Abstract
A multilocus microsatellite typing (MLMT) approach based on the analysis of 15 independent loci has been developed for the discrimination of strains belonging to different Viannia species. Thirteen microsatellite loci were isolated de novo from microsatellite-enriched libraries for both Leishmania braziliensis and L. guyanensis. Two previously identified markers, AC01 and AC16, were modified and added to our marker set. Markers were designed to contain simple dinucleotide repeats flanked by the minimal possible number of nucleotides in order to allow variations in repeat numbers to be scored as size variations of the PCR products. The 15 markers in total were amplified for almost all of the strains of Viannia tested; one marker did not amplify from the two L. peruviana strains included in the study. When 30 strains of L. braziliensis, 21 strains of L. guyanensis, and 2 strains of L. peruviana were tested for polymorphisms, all strains except two strains of L. guyanensis had individual MLMT types. Distance-based analysis identified three main clusters. All strains except one strain of L. guyanensis grouped together. Two clusters consisted of strains of L. braziliensis according to their geographical origins. The two strains of L. peruviana grouped together with strains of L. braziliensis from Peru and the adjacent Brazilian state of Acre. MLMT has proven capable of individualizing strains even from the same areas of endemicity and of detecting genetic structures at different levels. MLMT is thus applicable for epidemiological and population genetic studies of strains within the subgenus Viannia.
Cutaneous leishmaniasis (CL) is a serious but neglected public health problem that is endemic in 22 countries of Latin America, with Brazil and Colombia being among the six countries reporting 90% of the worldwide cases. Species of the subgenus Viannia cause the majority of cases of CL and mucocutaneous leishmaniasis (MCL) in South America and often overlap in their distribution (22). The subgenus is subdivided into species complexes representing Leishmania braziliensis, L. guyanensis, and L. naiffi (8). The L. guyanensis complex includes some named species, such as L. panamensis and L. shawi, which were found to be very similar to each other (7). On the other hand, L. peruviana was always considered to be closely related to L. braziliensis (14, 30). L. lainsoni was shown to be very distinct from the other species (8). In addition, numerous interspecies hybrids have been reported, such as L. braziliensis/L. peruviana (26), L. braziliensis/L. guyanensis (10), L. braziliensis/L. panamensis (4), and L. lainsoni/L. naiffi (35).
Many of the Leishmania (Viannia) species are capable of producing a wide spectrum of diseases, with the severity varying from self-limiting CL to severe MCL (9, 15). MCL is principally caused by L. braziliensis, but less than 5% of patients with a primary cutaneous lesion will develop metastatic mucosal involvement during their life course (17, 23, 24). Other species of the subgenus Leishmania (Viannia), such as L. guyanensis, may be associated with MCL (34), but the exact risk and frequency are unknown. Interestingly, L. peruviana, the species that is the closest to L. braziliensis, has not yet been reported to cause MCL.
The transmission of leishmaniasis in South America occurs through sand flies of the genus Lutzomyia, with each Leishmania species usually being transmitted by more than one sand fly species, and the preferred mammalian hosts also vary according to the Leishmania species (15). For some of the Leishmania (Viannia) species, such as L. guyanensis and L. panamensis, the patterns of transmission are well understood, but for other species they are not. Knowledge of the patterns of transmission are necessary, however, to develop disease control measures and surveillance activities.
Markers that are able to discriminate Leishmania (Viannia) organisms to the species and strain levels are needed to resolve the diversity and structure of Leishmania (Viannia) populations. Multilocus microsatellite typing (MLMT) has proven highly discriminatory in the typing of strains (5) and has been successfully used in population genetic studies of different Old World species of Leishmania (2, 19, 20, 32). This approach could be performed directly with clinical materials without prior cultivation of the parasites. Its results are reproducible, can be stored in databases, and can be exchanged between different laboratories. The first three microsatellite markers for the subgenus Leishmania (Viannia) were published in 1999 (30). Two of them, AC01 and AC52, were polymorphic in L. braziliensis, L. peruviana, L. guyanensis, and L. panamensis; however, marker AC16 was monomorphic for the seven strains of L. guyanensis analyzed. Marker AC52 had a composite repeat and needed to be sequenced because electrophoretic alleles of the same size could consist of different repeat compositions. This marker was also not very reliable when it was used to type strains of L. braziliensis and L. peruviana from Peru (26). Recently, an additional eight microsatellites designed by using genomic libraries of L. braziliensis genome project loci were found to be polymorphic in L. braziliensis (28). However, the variation of these markers in other Leishmania (Viannia) species was not tested.
The study described here aimed to develop an MLMT approach that could be used for epidemiological and population studies of different Leishmania (Viannia) species. For this purpose, 13 microsatellite loci were identified by searching microsatellite-enriched libraries for L. braziliensis and L. guyanensis. Two previously identified markers, AC01 and AC16, were modified and added to our marker set. Variations in repeat numbers should be scorable as size variations of the PCR products. The 15 markers in total were amplified for all Leishmania (Viannia) species tested and tested for polymorphisms in 31 strains of L. braziliensis, 20 strains of L. guyanensis, and 2 strains of L. peruviana.
MATERIALS AND METHODS
Parasite and DNA samples.
The sources and geographical origins of the Leishmania strains used in this study and the clinical manifestations that they caused are listed in Table 1. These included 31 strains of L. braziliensis from different endemic foci in Brazil, Paraguay, and Peru; 20 strains of L. guyanensis from Brazil, Peru, French Guiana, and Suriname; 2 strains of L. peruviana; and single strains each of L. shawi, L. naiffi, L. lainsoni, and L. panamensis. Most of the strains were retrieved from the authors' cryobanks. One strain of L. braziliensis (strain MHOM/BR/75/M2903) and three strains of L. guyanensis (strains MHOM/SR/87/TRUUS1, MHOM/SR/87/TRUUS3, and MHOM/SR/87/TRUUS4) were obtained from the cryobank of the Royal Tropical Institute, Amsterdam, The Netherlands; and an additional five strains of L. guyanensis (strains MHOM/FG/1984/H166, MHOM/FG/1985/H197, MHOM/FG/82/H60, MHOM/FG/87/H298, and MHOM/SR/1987/TRUUS2) were obtained from the cryobank of the Institute of Tropical Medicine, Antwerp, Belgium.
TABLE 1.
