Abstract
The objective of this study was to characterize by serotyping, pulsed-field gel electrophoresis (PFGE), and PCR amplification of virulence genes and markers of epidemic clones I, II, and III (ECI, ECII, and ECIII) 54 human isolates from apparently sporadic cases of infection occurring in the Lombardy region and in the province of Florence, Tuscany, Italy, in the years 1996 to 2007. Listeria monocytogenes isolates were provided by the clinical microbiology laboratories of the Lombardy region and the “Careggi” Hospital of Florence, Tuscany, Italy. Serotyping, PFGE after digestion with the AscI and ApaI enzymes, and PCR amplification for the inlA, inlC, and inlJ genes and ECI, ECII, and ECIII markers were performed according to procedures described previously. Twenty-five (46.3%) L. monocytogenes isolates were assigned to serotype 1/2a, 23 (42.6%) to serotype 4b, and 6 (11.1%) to serotype 1/2b. Thirty-one AscI pulsotypes were recognized among the 54 human isolates. Eleven molecular subtype clusters, of which eight included indistinguishable pulsotypes and three included closely related pulsotypes, were shared by two to seven isolates. Fifteen isolates exhibited unique AscI pulsotypes. Three groups of clustered isolates and two apparently sporadic isolates generated EC amplicons. All strains tested positive for the inlA, inlC, and inlJ genes. Based on the results of serotyping and molecular typing, there were 11 occasions when L. monocytogenes strains with the same subtype were isolated from more than one listeriosis case. A total of 39 out of 54 isolates (72.2%) were attributed to molecular subtype clusters. The results of the study suggest that routine subtyping of L. monocytogenes strains from human listeriosis cases could allow more-timely detection of outbreaks possibly caused by food-borne isolates from a common source and could lead to control of ongoing food exposure, thus preventing the occurrence of more cases.
Listeria monocytogenes is the etiological agent of listeriosis, a food-borne disease occurring primarily in immunocompromised individuals, causing septicemia and central nervous system infections, and in pregnant women, who may suffer preterm delivery, miscarriage, or stillbirth (8, 18). Healthy adults may suffer a febrile gastroenteritis after ingesting large numbers of L. monocytogenes cells (24).
Most industrialized countries, including those within the European Union (EU), have an annual listeriosis incidence of 2 to 10 reported cases per million people per year (13). However, listeriosis has a high case fatality rate that can exceed 30% (8, 28). Recently, several European countries have experienced apparently increasing incidences of listeriosis, mainly among persons aged 65 years and older (6, 13).
In Italy, notification of listeriosis cases has been mandatory since 1993, but data are transmitted to the national level quarterly. A second, syndrome-based surveillance system, which covers infections of the central nervous system, is in place. A National Reference Laboratory, which receives strains and epidemiological and clinical information on a voluntary basis, is also present. The incidence of cases reported by the Ministry of Health is lower than those in most EU countries; in recent years (2004 to 2006), it has been 0.8 case per million inhabitants annually (12; http://www.simi.iss.it/banca_dati_simi.htm). This low incidence must be considered with care because of the poor sensitivity of the universal passive surveillance system and the difficulties posed by L. monocytogenes infection epidemiology and natural history. Indeed, a recent report about a 1-year (2002-to-2003) period of enhanced laboratory-based surveillance showed an incidence of 1.3 cases per million inhabitants (12).
Throughout Europe and the United States, the majority of cases of listeriosis reported to public health authorities apparently are not linked to a common source and therefore are defined as sporadic (25, 27). However, because of the unique epidemiological and clinical characteristics of human food-borne listeriosis, traditional epidemiological surveillance systems alone are unable to detect most common-source outbreaks, particularly when a limited number of cases are scattered over a wide geographic area (25, 26, 29). Furthermore, when one is attempting to trace food exposures and transmission routes through food-processing chains, a further challenge is posed by the wide spectrum of L. monocytogenes strains, many of which are virulent and associated with significant morbidity and mortality while others are avirulent and unable to establish an infection in mammalian hosts (1, 19). Consequently, rapid and discriminatory subtyping methods, such as various DNA-based methods, e.g., pulsed-field gel electrophoresis (PFGE), PCR and restriction fragment length polymorphism analysis, or sequencing of some specific sequences, are essential for the epidemiological investigation of L. monocytogenes and the tracking of specific clones along food-processing chains (5, 26, 29). Recently, a rapid PCR-based method for the detection of epidemic clones I, II, and III (ECI, ECII, and ECIII) of L. monocytogenes has been reported by Chen and Knabel (2). Moreover, Liu et al. (20) have described a multiplex PCR assay where a combined application of inlA, inlC, and inlJ gene primers makes it possible to determine potential virulence rapidly.
