Abstract
Rotavirus infections can be diagnosed in stool samples by serological and molecular methods. We developed a novel reverse transcriptase PCR (RT-PCR) method for the amplification of rotavirus RNA and a reverse hybridization assay on a strip to detect amplimers and identify the specific G and P genotypes present in human stool specimens. An additional aim was to permit specific identification of the rotavirus G1P[8] strain, used in the Rotarix vaccine. Novel broad-spectrum PCR primers were developed for both VP4 and VP7, permitting the amplification of a wide range of rotavirus genotypes. Primer sets comprise mixtures of defined primer sequences. For the identification of G and P genotypes, two reverse hybridization strip assays were developed. Both the VP4 and the VP7 strip contain universal probes for the detection of VP4 and VP7 sequences, irrespective of the G or P genotype. The VP4 strip contains type-specific probes for P[4], P[6], P[8], P[9], and P[10]. The VP7 strip contains type-specific probes for G1, G2, G3, G4, G5, G6, G8, and G9. In addition, probes to distinguish between wild-type G1 and G1 vaccine strain sequences were present. Testing by analysis of multiple reference strains confirmed that both RT-PCR methods allowed the detection of a broad spectrum of genotypes. RT-PCR for VP7 was more sensitive than RT-PCR for VP4, but all samples identified as positive for rotavirus antigen by an enzyme-linked immunosorbent assay (ELISA) were also positive for both VP4 and VP7. The high specificity of the reverse hybridization method was confirmed by sequence analysis as well as by type-specific PCR, and the vaccine strain could also be specifically identified. The reverse hybridization method permits accurate identification of mixed infections with different genotypes. Rotavirus genotypes for which no type-specific probes were present on the strip were adequately identified by the universal detection probes. The assay was formally validated by analyses of specificity, sensitivity, precision, accuracy, and robustness. In a panel of 149 ELISA-positive stool samples, comparison with conventional type-specific RT-PCR methods revealed the superiority of the novel method, mainly in cases of mixed rotavirus infections. This novel method permits highly accurate detection and identification of human rotavirus infections in stool samples. This validated assay could be useful for large-scale epidemiological and clinical trials.
Rotavirus infection is one of the most important causes of severe diarrhea in infants and young children. Worldwide, an estimated 611,000 children below the age of 5 years die as a consequence of rotavirus disease, mainly in low-income countries, and rotavirus accounts for 39% of hospitalizations for childhood diarrhea (37).
Rotavirus-induced illnesses most commonly affect children between the ages of 6 and 24 months, and the peak prevalence of the disease generally occurs during the cooler months in temperate climates and year-round in tropical areas. Human rotaviruses (HRV) are typically transmitted from person to person by the fecal-oral route, with an incubation period of 1 to 3 days. The disease is characterized by vomiting, watery diarrhea, fever, and abdominal pain. In contrast to the severe disease normally encountered in young children, most adults are protected as a result of previous rotavirus infection, so most adult infections are mild or asymptomatic (2).
Rotaviruses, classified as a genus in the family Reoviridae, are nonenveloped, double-shelled viruses, about 75 nm in diameter, that have a characteristic wheel-like appearance. Typically, the double-shelled capsid structure surrounds an inner protein shell or core that contains the viral genome. The genome of rotaviruses consists of 11 segments of double-stranded RNA, encoding 6 structural and 5 nonstructural proteins (12). One of the nonstructural proteins of relevance, NSP4, is a transmembrane, endoplasmic-reticulum-specific glycoprotein with pleiotropic functions in viral replication and pathogenesis. NSP4, encoded by gene segment 10 of group A rotavirus, is an enterotoxin causing diarrhea and is described as an activator of a signal transduction pathway that increases intracellular calcium levels by mobilizing calcium from the endoplasmic reticulum, resulting in chloride secretion (51).