Designation and characteristics of Leishmania (Viannia) strains included in the studya
| Species | Country | Region | Clinical picture | ZymodemeZ | WHO code |
|---|---|---|---|---|---|
| L. shawi | Brazil | Pará | CL | Z-26 | MCEB/BR/1984/M8408R |
| L. naiffi | Brazil | Pará | VL | Z-36 | MDAS/BR/1979/M5533R |
| L. lainsoni | Brazil | Pará | CL | Z-15 | MHOM/BR/1981/M6426R |
| L. panamensis | Panama | Canal Zone | CL | Z-21 | MHOM/PA/1971/LS94R |
| L. peruviana | Peru | ND | ND | ND | MHOM/PE/1984/LC39R |
| L. peruviana | Peru | Central | CL | ND | MHOM/PE/2006/LH3667 |
| L. braziliensis | Brazil | Bahia | MCL | ND | MHOM/BR/2000/LTB300y |
| Brazil | Pará | CL | Z-27 | MHOM/BR/1975/M2903R,y | |
| Brazil | Pernambuco | CL | Z-72 | MMES/BR/1999/Sentinela I | |
| Brazil | Pernambuco | CL | Z-105 | MHOM/BR/2000/LMG | |
| Brazil | Pernambuco | CL | Z-78 | MHOM/BR/2001/TSS | |
| Brazil | Bahia | MCL | Z-27 | MHOM/BR/2001/LTCP14183 | |
| Brazil | Bahia | CL | Z-27 | MHOM/BR/2001/LTCP14214 | |
| Brazil | Bahia | CL | Z-27 | MHOM/BR/2001/LTCP14182 | |
| Brazil | Acre | CL | Z-78 | MHOM/BR/2002/NMT-RBO005 | |
| Brazil | Acre | CL | Z-83 | MHOM/BR/2002/NMT-RBO011 | |
| Brazil | Acre | CL | Z-80 | MHOM/BR/2002/NMT-RBO018 | |
| Paraguay | San Pedro | CL | ND | MHOM/PY/1997/L8y | |
| Paraguay | San Pedro | CL | ND | MHOM/PY/1997/L16 | |
| Paraguay | San Pedro | CL | ND | MHOM/PY/2000/SP9 | |
| Paraguay | Concepcion | CL | ND | MHOM/PY/2000/CO2 | |
| Paraguay | Kanindeyú | CL | ND | MHOM/PY/2000/KA2 | |
| Paraguay | Kanindeyú | CL | ND | MHOM/PY/2000/KA11y | |
| Paraguay | Kanindeyú | CL | ND | MHOM/PY/2000/KA-13y | |
| Paraguay | Kanindeyú | CL | ND | MHOM/PY/2000/KA-32 | |
| Paraguay | Caaguazu | CL | ND | MHOM/PY/2000/CA2 | |
| Paraguay | Caaguazu | CL | ND | MHOM/PY/2000/CA4y | |
| Paraguay | Caaguazu | CL | ND | MHOM/PY/2000/CA5y | |
| Paraguay | Amambay | CL | ND | MHOM/PY/2000/AM1y | |
| Paraguay | Presidente Hayes | MCL | ND | MHOM/PY/2000/PH1 | |
| Paraguay | Central | CanL | ND | MCAN/PY/2000/Rocky | |
| Peru | Central | MCL | ND | MHOM/PE/2002/LH2215 | |
| Peru | Central | MCL | ND | MHOM/PE/2003/LH2920 | |
| Peru | South | CL | ND | MHOM/PE/1991/LC2177 | |
| Peru | South | CL | ND | MHOM/PE/2006/CU00176 | |
| Peru | South | MCL | ND | MHOM/PE/2003/LH2946 | |
| Peru | South | MCL | ND | MHOM/PE/2002/LH2330 | |
| L. guyanensis | Suriname | ND | ND | ND | MHOM/SR/1987/TRUUS1y |
| Suriname | ND | ND | ND | MHOM/SR/1987/TRUUS3 | |
| Suriname | ND | ND | ND | MHOM/SR/1987/TRUUS4 | |
| Suriname | ND | ND | ND | MHOM/SR/1987/TRUUS2y | |
| French Guiana | ND | ND | ND | MHOM/FG/1984/H166 | |
| French Guiana | ND | ND | ND | MHOM/FG/1985/H197y | |
| French Guiana | ND | ND | ND | MHOM/FG/1982/H60y | |
| French Guiana | ND | ND | ND | MHOM/FG/1987/H298y | |
| Peru | Central | CL | ND | MHOM/PE/2006/LH3554 | |
| Peru | North | CL | ND | MHOM/PE/2006/LH3635 | |
| Peru | Central | CL | ND | MHOM/PE/2006/LH3640 | |
| Peru | Central | CL | ND | MHOM/PE/2006/LH3673 | |
| Peru | Central | CL | ND | MHOM/PE/2006/LH3668 | |
| Peru | Central | Sand fly | ND | IPRN/PE/1987/LP52 | |
| Brazil | Para | CL | Z-23 | MHOM/BR/1975/M4147R,y | |
| Brazil | Acre | CL | Z-23 | MHOM/BR/2002/NMT-RBO013 | |
| Brazil | Amazonas | CL | Z-23 | MHOM/BR/1997/249-Py | |
| Brazil | Amazonas | CL | Z-23 | MHOM/BR/1997/240-Py | |
| Brazil | Amazonas | CL | Z-23 | MHOM/BR/1997/215-P | |
| Brazil | Amazonas | CL | Z-23 | MHOM/BR/1997/243-P |
R, reference strain of the respective species; Z, zymodeme system (multilocus enzyme electrophoresis) according to Cupolillo et al. (8); ND, not defined; y, strains used to test for the presence of size variations in microsatellite sequences; VL, visceral leishmaniasis.
The parasites were maintained in Novy-Nicolle-McNeal medium and cultured in Schneider's Drosophila medium supplemented with 20% fetal calf serum. Promastigotes were harvested by centrifugation and washed twice in phosphate saline buffer. DNA was extracted by the phenol-chloroform method described previously (25), resuspended in Tris-EDTA buffer (pH 7.4), and stored at 4°C.
Microsatellite-enriched library.
