more diverse group of backgrounds; they distributed with CC8 and CC5 strains and with the European clone ST80-IV-agrIII (V5) (5). No isolate was characterized as one of the emerging clones observed in neighboring European countries, such as ST22-IV, ST45-IV, ST125-IV, and ST225-I (1, 10, 14, 20).
The first two luk-positive MRSA strains identified since 2000 differed according to their antimicrobial susceptibility profiles and PFGE patterns but belonged to the European or the Mediterranean clone CC80-IV-agr3 (5). This suggests that the current emergence of luk-positive strains in France is not associated with an epidemic spread such as that reported in Greece (28). In contrast, three of the five tst-positive MRSA strains identified in 2007 to 2008 were all ST8-IV-agr1 and shared similar PFGE patterns, suggesting the installation of these strains at a regional level.
Using a highly diverse set of MSSA and MRSA strains, VNTR analysis provided highly congruent results with data obtained by MLST, as all major pandemic lineages were clustered into distinct VNTR groups. These results support the use of these methods to easily genetically characterize MSSA and MRSA strains. VNTR data confirmed (i) the genetic diversity of NORSA strains, (ii) the homogenous population of tst-positive MSSA strains, and (iii) the homogenous population of luk-positive MRSA strains.
Conclusions.
Data reported here encourage us to continue the implementation of active control measures with all S. aureus BSI cases and raise questions about the efficiency of the measures currently applied in our HCIs facing the clonal spread of tst-harboring S. aureus strains. We confirm the trends observed in 2006 and emphasize the need to reinforce the prevention of transmission of bacteria, especially that of MSSA strains. Our findings demonstrate that our current infection control practices, focusing on the prevention of infection spreading from patients with symptomatic disease, mainly restricted to the hospital setting, may not be sufficient.
Acknowledgments
This work was supported by the Centre de Coordination de la Lutte contre les Infections Nosocomiales de l'Ouest de la France (CCLIN Ouest), the Agence Régionale de l'Hospitalization du Centre, and the Centre Universitaire de Tours, France. This study was also supported by grants 3100A0-112370/1 (awarded to J.S.) and 3100A0-116075 (awarded to P.F.) from the Swiss National Science Foundation; grants CI 70889, CI 70903, and CI 70897 from the University of Geneva Hospitals Quality Improvement Research Program; and the European Commission's 6th Framework Program (MagRSA project no. 37957).
We thank Myriam Girard for outstanding technical assistance.
The members of the Bloodstream Infection Study Group of the Relais des Hygiénistes du Centre (in France) are P. Amirault, Centre Hospitalier Vierzon; M. Archambault, Centre Hospitalier Pithiviers; J.-P. Arnould, Centre Hospitalier Chateaudun; M.-N. Bachelier, Centre Hospitalier Bourges; V. Baye, Clinique St. François Mainvilliers; Z. Benseddik, Centre Hospitalier Chartres; M. Blanchard, Clinique J. d'Arc St. Benoit la Foret; I. Blasi, Polyclinique des Longues Allées St. Jean de Braye; D. Bloc, Centre Hospitalier Régional Universitaire Tours; N. Boursier, Centre Hospitalier Nogent le Rotrou; M. Cahiez, Laboratoire Lescaroux-Camenen-Jamet Chateauroux; B. Cattier, Centre Hospitalier Inter-Communal Amboise; M. Chabaud-Mayer, Centre Hospitalier Chartres; C. Chandesris, Centre Hospitalier Amilly Montargis; V. Chevereau, Polyclinique de Blois la Chaussee St. Victor; V. Chieux, Centre Hospitalier Chartres; G. Courouble, Laboratoire Lescaroux-Camenen-Jamet Chateauroux, Centre Hospitalier Blois; M.-C. Courtin, Centre Hospitalier Inter-Communal Amboise; G. Delaporte, Centre Hospitalier Gien; C. Denis, Centre Hospitalier Loches; F. Deperrois, Centre Hospitalier Chinon; F. Dieu, Clinique St. François Mainvilliers; M.-C. Farcy, Polyclinique des Longues Allées St. Jean de Braye; C. Fievre, Centre Hospitalier le Blanc; P. Foloppe, Laboratoire Foloppe-Leclercq Loches; F. Fongauffier, Centre Hospitalier Chateaudun; K. Fontaine, Clinique de la Présentation Fleury les Aubrais; R. Fournier-Hoock, Centre Hospitalier Amilly Montargis; N. Girard, Centre Hospitalier Régional Universitaire Tours; T. Gourdet, Laboratoire Lamotte-Gourdet la Chaussee St. Victor; J.-L. Graveron, Clinique de la Présentation Fleury les Aubrais; F. Grobost, Laboratoire Selarl C+Bio la Ferte Bernard; M.