Our study is a retrospective subtyping analysis of 42 human isolates identified from apparently sporadic cases of infection in the Lombardy region and in the province of Florence, Tuscany, Italy, in the years 2006 to 2007. Twelve further isolates from human cases occurring in the same geographic areas since 1996 were available and were included in the study. The objectives were to assess the molecular clustering of L. monocytogenes isolates by using serotyping and PFGE and to evaluate the distribution of EC markers and genetic determinants of virulence among the human isolates under investigation.
MATERIALS AND METHODS
Bacterial isolates.
Between January 2006 and October 2007, 34 human isolates from cases of listeriosis were collected from the laboratories of diagnostic microbiology of the Lombardy region, Italy (Tables 1 and 2). Eight further isolates were recovered in the same period from the “Careggi” Hospital of Florence, Tuscany, Italy (Tables 1 and 2). Twelve sporadic human isolates of L. monocytogenes that had been identified by the same laboratories since 1996 were also included in the study (Tables 1 and 2). The two geographical areas were chosen based on the willingness of the regional health authorities of Lombardy and the laboratory microbiologists of the “Careggi” Hospital of Florence to participate in a collaborative study of L. monocytogenes epidemiology. In Lombardy, a strengthening of the surveillance system was implemented in the period of the study.
TABLE 1.
Characteristics of cluster-associated strains of L. monocytogenes from human sources
| Cluster | No. of strains | Place of isolation (region) | Date of isolation
|
Serotype | AscI pulsotype (no. of strains) | EC marker | |
|---|---|---|---|---|---|---|---|
| First case | Last case | ||||||
| I | 4 | Lombardy | 11 May 2006 | 6 February 2007 | 1/2b | 1A (3), 1B (1) | |
| II | 5 | Lombardy | 7 July 2006 | 2 August 2006 | 1/2a | 2 | |
| III | 6 | Lombardy/Tuscany | 17 November 2006 | 2 November 2007 | 4b | 3 | |
| IV | 2 | Lombardy/Tuscany | 12 January 1996 | 27 July 2005 | 1/2a | 4 | |
| V | 3 | Lombardy | 11 December 2006 | 31 October 2007 | 1/2a | 5 | |
| VI | 3 | Lombardy | 29 July 2006 | 14 September 2006 | 1/2a | 7A (2), 7B (1) | III |
| VII | 2 | Lombardy | 19 March 2005 | 21 March 2005 | 1/2a | 8 | |
| VIII | 7 | Lombardy/Tuscany | 25 July 2005 | 13 July 2007 | 4b | 6A (3), 6B (2), 6C (1), 6D (1) | I |
| IX | 3 | Lombardy | 19 July 2006 | 15 September 2007 | 4b | 19 | II |
| X | 2 | Lombardy | 11 October 2000 | 18 May 2007 | 1/2a | 20 | |
| XI | 2 | Lombardy | 18 May 2006 | 4 September 2007 | 4b | 10 | |
TABLE 2.