Isolation of the microsatellite loci was based on a modification of the method of Bloor et al. (http://www.genomics.liv.ac.uk/animal/Protocol1.html). First, 60 μg of genomic DNA from L. braziliensis reference strain MHOM/BR/84/LTB300 was digested with HaeIII, RsaI, and AluI (Promega) and ligated to a duplex linker (oligonucleotide A [5′-CTCTTGCTTACGCGTGGACTA-3′] and oligonucleotide B [5′-PO4-TAGTCCACGCGTAAGCAAGAGCACA-3′]). DNA fragments between 300 and 750 bp were excised and extracted from a 1.7% NuSieve GTG agarose gel (FMC Bioproducts). Enrichment for microsatellite-containing fragments was done by using a biotin-labeled (GT)12 capture oligonucleotide bound to M280 streptavidin-coated magnetic beads (Dynal, Norway). After differential stringency washes, the DNA was recovered, amplified by PCR, and ligated into plasmid vector pCR 2.1-TOPO (Invitrogen). Plasmids containing a microsatellite insert were identified by the presence of two or more amplified products after PCR with the linker and the (GT)12 oligonucleotides and subsequently by hybridization with a (GT)12 probe labeled with [32P]ATP by using T4 kinase (Promega). Plasmids positive by both PCR and hybridization were cycle sequenced by electrophoresis on an ABI 377 or CEQ 2000XL Beckman Coulter apparatus. Second, a library enriched for L. guyanensis MHOM/SR/87/TRUUS1 was prepared by a modification of the method described above (27).
Design of microsatellite primers.
PCR primers between 17 bp and 22 bp in length were designed against sequences flanking the microsatellites detected by the use of Primer3 software (29). Primers were deduced from sequences 1 to 40 nucleotides upstream and downstream of the microsatellite repeats. New primers were designed for microsatellites AC01 and AC16 published by Russell et al. (30) in order to avoid long flanking regions. These primers were named AC01R and AC16R, respectively. A BLAST search was conducted for all markers by using the nucleotide sequence information assembled by the Leishmania braziliensis MHOM/BR/1975/M2904 genome project (http://www.sanger.ac.uk/Projects/L_braziliensis) in order to determine the chromosome on which the amplified region was localized.
Analysis of microsatellite variation.
PCR assays with all primer pairs were optimized for the annealing temperature and the concentrations of Mg2+ used, and the DNA of the originally cloned strain was used as the template. Finally, each PCR mixture contained 200 μM of each deoxynucleoside triphosphate, 1 U AmpliTaq DNA polymerase (Applied Biosystems), and 10 mM Tris-HCl buffer (pH 8.3) containing 50 mM KCl and 1.5 mM MgCl2, ∼10 ng template DNA, and 5 pmol of each primer in a volume of 25 μl. All amplification reactions were performed in Robocycler Gradient 40 apparatus (Stratagene, La Jolla, CA). After an initial denaturation step of 5 min at 95°C, the samples were processed through 35 cycles consisting of 30 s at 95°C, 30 s at the annealing temperature indicated in Table 2, and 1 min at 72°C, followed by a terminal elongation step of 6 min at 72°C.
TABLE 2.
Microsatellite markers developed in this studya
| Primer | Primer sequence (5′-3′)
|
Fragment size (bp [reference strain]) | Repeat type of sequenced reference strain | Chromosome M2904S | Position strain M2904S | GenBank accession no. | TA (°C) | |
|---|---|---|---|---|---|---|---|---|
| Forward | Reverse | |||||||
| CSg46 | AAACGTGCAAAGGCACATC | TCTATTACCGCGCTCATGCT | 79 (GUA-02) | (AC)5(GC)(AC)4 (GUA-02) | 18 | 389543-389597b | FM865627 | 54 |
| CSg47 | GTGTTCGTGAAACGTCGAAA | AAAAGGCCGGTTTCAAATTC | 97 (GUA-02) | (TG)13 (GUA-02) | 29 | 537610-537720 | FM865628 | 56 |
| CSg48 | TTGACGTGTACACCGCTCTT | TTCTGAGAAAGGCAACCGATA | 76 (GUA-02) | (TG)7 (GUA-02) | 20 | 1190658-1190730 | FM865629 | 56 |
| CSg53 | CATGTAGGCATGCGGTTGTA | GCTCCTTTTCTCGTTTGAAC | 100 (GUA-02) | (AC)6(AT)(AC)8 (GUA-02) | 21 | 493063-493160 | FM865631 | 54 |
| CSg55 | GCTTTGCTTGGACTGGAGAG | GGAGGGAAAAGGAAGTCCAG | 97 (GUA-02) | (TG)13 (GUA-02) | 10 | 53298-53386 | FM865632 | 56 |
| CSg59 | CATTTGAGCTGCACGTGTCT | AGGGGAGAGTGTGTTTGGTG | 94 (GUA-02) | (TC)6 (GUA-02) | 25 | 573032-573127 | FM865634 | 56 |
| 7GN | CTCCTGGAACGGCTAACAC | TGATATGAGGCACATTCAGC | 115 (BRA-04) | (AC)11 (BRA-04) | 35 | 2357935-2358045 | FM877579 | 56 |
| 11H | CACACCTGCTACTGGTCCTC | TCTGTTTCATGACATGCCTTT | 92 (BRA-04) | (GT)10 (BRA-04) | 21 | 658499-658584 | FM877806 | 54 |
| 11C | GTGGGTATGCGTGTGTCTCT | ATTAAAGTTGCCACCCTCAC | 100 (BRA-04) | (TG)10 (BRA-04) | 33 | 355989-356086 | FM877805 | 58 |
| 6F | CAACAGCAAAGCACAAAGAA | CAGCAATGCCGATAAGAAAC | 91 (BRA-04) | (AC)11 (BRA-04) | 27 | 300255-300208c | FM877578 | 56 |
| 10F | TGCGAGTATACCTCTCCTTCA | CAACAACAACAACCGAGAGG | 99 (BRA-04) | (CA)16 (BRA-04) | 18 | 274048-274132 | FM877804 | 58 |
| B6F | CACCTCTTGCCTGCACTT | TTTAAACGTCGGTCTGTGTG | 95 (BRA-04) | (AC)14 (BRA-04) | 16 | 407879-407961 | FM877808 | 58 |
| B3H | GGTATGCGTGGATATGAAGC | CTCGGCATCGCAGTTTC | 77 (BRA-04) | (AC)13 (BRA-04) | 28 | 357912-357951b | FM877807 | 58 |
| AC01R | ACGTCAGCACACAAACGTC | CTTCTTCCTGCTTTGCCTCT | 99 (BRA-30) | (CA)8 (BRA-30) | 19 | 35871-35969 | AF139110 | 58 |
| AC16R | GGGTGTCGAGGATGAGGT | TAGTGCCATTAGGGGCTCA | 93 (BRA-30) | (TG)13 (BRA-30) | 11 | 91859-91951 | AF139112 | 58 |
M2904S, L. braziliensis strain MHOM/BR/1975/M2904, whose genome has been completely sequenced. The following strains were used to develop the microsatellite markers: L. (V.) guyanensis MHOM/SR/1987/TRUUS1 (GUA-02), L. (V.) braziliensis MHOM/BR/2000/LTB300 (BRA-04), and L. (V.) braziliensis MHOM/BR/1975/M2904 (BRA-30). TA, annealing temperature.