Two of the structural viral proteins, designated VP4 and VP7, are arranged on the exterior of the double-shelled capsid structure. VP4, a spike protein, is the translational product of genomic segment 4, whereas VP7 belongs to the outer capsid and is the translational product of genomic segment 7, 8, or 9, depending on the strain (48). The inner capsid of the rotavirus presents the protein designated VP6. This protein determines the antigenic group and subgroup classifications of a rotavirus. More specifically, antigenicity is classified into five major VP6 groups (A to E) and four subgroups within VP6 group A (I, II, I plus II, neither I nor II). Furthermore, the VP4 and VP7 proteins are the determinants of the serotype specificity of group A rotaviruses. VP4 protein, designated P, is a nonglycosylated protease-sensitive protein of approximately 88 kDa. VP7 protein, a glycoprotein of 38 kDa (34 kDa when nonglycosylated) designated G, stimulates the formation of the major neutralizing antibody following rotavirus infection.
To date, at least 19 G serotypes and 27 P serotypes have been defined. In general, strains sharing more than 89% amino acid identity are considered to belong to the same P genotype. The nomenclatures for G genotypes and serotypes are identical (G followed by a number), but the numbers indicating P genotypes are enclosed in brackets, while those for serotypes are not (34). Since the VP4 and VP7 genes are independently segregated, different G and P combinations have been observed in natural infections. Based on global epidemiology data, G1P[8], G2P[4], G3P[8], G4P[8], G9P[6], and G9P[8] are the most prevalent genotypic combinations found in humans (21, 44). Other genotypes are often found in animals, although transmission to humans is possible, and the spectrum of genotypes appearing in humans is increasing (15, 27, 30, 31, 38).
Routine diagnosis is based on rapid detection of group A antigen in feces, generally by latex agglutination or an enzyme immunoassay (11, 13). However, the abilities of serological assays to distinguish accurately between the different rotavirus serotypes are limited (23). Therefore, analysis of the viral RNA is more suited for the genotyping of rotavirus isolates. Rotavirus genomic RNA can be detected by reverse transcriptase PCR (RT-PCR). Despite the high degrees of sequence heterogeneity within the VP4 and VP7 genes, distinct phylogenetic clades can be clearly recognized, permitting the identification of P genotypes and G genotypes in the VP4 and VP7 genes, respectively (34).
Various methods have been developed to detect rotavirus RNA in human stool samples and to identify specific genotypes. These include type-specific PCR (20), restriction fragment length polymorphism (24), sequence analysis (3, 5, 10), capture and primer extension (32), and hybridization to oligonucleotide probes (7, 43, 44).
An attenuated G1P[8] HRV vaccine has been developed and tested on a large group of infants (41). Two doses of this Rotarix vaccine were highly effective in protecting infants against severe rotavirus gastroenteritis.
In order to detect the presence of rotavirus in human stool samples and to identify the VP4 and VP7 genotypes (including those of the Rotarix vaccine strain), a novel diagnostic method was developed. This assay is based on RT-PCR amplifying a broad-spectrum of G and P genotypes, followed by identification of the amplimers by reverse hybridization to a strip containing both universal and type-specific oligonucleotide probes. The present report describes the development and analytical validation of this novel assay.
MATERIALS AND METHODS
Reference rotavirus strains.
Reference strains of HRV used in this study are shown in Table 1 (13).
TABLE 1.
Rotavirus reference strains
| Strain | G and P genotypes | FFU/ml | Genotype for which the strain is used as a reference
|
|
|---|---|---|---|---|
| VP7 | VP4 | |||
| RIX4414 | G1P[8]vac | 2.00 × 107 | G1vac | P[8]vac |
| WA | G1P[8]WT | 6.94 × 106 | G1WT | P[8]WT |
| WI61 | G9P[6] | 1.43 × 107 | G9 | P[6] |
| DS-1 | G2P[4] | 106 | G2 | P[4] |
| P | G3P[8] | 106 | G3 | |
| K8 | G1P[9] | 5 × 107 | P[9] | |
| 69M | G8P[10] | 2.51 × 107 | G8 | P[10] |
| ST3 | G4P[6] | 3.16 × 106 | G4 | |
| NCDV | G6bovine | 1.86 × 108 | G6 | |
| TF4-41 | G5porcine | 106 | G5 | |
A live attenuated RIX4414 G1P[8] HRV vaccine strain was used for the preparation of the Rotarix vaccine (Rotarix is a trademark of the GlaxoSmithKline group of companies) (41). The RIX4414 strain has been deposited at the European Collection of Cell Cultures under accession number ECACC 99081301.