Characteristics of non-cluster-associated strains of L. monocytogenes from human sources
| Place of isolation (region) | Date of isolation | Serotype | AscI pulsotype | EC marker |
|---|---|---|---|---|
| Lombardy | 2 August 2006 | 1/2b | 9 | |
| Lombardy | 16 August 2006 | 4b | 11 | |
| Tuscany | 5 July 2004 | 4b | 12 | II |
| Tuscany | 11 July 2006 | 1/2b | 13 | |
| Tuscany | 27 February 2006 | 4b | 14 | |
| Lombardy | 2 February 2006 | 1/2a | 15 | |
| Lombardy | 10 October 2006 | 1/2a | 16 | |
| Lombardy | 10 August 2005 | 1/2a | 17 | |
| Tuscany | 1 August 2002 | 1/2a | 18 | |
| Tuscany | 23 November 2006 | 1/2a | 26 | |
| Tuscany | 26 May 2004 | 4b | 21 | II |
| Lombardy | 14 April 2007 | 1/2a | 22 | |
| Lombardy | 15 September 2007 | 4b | 23 | |
| Lombardy | 18 September 2007 | 1/2a | 24 | |
| Lombardy | 3 October 2007 | 1/2a | 25 |
Most strains had been isolated from the blood or cerebrospinal fluid of immunocompromised or elderly patients. Only three isolates were from maternal-neonatal infections. Each isolate in this study represents a single human listeriosis case.
No case had been formally recognized as belonging to a food-borne outbreak or as associated with a specific food vehicle during the conventional epidemiological investigations conducted by the public health departments.
The International Life Sciences Institute North America L. monocytogenes outbreak and diversity strain sets were used as reference strains (11).
Serotyping.
All strains tested were serotyped by using commercial specific antisera according to the manufacturer's instructions (Denka Seiken, Tokyo, Japan).
PFGE analysis.
PFGE was performed according to the PulseNet protocol with the AscI and ApaI enzymes (14). Bacterial cultures were embedded in agarose, lysed, washed, digested with the restriction enzyme, and electrophoresed on a Chef Mapper XA system (Bio-Rad Laboratories, Hercules, CA) at 6 V/cm for 22 h with switch times of 4 s to 40.01 s. XbaI-digested DNA from Salmonella enterica serotype Braenderup H9812 was used as the size reference standard. The profiles obtained were compared using the BioNumerics software package (version 5.1; Applied Maths, Saint-Martins-Latem, Belgium). Pattern clustering was performed by the unweighted-pair group algorithm and the Dice correlation coefficient with a tolerance of 1.5%. The results of the clustering analysis were confirmed by visual comparison of the PFGE profiles. In a first round of typing, AscI was used. Two profiles obtained by AscI were classified as indistinguishable if the DNA fragment patterns matched each other completely, as closely related if they differed by one to three bands, and as unrelated if they differed by more than three bands (25). Numbers were used to designate the AscI profiles. Closely related patterns were assigned an additional capital letter. Indistinguishable or closely related strains were subsequently cleaved with ApaI for clustering confirmation.
Multiplex PCR for detection of epidemic clones.
The multiplex PCR protocol described by Chen and Knabel (2), with minor modifications, was used for the identification of isolates belonging to L. monocytogenes ECI, ECII, and ECIII. The primer sequences have been described previously (2). In the PCR mixture, 50 nM ECI marker, 90 nM ECII marker, and 30 nM ECIII marker were added. PCR was performed in a volume of 25 μl containing 1.0 U Taq DNA polymerase (Promega), 1× PCR buffer, 200 μM deoxynucleoside triphosphates, and 10 ng of DNA from each L. monocytogenes strain. The cycling program and electrophoresis conditions have been described previously (2). A 1-kbp DNA ladder (Promega) was used as a molecular size marker.
Multiplex PCR for the detection of virulence-specific genes.
A multiplex PCR including inlA, inlC, and inlJ gene primers was performed as described by Liu et al. (20) with minor modifications. Briefly, genomic DNA from L. monocytogenes strains was obtained after treatment with lysozyme and proteinase K, resuspended in sterile distilled water, and either used immediately for PCR or stored at −20°C until use. Oligonucleotide primers for the L. monocytogenes internalin genes inlA, inlC, and inlJ were used in a multiplex PCR format by using the GeneAmp PCR system 2400 (Perkin-Elmer, Boston, MA) in a volume of 25 μl containing 1.0 U Taq DNA polymerase (Promega, Madison, WI), 1× PCR buffer, 200 μM deoxynucleoside triphosphates, and 10 ng of DNA from each L. monocytogenes strain, together with 40 pmol each inlA primer, 30 pmol each inlC primer, and 20 pmol each inlJ primer. The cycling program and electrophoresis conditions have been described previously (20). A 1-kbp DNA ladder (Promega) was used as a molecular size marker.