Only the 3′ end of the marker fragment.
Only the 5′ end of the marker fragment.
Screening of the length variations of the amplified markers was performed by automated fragment analysis. PCRs were performed with fluorescence-conjugated forward primers by using 6-carboxyfluorescein and 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein (Sigma-Aldrich Co., St. Louis, MO) as two different labels. The amplified products were commercially analyzed on an automated capillary sequencer (SMB Services in Molecular Biology, Berlin, Germany) with an ABI Prism GeneMapper apparatus (Applied Biosystems, Foster City, CA).
Microsatellite-based genetic distances were calculated with MSA software (12) and POPULATIONS software (http://www.legs.cnrs-gif.fr/bioinfo/populations) by applying the Chord distance measure (6). Neighbor-joining trees, which included the test for confidence intervals by bootstrapping (100 replicates) and which were based on the resulting distance matrix, were constructed with the programs POPULATIONS and MEGA (21).
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers of the microsatellite markers developed in this study are given in Table 2.
RESULTS
A total of 74 clones containing microsatellite structures were obtained from the L. (V.) braziliensis (CA)n-enriched genomic library. Poor sequences, redundancies, and complex structures reduced the number of suitable sequences to 24. From the L. (V.) guyanensis-enriched library, 150 microsatellite-containing clones were sequenced and primers flanking 17 of the microsatellite sequences that were confirmed were designed. Primers were developed to align closely to the CA/GT repeats for two reasons: (i) to minimize biases due to additional insertion/deletion events in the flanking regions and (ii) because size differences in short PCR products can be detected more easily if MetaPhor agarose gels are used (27). For the same reasons, we have also redesigned primers AC01R and AC16R for microsatellites AC01 and AC16, respectively (30); they then annealed much closer to the CA and TG repeats, respectively. The 24 microsatellite markers from the L. braziliensis library and the 17 markers from the L. guyanensis library were amplified for 9 and 18 strains of the Leishmania (Viannia) subgenus, respectively, including the strains used for the construction of the libraries as controls (Table 1). Sixteen of these markers, eight from each library, yielded PCR products of the expected size which were polymorphic in 4% MetaPhor agarose gels (results not shown) for strains of both L. (V.) braziliensis and L. (V.) guyanensis. Of these, after automated fragment analysis was performed, 13 markers (Table 2), in addition to markers AC01R and AC16R, were finally selected for further analysis. The 15 markers are located on 13 chromosomes. Markers CSg46 and 10F are both located on chromosome 18, and markers CSg53 and 11H are both located on chromosome 21, albeit far enough apart to be considered independent.
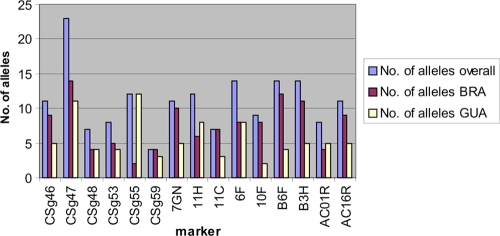
The set of 15 microsatellite markers was tested with 31 strains of L. (V.) braziliensis, 20 strains of L. (V.) guyanensis, and 2 strains of L. (V.) peruviana from various areas of endemicity for CL and MCL in South America. In addition, WHO reference strains of other species of the subgenus Leishmania (Viannia) were included, such as L. shawi, L. naiffi, L. lainsoni, and L. panamensis (Table 1). With the exception of marker AC16R, which did not amplify the two strains of L. peruviana investigated in the present study, one or two PCR products of the expected size were obtained for all strains and markers. The size of the amplified fragment (and thus the number of repeats) was always compared to the size of the fragments from cloned strains MHOM/BR/84/LTB300 and MHOM/SR/87/TRUUS1. All 15 markers showed polymorphisms within strains of L. braziliensis as well as strains of L. guyanensis (Table 3). Strains of L. braziliensis presented more variation in most of the markers (Fig. 1). Six of the markers differed in repeat numbers for the two strains of L. peruviana tested.
TABLE 3.