-F. Guillon, Clinique St. François Chateauroux; F. Guinard, Laboratoire Nuret Guinard Bourges; S. Guittet, Pôle Santé Léonard da Vinci Chambray; P. Harriau, Laboratoire Harriau-Lardy-Ruffel St. Amand Montrond; C. Hombrouck-Alet, Centre Hospitalier Blois, Centre Hospitalier Vendome, and Centre Hospitalier Romorantin; S. Huault, Clinique Velpeau Tours; D. Imbault, Laboratoire Salaun-Imbault Vendome; M. Jamain, Clinique de l'Alliance St. Cyr/Loire; D. Jehanno, Clinique de la Présentation Fleury les Aubrais; P. Laudat, Laboratoire Arnaud Tours; O. Lehiani, Centre Hospitalier Vierzon, Centre Hospitalier Bourges, and Centre Hospitalier St. Amand Montrond; A. Lepineux da Rocha, Clinique Chirurgicale les Grainetières St. Amand Montrond; A.-L. Lesimple, Laboratoire Vendome; V. Morange, Centre Hospitalier Régional Universitaire Tours; E. Morel-Desjardins, Clinique Chirurgicale Marie Immaculée Bourges, Maison le Blaudy St. Doulchard; E. Morin, Clinique de la Reine Blanche Orleans; C. Naudion, Laboratoire Romorantin; C. Neveu, Centre Hospitalier Dreux; O. Paba, Clinique St. Cœur Vendome; F. Perigois, Laboratoire le Blanc; G. Petit le Gouas, Centre Hospitalier Nogent le Rotrou; J.-M. Pone, Centre Hospitalier Pithiviers; M. Prevost-Oussar, Centre Hospitalier Pithiviers; R. Quentin, CHU de Tours; C. Querrien, Centre Hospitalier Vendome; D. Ratovohery, Centre Hospitalier Chateauroux; C. Renaud, Clinique de la Reine Blanche Orleans; B. Rousseau, Centre Hospitalier Gien; A. Roussin, Clinique de la Reine Blanche Orleans; V. Salaün, Clinique St. Gatien Tours; H. Schill, Clinique de Montargis Montargis; N. Schlienger, Crf les Ormes Ouzouer des Champs; A. Secher, Centre Hospitalier Dreux; J.-F. Theron le Gargasson, Laboratoire Cazala Chateauroux; N. van der Mee, Centre Hospitalier Régional Universitaire Tours; R. Vergez-Pascal, Laboratoire de Chartres; and S. Watt, Laboratoire Jacquet Chinon.
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.Aires-de-Sousa, M., B. Correia, H. de Lencastre, and the Multilaboratory Project Collaborators. 2008. Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J. Clin. Microbiol. 462912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comité Technique des Infections Nosocomiales. 1999. 100 recommandations pour la surveillance et la prévention des infections nosocomiales, 2nd ed. Ministère de l'Emploi et de la Solidarité, Secrétariat d'Etat à la Santé et à l'Action Sociale, Paris, France.
- 3.Crisóstomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 989865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crum, N. F., R. U. Lee, S. A. Thornton, O. C. Stine, M. R. Wallace, C. Barrozo, A. Keefer-Norris, S. Judd, and K. L. Russell. 2006. Fifteen-year study of the changing epidemiology of methicillin-resistant Staphylococcus aureus. An. J. Med. 119943-951. [DOI] [PubMed] [Google Scholar]
- 5.Dauwalder, O., G. Lina, G. Durand, M. Bes, H. Meugnier, V. Jarlier, B. Coignard, F. Vandenesch, J. Etienne, and F. Laurent. 2008. Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J. Clin. Microbiol. 463454-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Miranda, O. P., M. C. Silva-Carvalho, A. Ribeiro, F. Portela, R. P. Cordeiro, N. Caetano, C. F. Vidal, and A. M. Figueiredo. 2007. Emergence in Brazil of methicillin-resistant Staphylococcus aureus isolates carrying SCCmecIV that are related genetically to the USA800 clone. Clin. Microbiol. Infect. 131165-1172. [DOI] [PubMed] [Google Scholar]
- 7.Durand, G., M. Bes, H. Meugnier, M. C. Enright, F. Forey, N. Liassine, A. Wenger, K. Kikuchi, G. Lina, F. Vandenesch, and J. Etienne. 2006. Detection of new methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital- and community-acquired infections in France. J. Clin. Microbiol. 44847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ender, M., N. McCallum, R. Adhikari, and B. Berger-Bächi. 2004. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob. Agents Chemother. 482295-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fossum, A. E., and G. Bukholm. 2006. Increased incidence of methicillin-resistant Staphylococcus aureus ST80, novel ST125 and SCCmecIV in the south-eastern part of Norway during a 12-year period. Clin. Microbiol. Infect. 12627-633. [DOI] [PubMed] [Google Scholar]