RESULTS
A total of 25 clinical isolates (46.3%) were assigned to serotype 1/2a, 23 (42.6%) to serotype 4b, and 6 (11.1%) to serotype 1/2b.
Thirty-one AscI pulsotypes were recognized among the 54 human isolates. Fifteen isolates showed unique AscI pulsotypes, and the remaining AscI pulsotypes were assigned to 11 clusters. Eight clusters—II, III, IV, V, VII, IX, X, and XI—included isolates with indistinguishable pulsotypes, and three clusters—I, VI, and VIII-consisted of isolates with closely related pulsotypes (Table 1). Each cluster contained two to seven isolates (Table 1). The ApaI profiles of the strains with indistinguishable AscI profiles showed no difference in the number or position of bands in all clusters except cluster I, where the ApaI profiles of two isolates differed by two bands. Isolates that were closely related by AscI differed by one to three bands by ApaI.
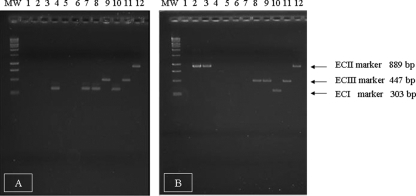
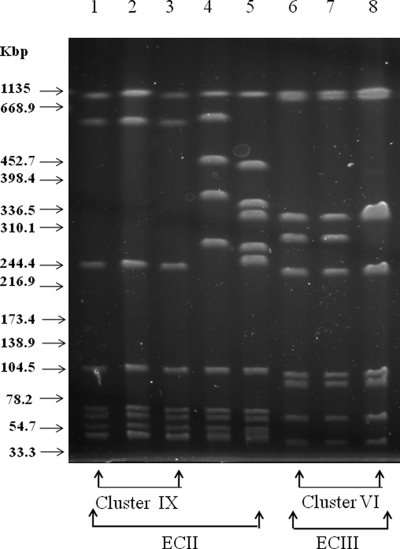
A total of 15 L. monocytogenes isolates tested positive for the EC markers. Three groups of clustered isolates, including three isolates of serotype 1/2a, seven isolates of serotype 4b, and three isolates of serotype 4b, generated ECIII, ECI, and ECII amplicons, respectively (Table 1). Furthermore, two apparently sporadic isolates showing serotype 4b yielded the ECII-specific band (Table 2). Figure 1 shows the PCR products of some representative isolates of L. monocytogenes and the reference strains FSL J1-110 (ECI), FSL N1-225 (ECII), and FSL R2-499 (ECIII). The AscI PFGE patterns of the five strains positive for the ECII marker and the three strains positive for the ECIII marker are shown in Fig. 2. The two sporadic isolates belonging to ECII were characterized by unique pulsotypes differing from each other and from the clustered isolates by more than three bands; consequently, they were classified as unrelated according to the interpretative criteria adopted in this study.
FIG. 1.
Agarose gel electrophoresis of EC markers detected in representative human isolates of L. monocytogenes. Panels A and B represent two different agarose gels. Lanes: MW (molecular weight), 1-kbp DNA ladder (Promega); 1 to 9, PCR products from human isolates analyzed in this study; 10 to 12, positive-control strains FSL J1-110 (ECI), FSL R2-499 (ECIII), and FSL N1-225 (ECII) (International Life Sciences Institute North America L. monocytogenes strain collection [11]). Genetic markers and molecular sizes corresponding to the amplified fragments are indicated on the right.
FIG. 2.
AscI PFGE profiles of the ECII and ECIII strains. Lanes: 1 to 3, profile 19, detected in the three ECII isolates included in cluster IX; 4 and 5, profiles 12 and 21, detected in the ECII sporadic isolates; 6 to 8, profiles 7A (lanes 6 and 7) and 7B (lane 8), detected in the three ECIII isolates included in cluster VI. Molecular sizes corresponding to XbaI-digested Salmonella serovar Braenderup H9812 DNA are given on the left.