Characterization of the 15 microsatellite markers developed for population analysis of Viannia speciesa
| Marker | Species | No. of isolates | Repeat array | Fragment size (bp) | A | He | Ho | FIS |
|---|---|---|---|---|---|---|---|---|
| CSg46 | L. braziliensis | 31 | (AC) 10-20 | 79-99 | 9 | 0.783 | 0.419 | 0.468 |
| L. guyanensis | 20 | (AC) 10-16 | 79-91 | 5 | 0.719 | 0.400 | 0.450 | |
| CSg47 | L. braziliensis | 31 | (TG) 7-31 | 85-133 | 14 | 0.887 | 0.710 | 0.203 |
| L. guyanensis | 20 | (TG) 3-16 | 77-103 | 11 | 0.893 | 0.700 | 0.221 | |
| CSg48 | L. braziliensis | 31 | (TG) 4-8 | 70-78 | 4 | 0.524 | 0.167 | 0.685 |
| L. guyanensis | 20 | (TG) 5-9 | 72-80 | 4 | 0.568 | 0.150 | 0.741 | |
| CSg53 | L. braziliensis | 31 | (AC) 7-15 | 84-100 | 5 | 0.668 | 0.387 | 0.425 |
| L. guyanensis | 20 | (AC) 7-15 | 84-100 | 4 | 0.706 | 0.250 | 0.652 | |
| CSg55 | L. braziliensis | 31 | (TG) 11,15 | 93-101 | 2 | 0.151 | 0.097 | 0.362 |
| L. guyanensis | 20 | (TG) 11-24 | 93-119 | 12 | 0.876 | 0.600 | 0.320 | |
| CSg59 | L. braziliensis | 31 | (TC) 6-9 | 94-100 | 4 | 0.591 | 0.548 | 0.073 |
| L. guyanensis | 20 | (TC) 6-8 | 94-98 | 3 | 0.656 | 0 | 1.000 | |
| 7GN | L. braziliensis | 31 | (AC) 7-19 | 92-116 | 10 | 0.736 | 0.387 | 0.478 |
| L. guyanensis | 20 | (AC) 5-10 | 88-98 | 5 | 0.786 | 0.150 | 0.813 | |
| 11H | L. braziliensis | 31 | (GT) 7-12 | 86-96 | 6 | 0.637 | 0.323 | 0.498 |
| L. guyanensis | 20 | (GT) 7-15 | 86-102 | 8 | 0.788 | 0.250 | 0.688 | |
| 11C | L. braziliensis | 31 | (TG) 5-11 | 90-102 | 7 | 0.742 | 0.645 | 0.133 |
| L. guyanensis | 20 | (TG) 6-8 | 92-96 | 3 | 0.376 | 0.250 | 0.340 | |
| 6F | L. braziliensis | 31 | (AC) 10-22 | 89-113 | 8 | 0.779 | 0.613 | 0.216 |
| L. guyanensis | 20 | (AC) 6-16 | 81-101 | 8 | 0.824 | 0.250 | 0.702 | |
| 10F | L. braziliensis | 31 | (CA) 12-21 | 91-109 | 8 | 0.818 | 0.581 | 0.294 |
| L. guyanensis | 20 | (CA) 12-21 | 91-109 | 2 | 0.296 | 0.150 | 0.500 | |
| B6F | L. braziliensis | 31 | (AC) 6-20 | 79-107 | 12 | 0.904 | 0.800 | 0.117 |
| L. guyanensis | 20 | (AC) 8-11 | 83-89 | 4 | 0.744 | 0.350 | 0.536 | |
| B3H | L. braziliensis | 31 | (AC) 5-19 | 61-89 | 11 | 0.877 | 0.710 | 0.194 |
| L. guyanensis | 20 | (AC) 7-12 | 65-75 | 5 | 0.681 | 0.450 | 0.345 | |
| AC01R | L. braziliensis | 31 | (CA) 7-11 | 97-105 | 4 | 0.290 | 0.258 | 0.111 |
| L. guyanensis | 20 | (CA) 7-16 | 97-115 | 5 | 0.718 | 0.550 | 0.239 | |
| AC16R | L. braziliensis | 31 | (TG) 8-24 | 83-115 | 9 | 0.721 | 0.484 | 0.333 |
| L. guyanensis | 20 | (TG) 11-15 | 89-97 | 5 | 0.673 | 0.300 | 0.561 | |
| Mean | All strainsb | 57 | 11.0 | 0.786 | 0.401 | 0.492 | ||
| L. braziliensis | 31 | 7.5 | 0.674 | 0.475 | 0.298 | |||
| L. guyanensis | 20 | 5.6 | 0.687 | 0.320 | 0.541 |
A, number of alleles; He, expected heterozygosity; Ho, observed heterozygosity; FIS, inbreeding coefficient.
Including the two strains of L. peruviana and the single strains of L. shawi, L. naiffi, L. lainsoni, and L. panamensis.
FIG. 1.
Numbers of alleles calculated for the 15 microsatellite markers and the 31 strains of L. braziliensis (BRA) and 20 strains of L. guyanensis (GUA) tested in this study.
The genetic diversity within the two main groups of strains, L. braziliensis and L. guyanensis, was estimated by calculating the number of alleles per locus and the observed and expected heterozygosities (Table 3). The number of microsatellite alleles ranged from 4 to 23 (mean, 11) for all strains, from 2 to 14 (mean, 7.5) for the strains of L. braziliensis, and from 2 to 12 (mean, 5.6) for the strains of L. guyanensis. The most variable marker was CSg47, and the least variable one CSg59. The observed levels of heterozygosity varied between 0.097 and 0.800 for the L. braziliensis strains and 0 and 0.700 for the L. guyanensis strains, with a clear tendency to heterozygosity for most microsatellite loci. The levels of heterozygosity expected under Hardy-Weinberg equilibrium, a measure of genetic diversity, were between 0.151 and 0.904 for the L. braziliensis strains and 0.296 and 0.893 for the L. guyanensis strains and for all but two markers were higher than the mean observed heterozygosity (0.475 for the L. braziliensis strains and 0.320 for the L. guyanensis strains). This discrepancy can be explained by the inbreeding of individuals due to deviation from random mating. The value of the inbreeding coefficient can vary by −1 (all individuals are heterozygous for the same allele pair), 0 (random distribution of alleles), and +1 (all individuals are homozygous) (11). Here, the mean inbreeding coefficients were 0.298 and 0.541 for the L. braziliensis and L. guyanensis strains, respectively, pointing to a predominance of homozygotes, which is in agreement with previous findings (28).
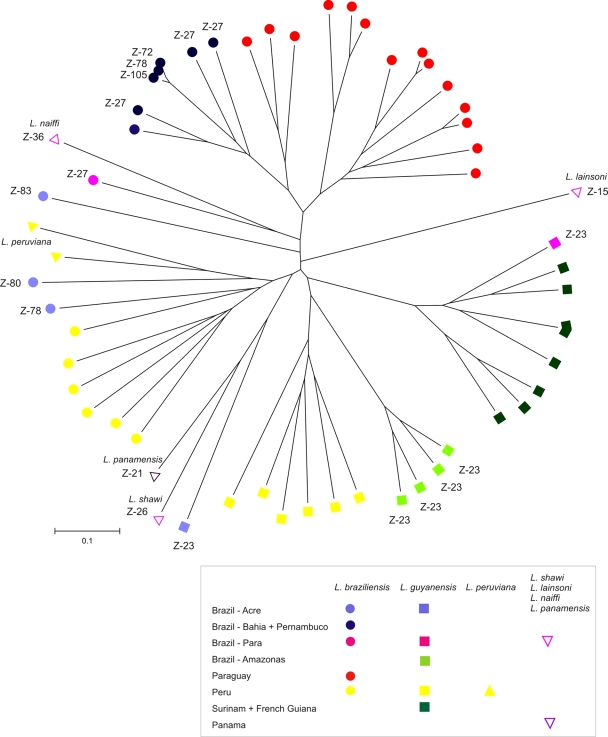
Fifty-six different MLMT profiles comprising the number of repeats in each of the 15 markers were revealed among the 57 strains analyzed in this study. All profiles except one were represented by a single strain; only two strains of L. guyanensis from Suriname (strains MHOM/SR/1987/TRUUS2 and MHOM/SR/1987/TRUUS3) shared an identical MLMT profile. For the microsatellite genotypes obtained for all 57 strains included in this study (Table 1) a neighbor-joining phylogram was constructed on the basis of the Chord distance matrix obtained (Fig. 2). The phylogram falls into three main clusters: cluster 1 was the L. guyanensis cluster, which comprised all but one strain of this species; cluster 2 was the L. braziliensis cluster, which consisted of all strains from Paraguay and from the Bahia and Pernambuco areas in Brazil; and cluster 3 was the L. braziliensis/L. peruviana cluster, which included strains from Peru and Brazil (state of Acre). Further substructuring according to the geographical origin of the strains is apparent, especially in clusters 1 and 2. The two single strains of L. shawi and L. panamensis are located on a common branch, together with L. guyanensis strain MHOM/BR/2002/NMT-RBO013 from Acre, Brazil, and are most closely related to the strains of L. braziliensis and L. peruviana in cluster 3. The strain of L. naiffi is found on a separate branch together with L. braziliensis strain MHOM/BR/1975/M2903 from Pará, Brazil. The strains of L. lainsoni and L. braziliensis MHOM/BR/2002/NMT-RBO011 have completely unique positions in the tree.