- 11.François, P., A. Huyghe, Y. Charbonnier, M. Bento, S. Herzig, I. Topolski, B. Fleury, D. Lew, P. Vaudaux, S. Harbarth, W. van Leeuwen, A. van Belkum, D. S. Blanc, D. Pittet, and J. Schrenzel. 2005. Use of an automated multiple-locus, variable-number tandem repeat-based method for rapid and high-throughput genotyping of Staphylococcus aureus isolates. J. Clin. Microbiol. 433346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.François, P., S. Harbarth, A. Huyghe, G. Renzi, M. Bento, A. Gervaix, D. Pittet, and J. Schrenzel. 2008. Methicillin-resistant Staphylococcus aureus, Geneva, Switzerland, 1993-2005. Emerg. Infect. Dis. 14304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.François, P., T. Koessler, A. Huyghe, S. Harbarth, M. Bento, D. Lew, J. Etienne, D. Pittet, and J. Schrenzel. 2006. Rapid Staphylococcus aureus agr type determination by a novel multiplex real-time quantitative PCR assay. J. Clin. Microbiol. 441892-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghebremedhin, B., W. König, and B. König. 2005. Heterogeneity of methicillin-resistant Staphylococcus aureus strains at a German university hospital during a 1-year period. Eur. J. Clin. Microbiol. Infect. Dis. 24388-398. [DOI] [PubMed] [Google Scholar]
- 15.Haley, R. W., D. H. Culver, J. W. White, W. M. Morgan, T. G. Emori, and V. P. Munn. 1985. The efficacy of infection surveillance and control programs in preventing nosocomial infections in US hospitals. Am. J. Epidemiol. 121182-205. [DOI] [PubMed] [Google Scholar]
- 16.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins, T. C., B. D. McCollister, R. Sharma, K. K. McFann, N. E. Madinger, M. Barron, M. Bessesen, C. S. Price, and W. J. Burman. 2009. Epidemiology of healthcare-associated bloodstream infection caused by USA300 strains of methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Infect. Control Hosp. Epidemiol. 30233-241. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J. H., J. M. Jeong, Y. H. Park, S. S. Choi, Y. H. Kim, J. S. Chae, J. S. Moon, H. Park, S. Kim, and S. K. Eo. 2004. Evaluation of the methicillin-resistant Staphylococcus aureus (MRSA)-Screen latex agglutination test for detection of MRSA of animal origin. J. Clin. Microbiol. 422780-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nulens, E., E. E. Stobberingh, H. van Dessel, S. Sebastian, F. H. van Tiel, P. S. Beisser, and R. H. Deurenberg. 2008. Molecular characterization of Staphylococcus aureus bloodstream isolates collected in a Dutch university hospital between 1999 and 2006. J. Clin. Microbiol. 462438-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nulens, E., E. E. Stobberingh, E. Smeets, H. van Dessel, M. A. Welling, S. Sebastian, F. H. van Tiel, P. S. Beisser, and R. H. Deurenberg. 2009. Genetic diversity of methicillin-resistant Staphylococcus aureus in a tertiary hospital in The Netherlands between 2002 and 2006. Eur. J. Clin. Microbiol. Infect. Dis. 28631-639. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 462155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul, J. 2006. Surveillance and management of all types of Staphylococcus aureus bacteraemia. BMJ 333269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popovich, K. J., R. A. Weinstein, and B. Hota. 2008. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains. Clin. Infect. Dis. 46787-794. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Bano, J., M. Angeles Dominguez, A. Blas Millan, C. Borraz, M. Pau Gonzales, B. Almirante, E. Cercenado, B. Padilla, and M. Pujol on behalf of GEIH/GEMARA (SEIMC), REIPI. 15 May 2009. Clinical and molecular epidemiology of community-acquired, healthcare-associated and nosocomial methicillin-resistant Staphylococcus aureus in Spain. Clin. Microbiol. Infect. [Epub ahead of print.] doi: 10.1111/j.1469-0691.2009.02717.x. [DOI] [PubMed]
- 25.Société Française de Microbiologie. 2009. Recommandations du Comité de l'Antibiogramme de la Société Française de Microbiologie. Société Française de Microbiologie, Paris, France. www.sfm.asso.fr.
- 26.van der Mee-Marquet, N., A. S. Domelier, N. Girard, R. Quentin, and the Bloodstream Infection Study Group of the Relais d'Hygiene du Centre. 2004. Epidemiology and typing of Staphylococcus aureus strains isolated from bloodstream infections. J. Clin. Microbiol. 425650-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Mee-Marquet, N., C. Epinette, J. Loyau, L. Arnault, A. S. Domelier, B. Losfelt, N. Girard, R. Quentin, and the Bloodstream Infection Study Group of the Relais d'Hygiene du Centre. 2007. Staphylococcus aureus strains isolated from bloodstream infections changed significantly in 2006. J. Clin. Microbiol. 45851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vourli, S., H. Vagiakou, G. Ganteris, M. Orfanidou, M. Polemis, A. Vatopoulos, and H. Malamou-Ladas. 15 January 2009. High rates of community-acquired, Panton-Valentine leukocidin (PVL)-positive methicillin-resistant S. aureus (MRSA) infections in adult outpatients in Greece. Euro Surveill. vol. 14, issue 2, article 5. [DOI] [PubMed]