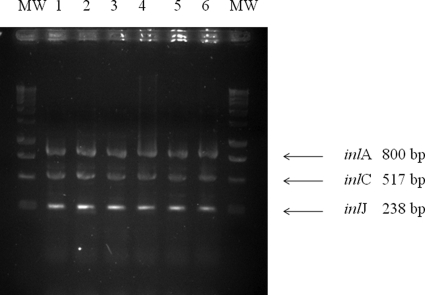
PCR of the virulence genes inlA, inlC, and inlJ identified the expected bands of 800, 517, and 238 bp in all strains under study (Fig. 3).
FIG. 3.
Agarose gel electrophoresis of inlA, inlC, and inlJ amplicons obtained from representative human isolates of L. monocytogenes. Lanes: MW (molecular weight), 1-kbp DNA ladder (Promega); 1 to 6, PCR products from human isolates analyzed in this study. Molecular sizes corresponding to the amplified fragments are given on the right.
Clustering of L. monocytogenes isolates.
Based on the results of PFGE, 11 occasions were found when strains of L. monocytogenes with the same subtype were isolated from more than one listeriosis case. A total of 39 out of 54 isolates (72.2%) were attributed to molecular subtype clusters, with each cluster consisting of two to seven isolates. Eight clusters comprised isolates from the Lombardy region only, whereas three clusters contained isolates from the two different geographic areas under study, i.e., Lombardy and the province of Florence, Tuscany. With the exception of clusters IV and X, the strains of L. monocytogenes within a single cluster had been isolated over an interval ranging from a minimum of zero days (i.e., the same day) to a maximum of 2 years. Cluster IV grouped together isolates identified in 1996 and 2005, and cluster X contained isolated identified in 2000 and 2007.
No epidemiological evidence was apparent for any putative cluster.
Tables 1 and 2 summarize the characteristics of cluster-associated and non-cluster-associated L. monocytogenes isolates, respectively.
DISCUSSION
Because of the complex epidemiology of L. monocytogenes, routine adoption of subtyping tools for the identification and tracking of epidemic clones and outbreak strains can contribute greatly to prevention and control activities. Indeed, timely identification of food-borne outbreaks/epidemics and transmission routes will likely continue to be a challenging task for public health departments due to the ubiquitous nature of this organism, its ability to persistently colonize food-processing plants, and its ability to multiply in foods at refrigeration temperatures. The epidemiological picture is often intricate because of the increasingly wider scale of commercialization of many food products and the evolving food habits associated with demographic, social, and economic changes. Finally, further difficulties arise from the protracted period of incubation of the listeriosis agent and the possibly space-time-scattered distribution of cases (7, 17, 22, 23).
PFGE typically provides high sensitivity for the identification of differences in molecular subtypes. In our experience, overall, 11 clusters were identified among 54 human isolates from two different geographic areas of Italy based on serotyping and AscI pulsotypes and clustering confirmation by ApaI. In previous studies, AscI PFGE has provided evidence to accurately cluster epidemiologically related isolates and to separate isolates from different outbreaks, whereas the use of more-discriminatory ApaI PFGE as a primary approach led in some instances to inaccurate separation of epidemiologically related isolates (15). Consequently, a large proportion of the human listeriosis cases under investigation could be grouped into molecular subtype clusters, some of which could represent common-source outbreaks. Furthermore, the geographic scale of these clusters appeared in some cases to be limited to a single region but in others to be multiregional, according to previous reports documenting that human listeriosis clusters can be widely distributed geographically (17, 22, 23). Two clusters, IV and X, included strains isolated over periods of approximately 8 and 7 years, respectively. The occurrence of two indistinguishable subtypes by chance only should be considered first as a possible explanation. However, considering that L. monocytogenes colonization can persist in food-processing plants for some years, the prolonged shelf lives of some food products, and the combined effects of these two factors, alternative hypotheses should not be discarded a priori (17). In any case, additional information apart from molecular subtyping data must be gathered before more-reliable epidemiological inferences can be drawn. A previous report on human listeriosis cases from eight different regions of Italy by Gianfranceschi et al. (12) also identified isolates with identical restriction patterns from different geographic sources but did not interpret this finding as evidence of correlation between them. However, such a conservative interpretation appears to be questionable in light of the frequent nationwide and often worldwide dissemination of food-borne pathogens, including L. monocytogenes.