FIG. 2.
Neighbor-joining tree (rooted at the midpoint) inferred from the Chord distances calculated for the microsatellite data for 57 strains of Viannia species. The three populations consisted of one population of strains of only L. guyanensis, another one of strains of L. braziliensis from Paraguay and Brazil, and the last of strains of L. Braziliensis and L. peruviana. Reference strains of L. lainsoni, L. naiffi, L. panamensis, and L. shawi are found on separate branches. The origins of the strains are indicated by colored circles, rectangles, and triangles.
DISCUSSION
An important impetus for this study was the establishment of an MLMT protocol for use in future epidemiological and population genetic studies of strains belonging to the subgenus Leishmania (Viannia). Altogether, 15 hypervariable and independent microsatellite markers have been identified, and their discriminative powers were tested with 57 strains of different Leishmania (Viannia) species. Of these markers, 13 were isolated de novo from the L. braziliensis and L. guyanensis genomic libraries and 2 were described previously (30) and modified for the MLMT approach described here. PCR products were obtained for almost all markers and Leishmania (Viannia) species tested in the present study. The only exception was L. peruviana, which did not amplify with marker AC16R. Size variations were detected for all markers when the 31 strains of L. braziliensis and 20 strains of L. guyanensis were analyzed, with those for L. braziliensis being more variable. The latter finding is in congruence with the findings of previous studies performed in Brazil on the basis of multilocus enzyme electrophoresis (8) and polymorphisms in the internal transcribed spacer and hsp70 sequences (our unpublished data). The two strains of L. peruviana included in this study were polymorphic for six markers. Experiments with more strains of this species, which have not yet been completed, suggest that the MLMT approach described here is also able to differentiate strains of L. peruviana (data not shown). In order to elucidate their degrees of microsatellite variation, more strains of L. panamensis, L. shawi, L. naiffi, and L. lainsoni need to be analyzed because only reference strains have been investigated so far. Individual microsatellite patterns were observed for all strains tested except two strains of L. guyanensis that shared a common pattern. MLMT thus offers a possible means of differentiating strains, at least those of L. braziliensis and L. guyanensis, and investigating the population dynamics of these species.
Distance-based analysis identified three main clusters within the 57 strains typed in this study. In the first cluster, all strains of L. guyanensis except one grouped together. The other two clusters consisted of strains of L. braziliensis that grouped according to their geographical origins. Of these, one cluster comprised strains from Paraguay and strains isolated in eastern Brazil; and the other cluster comprised strains from Peru, the adjacent Brazilian state of Acre, and the two strains of L. peruviana. The latter had been identified by using a multilocus PCR-restriction fragment length polymorphism approach and were found to be the most closely related to the strains of L. braziliensis analyzed in that study (14). The three main clusters in the phylogram were further subdivided according to the geographical origins of the strains, especially the clusters formed by strains of L. guyanensis and Paraguayan and Brazilian strains of L. braziliensis. A hierarchical population structure of Leishmania (Viannia) possibly exists in the study area, and populations that are further divided into subpopulations may exist. This needs to be verified by analyzing larger sets of samples from the different areas and by comparing the findings of distance-based analyses of MLMT data with the findings of analyses based on Bayesian statistics. The topologies of microsatellite distance trees always suffer from poor statistical support; however, in previous studies, the main clusters were always confirmed when a Bayesian model-based clustering method for inferring population structure was used (2, 19, 20, 32). In order to evaluate the discriminatory power of our MLMT approach, we used only a limited number of Leishmania (Viannia) strains of different species and from different areas of endemicity. The application of MLMT to this restricted sample set nevertheless demonstrated the great potential of this approach for the differentiation of strains, with the yield being individual profiles for strains within regions of endemicity, and for population genetic studies of the subgenus Leishmania (Viannia).
As in other studies of microsatellite variation in Old World and New World Leishmania strains (2, 19, 20, 26, 28, 32), double bands indicating the heterozygosity of the locus were often seen in our study. Because the strains tested were not cloned, the possibility that there were mixtures of strains cannot be excluded. Since no loci presented three or four peaks, aneuploidy is not likely for the loci used in our MLMT approach. The homozygote excess indicated by the inbreeding coefficient measures probably results from a population substructure (the Wahlund effect), as can be assumed from the neighbor-joining tree shown in Fig. 2. More strains of L. braziliensis and L. guyanensis from different foci should be analyzed in the future in order to get insights into the population genetics of these parasites.
A high number of independent microsatellite markers developed by us and other authors (28, 30) are now available for microsatellite typing of strains belonging to different Leishmania (Viannia) species. More microsatellite markers for L. braziliensis can be developed by using a searchable database of microsatellite loci within the genome at http://www.genomics.liv.ac.uk/tryps/Microsatellites.V1.html (13). Microsatellite sequence variation resulting from the gain and the loss of single repeat units can be screened by polyacrylamide gel electrophoresis and MetaPhor gel electrophoresis, automated sequencing, and automated fragment analysis. All these methods were shown to produce comparable and reproducible results (27). Sequencing is, however, indispensable for the analysis of fragments containing more than one microsatellite, as described by Russell et al. (30) and Rougeron et al. (28).