Food vehicles had not been identified for any case of listeriosis, but cluster I, including four human isolates from Lombardy, retrospectively proved to contain isolates indistinguishable by serotyping and DNA-based methods from L. monocytogenes strain DUP-1034, previously detected on the rinds of Taleggio cheeses produced by an Italian plant (5, 21).
The detection of EC markers in both clustered and sporadic isolates is an intriguing finding. These clones, indeed, have been associated with major nationwide and international food-borne outbreaks, involving both the United States and some European countries (16). It seems reasonable to assume that previously identified epidemic clones will likely be involved in future food-borne events, perhaps because of their enhanced transmissibility, virulence, and/or persistence. Therefore, a rapid and reliable method to identify EC markers, which can provide useful clues to surveillance systems and public health policy makers, warrants further consideration. However, only 3 of the 11 clusters of strains possessed ECI, ECII, and ECIII markers, proving that these markers can be associated with several different subtypes. The detection of the ECII marker in three clustered and two unrelated strains by AscI PFGE warrants consideration, because strains of serotype 4b belonging to this clonal group have been associated with multistate outbreaks in 1998 to 1999 and in 2002 in the United States, but never in Europe, unlike the cosmopolitan clone ECI (4). ECII strains belonging to cluster IX appear to be very similar in their AscI PFGE patterns to the 1998 outbreak clone (3). This finding also confirms the specificity of detection of the ECII genetic marker by the multiplex PCR protocol of Chen and Knabel (2). On the other hand, our results confirm the observation about the heterogeneity of serotype 4b strains within the ECII clonal group published by Franciosa et al. (10). Indeed, that report, which describes the distribution of selected ECI and ECII markers among L. monocytogenes serotype 4b strains isolated in Italy from human, food, and environmental sources, discloses higher heterogeneity among strains exhibiting at least one of the three ECII markers SF7, 1365-66, and SF18 than among strains carrying ECI markers.
Attempts to improve the discrimination of virulent versus avirulent strains have been made for many years, although current understanding of the genome of L. monocytogenes does not allow for straightforward differentiation. The internalin genes inlC and inlJ, which are involved in passage through the intestinal barrier and the postintestinal stages of infection, performed well in discriminating strains that can or cannot cause mouse mortality via the intraperitoneal route (19). Although this does not automatically relate to the ability to produce disease in humans via the oral route, detection of the inlC and inlJ genes in all human isolates under study is an interesting preliminary finding that should be complemented by comparison with food and environmental isolates from the same geographic context.
This study has some limits. Because of the limited geographical sources of L. monocytogenes strains, any generalization of the results is questionable, and an overestimate of clustering has to be considered. Moreover, the lack of epidemiological information about possible food vehicles or food exposures that place consumers at risk makes it impossible to compare human and food isolates and to track their transmission chains.
The Scientific Panel on Biological Hazards of the European Food Safety Authority recently recommended that efforts to reduce risks to human health should focus on risk reduction practices both during the process of production of ready-to-eat foods and at home by consumers (9). The report also recommended further investigation of listeriosis cases and the generation and analysis of data on foods where L. monocytogenes is most commonly found. Additional areas for attention may include changes to food formulations, such as the addition of salt or other preservatives (13).
The recent increase in the number of listeriosis cases in Europe emphasizes the need for enhanced surveillance at the EU level to better estimate the burden of disease and the prevalence of this bacterium in the food chain. The results obtained in this study, despite the limits described above, may be helpful in reinterpreting the epidemiology of L. monocytogenes in Italy.
Complementing conventional with molecular epidemiology both in investigations of food-borne events and in surveillance systems is essential for the prevention and control of human listeriosis.
Acknowledgments
We are sincerely grateful to Martin Wiedmann, Cornell University, Ithaca, NY, for providing the diversity and outbreak strain sets of L. monocytogenes. We also thank the personnel of the hospital microbiological laboratories of Lombardy for facilitating the collection of isolates and providing clinical information.