The significance of microsatellite typing is influenced by the number of loci tested (18). Microsatellite sequences, in general, tend to gain repeat units. Natural selection, however, acts against the formation of very long repeats, which will result in homoplasy caused by alleles identical in size but not by descent (33). To minimize the effect on homoplasy, it is recommended that a panel of 10 to 20 unlinked microsatellite markers should be used. The fewer microsatellite loci that are analyzed, the more that the differences estimated between the strains depend upon the markers selected. Consequently, whether the analysis of more loci will improve the discriminatory power of MLMT significantly always needs to be examined (18). The 15 markers developed in this study individualized all but two of the strains tested and indicated the existence of genetic structures at different levels. Thus, MLMT with these markers is potentially useful for epidemiological and population genetic studies of strains within the subgenus Leishmania (Viannia) in order to investigate the structure and dynamics of the corresponding natural foci. It will also help to answer specific clinical questions, such as the role of parasite persistence after subclinical infection; whether endogenous versus exogenous reinfection is associated with immunosuppression; and whether transmission is vector independent, such as via blood transfusion or organ transplantation. In Leishmania species, microsatellite markers are specific at the species or species complex level (16, 31). When DNA from other species of Leishmania is used, in most cases no PCR product is obtained or the products differ significantly in size. Therefore, the infecting agents first need to be identified, at least to the Viannia versus non-Viannia level in this study, before they are typed by MLMT. The ability to amplify microsatellite markers directly from clinical samples containing small amounts of parasites, as shown by an MLMT approach that differentiates strains of the L. donovani complex (1, 3), will be of added value for these studies.
Acknowledgments
We thank Juan D. Maciel, Andrés Canese, and Cynthia Céspedes for their exhaustive collaboration during the isolation of strains in Paraguay. We are grateful to Manal Jamjoom of the Liverpool School of Tropical Medicine for her assistance with the preparation of the microsatellite libraries. We thank Henk Schallig of the Royal Tropical Institute in Amsterdam, The Netherlands, and Jean-Claude Dujardin of the Institute of Tropical Medicine in Antwerp, Belgium, for providing strains of Leishmania (Viannia) species.
We acknowledge the Brazilian financial agencies NPq, FAPERJ, and FINEP for giving constant support to the field work (isolation of the parasites) and maintenance of the Leishmania collection. We also acknowledge the support given by Antonieta Rojas de Arias and the European Commission Alfa Project, contract no. 6-0008-9. This research was supported by a grant (grant INCO-CT2005-015407) from the European Union.
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Alam, M. Z., K. Kuhls, C. Schweynoch, S. Sundar, S. Rijal, A. K. Shamsuzzaman, B. V. Raju, P. Salotra, J. C. Dujardin, and G. Schonian. 2009. Multilocus microsatellite typing (MLMT) reveals genetic homogeneity of Leishmania donovani strains in the Indian subcontinent. Infect. Genet. Evol. 924-31. [DOI] [PubMed] [Google Scholar]
- 2.Al-Jawabreh, A., S. Diezmann, M. Mueller, T. Wirth, L. F. Schnur, M. V. Strelkova, D. A. Kovalenko, S. A. Razakov, J. Schwenkenbecher, K. Kuhls, and G. Schoenian. 2008. Identification of geographically distributed sub-populations of Leishmania (Leishmania) major by microsatellite analysis. BMC Evol. Biol. 8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amro, A., G. Schonian, M. B. Al-Sharabati, K. Azmi, A. Nasereddin, Z. Abdeen, L. F. Schnur, G. Baneth, C. L. Jaffe, and K. Kuhls. 2009. Population gentics of Leishmania infantum in Israel and Palestinian Authority through microsatellite analysis. Microbes Infect. 11484-492. [DOI] [PubMed] [Google Scholar]
- 4.Belli, A. A., M. A. Miles, and J. M. Kelly. 1994. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology 109(Pt. 4)435-442. [DOI] [PubMed] [Google Scholar]
- 5.Botilde, Y., T. Laurent, W. Quispe Tintaya, C. Chicharro, C. Canavate, I. Cruz, K. Kuhls, G. Schonian, and J. C. Dujardin. 2006. Comparison of molecular markers for strain typing of Leishmania infantum. Infect. Genet. Evol. 6440-446. [DOI] [PubMed] [Google Scholar]
- 6.Cavalli-Sforza, L. L., and A. W. Edwards. 1967. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 19233-257. [PMC free article] [PubMed] [Google Scholar]
- 7.Cupolillo, E., F. Aguiar Alves, L. R. Brahim, M. F. Naiff, L. O. Pereira, M. P. Oliveira-Neto, A. Falqueto, and G. Grimaldi, Jr. 2001. Recent advances in the taxonomy of the New World leishmanial parasites. Med. Microbiol. Immunol. 19057-60. [DOI] [PubMed] [Google Scholar]
- 8.Cupolillo, E., G. Grimaldi, Jr., and H. Momen. 1994. A general classification of New World Leishmania using numerical zymotaxonomy. Am. J. Trop. Med. Hyg. 50296-311. [DOI] [PubMed] [Google Scholar]
- 9.Cupolillo, E., H. Momen, and G. Grimaldi, Jr. 1998. Genetic diversity in natural populations of New World Leishmania. Mem. Inst. Oswaldo Cruz 93663-668. [DOI] [PubMed] [Google Scholar]
- 10.Delgado, O., E. Cupolillo, R. Bonfante-Garrido, S. Silva, E. Belfort, G. Grimaldi, Jr., and H. Momen. 1997. Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis. Mem. Inst. Oswaldo Cruz 92581-582. [DOI] [PubMed] [Google Scholar]
- 11.De Meeus, T., L. Lehmann, and F. Balloux. 2006. Molecular epidemiology of clonal diploids: a quick overview and a short DIY (do it yourself) notice. Infect. Genet. Evol. 6163-170. [DOI] [PubMed] [Google Scholar]