This work was supported by a fund from the Italian University Minister (MIUR), Programmi di ricerca di interesse nazionale (PRIN), year 2005.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Bierne, H., C. Sabet, N. Personnic, and P. Cossart. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 91156-1166. [DOI] [PubMed] [Google Scholar]
- 2.Chen, Y., and S. J. Knabel. 2007. Multiplex PCR for simultaneous detection of bacteria of the genus Listeria, Listeria monocytogenes, and major serotypes and epidemic clones of L. monocytogenes. Appl. Environ. Microbiol. 736299-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Y., and S. J. Knabel. 2008. Prophages in Listeria monocytogenes contain single-nucleotide polymorphisms that differentiate outbreak clones within epidemic clones. J. Clin. Microbiol. 461478-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y., W. Zhang, and S. J. Knabel. 2005. Multi-virulence-locus sequence typing clarifies epidemiology of recent listeriosis outbreaks in the United States. J. Clin. Microbiol. 435291-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Cesare, A., G. Manfreda, M. Macrì, and C. Cantoni. 2007. Application of automated ribotyping to support the evaluation of Listeria monocytogenes sources in a Taleggio cheese producing plant. J. Food Prot. 701116-1121. [DOI] [PubMed] [Google Scholar]
- 6.Denny, J., and J. McLauchlin. 2008. Human Listeria monocytogenes infections in Europe—an opportunity for improved European surveillance. Euro Surveill. 13(13)pii=8082. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8082. [PubMed] [Google Scholar]
- 7.de Valk, H., V. Vaillant, C. Jacquet, J. Rocourt, F. Le Querrec, F. Stainer, N. Quelquejeu, O. Pierre, V. Pierre, J. C. Desenclos, and V. Goulet. 2001. Two consecutive nationwide outbreaks of listeriosis in France, October 1999-February 2000. Am. J. Epidemiol. 154944-950. [DOI] [PubMed] [Google Scholar]
- 8.Drevets, D. A., and M. S. Bronze. 2008. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 53151-165. [DOI] [PubMed] [Google Scholar]
- 9.EFSA Panel on Biological Hazards (BIOHAZ). 2007. Request for updating the former SCVPH opinion on Listeria monocytogenes risk related to ready-to-eat foods and scientific advice on different levels of Listeria monocytogenes in ready-to-eat foods and the related risk for human illness. Scientific Opinion of the Panel on Biological Hazards. http://www.efsa.europa.eu/EFSA/efsa_locale-1178620753812_1178680093176.htm.
- 10.Franciosa, G., C. Scalfaro, A. Maugliani, F. Floridi, A. Gattuso, S. Hodzic, and P. Aureli. 2007. Distribution of epidemic clonal genetic markers among Listeria monocytogenes 4b isolates. J. Food Prot. 70574-581. [DOI] [PubMed] [Google Scholar]
- 11.Fugett, E., E. Fortes, C. Nnoka, and M. Wiedmann. 2006. International Life Sciences Institute North America Listeria monocytogenes strain collection: development of standard Listeria monocytogenes strain sets for research and validation studies. J. Food Prot. 692929-2938. [DOI] [PubMed] [Google Scholar]
- 12.Gianfranceschi, M., A. Gattuso, M. D'Ottavio, S. Fokas, and P. Aureli. 2007. Results of a 12-month long enhanced surveillance of listeriosis in Italy. Euro Surveill. 12(11)E7-E8. [DOI] [PubMed] [Google Scholar]
- 13.Goulet, V., C. Hedberg, A. Le Monnier, and H. de Valk. 2008. Increasing incidence of listeriosis in France and other European countries. Emerg. Infect. Dis. 14734-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 6555-62. [DOI] [PubMed] [Google Scholar]