- 12.Dieringer, D., and C. Schlötterer. 2003. Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite sets. Mol. Ecol. Notes 3167-169. [Google Scholar]
- 13.Fakhar, M., M. H. Motazedian, D. Daly, C. D. Lowe, S. J. Kemp, and H. A. Noyes. 2008. An integrated pipeline for the development of novel panels of mapped microsatellite markers for Leishmania donovani complex, Leishmania braziliensis and Leishmania major. Parasitology 135567-574. [DOI] [PubMed] [Google Scholar]
- 14.Garcia, A. L., A. Kindt, K. W. Quispe-Tintaya, H. Bermudez, A. Llanos, J. Arevalo, A. L. Banuls, S. De Doncker, D. Le Ray, and J. C. Dujardin. 2005. American tegumentary leishmaniasis: antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect. Genet. Evol. 5109-116. [DOI] [PubMed] [Google Scholar]
- 15.Grimaldi, G., Jr., and R. B. Tesh. 1993. Leishmaniases of the New World: current concepts and implications for future research. Clin. Microbiol. Rev. 6230-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamjoom, M. B., R. W. Ashford, P. A. Bates, S. J. Kemp, and H. A. Noyes. 2002. Towards a standard battery of microsatellite markers for the analysis of the Leishmania donovani complex. Ann. Trop. Med. Parasitol. 96265-270. [DOI] [PubMed] [Google Scholar]
- 17.Jones, T. C., W. D. Johnson, Jr., A. C. Barretto, E. Lago, R. Badaro, B. Cerf, S. G. Reed, E. M. Netto, M. S. Tada, T. F. Franca, et al. 1987. Epidemiology of American cutaneous leishmaniasis due to Leishmania braziliensis braziliensis. J. Infect. Dis. 15673-83. [DOI] [PubMed] [Google Scholar]
- 18.Koskinen, M. T., H. Hirvonen, P. A. Landry, and C. R. Primmer. 2004. The benefits of increasing the number of microsatellites utilized in genetic population studies: an empirical perspective. Hereditas 14161-67. [DOI] [PubMed] [Google Scholar]
- 19.Kuhls, K., C. Chicharro, C. Canavate, S. Cortes, L. Campino, C. Haralambous, K. Soteriadou, F. Pratlong, J. P. Dedet, I. Mauricio, M. Miles, M. Schaar, S. Ochsenreither, O. A. Radtke, and G. Schonian. 2008. Differentiation and gene flow among European populations of Leishmania infantum MON-1. PLoS Negl. Trop. Dis. 2e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhls, K., L. Keilonat, S. Ochsenreither, M. Schaar, C. Schweynoch, W. Presber, and G. Schonian. 2007. Multilocus microsatellite typing (MLMT) reveals genetically isolated populations between and within the main endemic regions of visceral leishmaniasis. Microbes Infect. 9334-343. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 22.Lainson, R., and J. J. Shaw. 1987. Evolution, classification and geographical distribution, p. 1-120. In W. Peters and R. Killick-Kendrick (ed.), The leishmaniases in biology and medicine. Academic Press, New York, NY.
- 23.Lessa, M. M., H. A. Lessa, T. W. Castro, A. Oliveira, A. Scherifer, P. Machado, and E. M. Carvalho. 2007. Mucosal leishmaniasis: epidemiological and clinical aspects. Braz. J. Otorhinolaryngol. 73843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsden, P. D. 1986. Mucosal leishmaniasis (“espundia” Escomel, 1911). Trans. R. Soc. Trop. Med. Hyg. 80859-876. [DOI] [PubMed] [Google Scholar]
- 25.Meredith, S. E., E. E. Zijlstra, G. J. Schoone, C. C. Kroon, G. J. van Eys, K. U. Schaeffer, A. M. el-Hassan, and P. G. Lawyer. 1993. Development and application of the polymerase chain reaction for the detection and identification of Leishmania parasites in clinical material. Arch. Inst. Pasteur Tunis 70419-431. [PubMed] [Google Scholar]
- 26.Nolder, D., N. Roncal, C. R. Davies, A. Llanos-Cuentas, and M. A. Miles. 2007. Multiple hybrid genotypes of Leishmania (Viannia) in a focus of mucocutaneous leishmaniasis. Am. J. Trop. Med. Hyg. 76573-578. [PubMed] [Google Scholar]
- 27.Ochsenreither, S., K. Kuhls, M. Schaar, W. Presber, and G. Schonian. 2006. Multilocus microsatellite typing as a new tool for discrimination of Leishmania infantum MON-1 strains. J. Clin. Microbiol. 44495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rougeron, V., E. Waleckx, M. Hide, T. De Meeûs, J. Arevalo, A. Llanos-Cuentas, and A. L. Banuls. 2008. A set of 12 microsatellite loci for genetic studies of Leishmania braziliensis. Mol. Ecol. Res. 8351-353. [DOI] [PubMed] [Google Scholar]
- 29.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132365-386. [DOI] [PubMed] [Google Scholar]
- 30.Russell, R., M. P. Iribar, B. Lambson, S. Brewster, J. M. Blackwell, C. Dye, and J. W. Ajioka. 1999. Intra and inter-specific microsatellite variation in the Leishmania subgenus Viannia. Mol. Biochem. Parasitol. 10371-77. [DOI] [PubMed] [Google Scholar]
- 31.Schwenkenbecher, J. M., C. Frohlich, F. Gehre, L. F. Schnur, and G. Schonian. 2004. Evolution and conservation of microsatellite markers for Leishmania tropica. Infect. Genet. Evol. 499-105. [DOI] [PubMed] [Google Scholar]
- 32.Schwenkenbecher, J. M., T. Wirth, L. F. Schnur, C. L. Jaffe, H. Schallig, A. Al-Jawabreh, O. Hamarsheh, K. Azmi, F. Pratlong, and G. Schonian. 2006. Microsatellite analysis reveals genetic structure of Leishmania tropica. Int. J. Parasitol. 36237-246. [DOI] [PubMed] [Google Scholar]
- 33.Taylor, J. S., J. S. Sanny, and F. Breden. 1999. Microsatellite allele size homoplasy in the guppy (Poecilia reticulata). J. Mol. Evol. 48245-247. [DOI] [PubMed] [Google Scholar]
- 34.Thomaz-Soccol, V., I. D. Velez, F. Pratlong, S. Agudelos, G. Lanotte, and J. A. Rioux. 2000. Enzymatic polymorphism and phylogenetic relationships in Leishmania Ross, 1903 (Sarcomastigophora: Kinetoplastida): a case study in Colombia. Syst. Parasitol. 4659-68. [DOI] [PubMed] [Google Scholar]
- 35.Tojal da Silva, A. C., E. Cupolillo, A. C. Volpini, R. Almeida, and G. A. Romero. 2006. Species diversity causing human cutaneous leishmaniasis in Rio Branco, state of Acre, Brazil. Trop. Med. Int. Health 111388-1398. [DOI] [PubMed] [Google Scholar]