- 15.Graves, L. M., S. B. Hunter, A. R. Ong, D. Schoonmaker-Bopp, K. Hise, L. Kornstein, W. E. DeWitt, P. S. Hayes, E. Dunne, P. Mead, and B. Swaminathan. 2005. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J. Clin. Microbiol. 432350-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 651811-1829. [DOI] [PubMed] [Google Scholar]
- 17.Keto-Timonen, R., R. Tolvanen, J. Lundén, and H. Korkeala. 2007. An 8-year surveillance of the diversity and persistence of Listeria monocytogenes in a chilled food processing plant analyzed by amplified fragment length polymorphism. J. Food Prot. 701866-1873. [DOI] [PubMed] [Google Scholar]
- 18.Lavi, O., Y. Louzoun, and E. Klement. 2008. Listeriosis: a model for the fine balance between immunity and morbidity. Epidemiology 19581-587. [DOI] [PubMed] [Google Scholar]
- 19.Liu, D., M. L. Lawrence, A. J. Ainsworth, and F. W. Austin. 2007. Toward an improved laboratory definition of Listeria monocytogenes virulence. Int. J. Food Microbiol. 118101-115. [DOI] [PubMed] [Google Scholar]
- 20.Liu, D., M. L. Lawrence, F. W. Austin, and A. J. Ainsworth. 2007. A multiplex PCR for species- and virulence-specific determination of Listeria monocytogenes. J. Microbiol. Methods 71133-140. [DOI] [PubMed] [Google Scholar]
- 21.Mammina, C., G. Manfreda, A. Aleo, A. De Cesare, V. Ferretti, N. Pellissier, C. Romani, A. Nastasi, and M. Pontello. 2008. Cluster of human listeriosis cases traced by molecular typing to taleggio cheese, Italy, abstr. P1692. 18th Eur. Cong. Clin. Microbiol. Infect. Dis. (ESCMID), Barcelona, Spain, 19 to 22 April 2008. http://www.blackwellpublishing.com/eccmid18/abstract.asp?id=69590.
- 22.Mead, P. S., E. F. Dunne, L. Graves, M. Wiedmann, M. Patrick, S. Hunter, E. Salehi, F. Mostashari, A. Craig, P. Mshar, T. Bannerman, B. D. Sauders, P. Hayes, W. Dewitt, P. Sparling, P. Griffin, D. Morse, L. Slutsker, and B. Swaminathan for the Listeria Outbreak Working Group. 2006. Nationwide outbreak of listeriosis due to contaminated meat. Epidemiol. Infect. 134744-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen, S. J., M. Patrick, S. B. Hunter, V. Reddy, L. Kornstein, W. R. MacKenzie, K. Lane, S. Bidol, G. A. Stoltman, D. M. Frye, I. Lee, S. Hurd, T. F. Jones, T. N. LaPorte, W. Dewitt, L. Graves, M. Wiedmann, D. J. Schoonmaker-Bopp, A. J. Huang, C. Vincent, A. Bugenhagen, J. Corby, E. R. Carloni, M. E. Holcomb, R. F. Woron, S. M. Zansky, G. Dowdle, F. Smith, S. Ahrabi-Fard, A. R. Ong, N. Tucker, N. A. Hynes, and P. Mead. 2005. Multistate outbreak of Listeria monocytogenes infection linked to delicatessen turkey meat. Clin. Infect. Dis. 40962-967. [DOI] [PubMed] [Google Scholar]
- 24.Ooi, S. T., and B. Lorber. 2005. Gastroenteritis due to Listeria monocytogenes. Clin. Infect. Dis. 401327-1332. [DOI] [PubMed] [Google Scholar]
- 25.Sauders, B. D., E. D. Fortes, D. L. Morse, N. Dumas, J. A. Kiehlbauch, Y. Schukken, J. R. Hibbs, and M. Wiedmann. 2003. Molecular subtyping to detect human listeriosis clusters. Emerg. Infect. Dis. 9672-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauders, B. D., Y. Schukken, L. Kornstein, V. Reddy, T. Bannerman, E. Salehi, N. Dumas, B. J. Anderson, J. P. Massey, and M. Wiedmann. 2006. Molecular epidemiology and cluster analysis of human listeriosis cases in three U.S. states. J. Food Prot. 691680-1689. [DOI] [PubMed] [Google Scholar]
- 27.Schlech, W. F., III. 2000. Foodborne listeriosis. Clin. Infect. Dis. 31770-775. [DOI] [PubMed] [Google Scholar]
- 28.Swaminathan, B., and P. Gerner-Smidt. 2007. The epidemiology of human listeriosis. Microbes Infect. 91236-1243. [DOI] [PubMed] [Google Scholar]
- 29.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85524-531. [PubMed] [Google Scholar]