Clinical samples.
A total of 149 stool samples were obtained from participants in a phase II vaccine study. Rotavirus was isolated from the stool samples as follows. One gram of stool was mixed with 4 ml Earle's balanced salt solution (EBSS) by vigorous vortexing followed by shaking for 20 min. The stool suspension was centrifuged at 12,500 × g at 4°C for 5 min; the supernatant was transferred to an empty tube; and the pellet was discarded. The supernatant, containing the rotavirus, was stored for the short term (as long as 90 days) at 4°C and for the long term at −20°C.
RNA isolation.
Total nucleic acid was extracted from 200 μl EBSS supernatant using the Roche MagNAPure LC automated system and the MagNAPure LC DNA isolation kit III. Briefly, viruses were first lysed by warming at 65°C and addition of lysis buffer to the sample before it was mixed with silica-coated paramagnetic particles. These particles selectively bind nucleic acids. After separation of the paramagnetic particles from the extract by using a magnet, followed by several washing steps, RNA was eluted from the paramagnetic particles. A rotavirus-positive sample (genotype G8P[10]) was used as a positive control.
RT-PCR.
Ten microliters of isolated VP4 and VP7 RNA was reverse transcribed into cDNA using the OneStep RT-PCR kit (Qiagen).
For the amplification of VP4 sequences, the VP4 primer set, a cocktail of six forward and three reverse primers, generating a fragment of 500 bp, was used (Table 2). For the amplification of VP7 sequences, the VP7 primer set, a cocktail of six forward and five reverse primers, resulting in an amplification product of 400 bp, was used (Table 3). After denaturation of the RNA at 80°C for 5 min, the RT-PCR comprised a reverse transcription step at 50°C for 30 min, a denaturation step of 15 min at 95°C, 40 cycles of PCR (94°C for 30 s, 52°C for 45 s, and 72°C for 45 s), and a final incubation at 72°C for 10 min. All reverse primers carry a biotin moiety at the 5′ end, allowing labeling of the amplified material.
TABLE 2.
Primers used for amplification of the VP4 gene fragment of HRV
| Primer designation | Sequence (5′→3′)a | Positionb |
|---|---|---|
| ROT4FOR1A | AGATGGTCCTTATCAGCCAAC | 204-224 |
| ROT4FOR1B | GANGGTCCTTATCAGCCTAC | 205-224 |
| ROT4FOR1C | AGATGGTCCTTATCAACCTAC | 204-224 |
| ROT4FOR1D | CGATGGTCCTTATCAACCAAC | 204-224 |
| ROT4FOR1E | GACGGACCATATCAACCGAC | 205-224 |
| ROT4FOR1F | GATGGTCCNTATCAACCNAC | 205-224 |
| ROT4REV1B | TTNCTTGTATTCTGNATTGGTG | 701-680 |
| ROT4REV1A | CATTTCTAGTATTTTGAATTGGTG | 703-680 |
| ROT4REV1C | CATTTCTAGTGTTTTGTATCGGT | 703-681 |
The letter N stands for inosine.
All sequence positions are given according to the sequence of the P[8] strain WA (GenBank accession number L34161).
TABLE 3.
Primers used for amplification of the VP7 gene fragment of HRV
| Primer designation | Sequence (5′→3′)a | Positionb |
|---|---|---|
| ROTFOR1A | AATGAATGGTTATGTAATCCAATGG | 529-553 |
| ROTFOR1B | AANGAATGGCTGTGCAATCCNATGG | 529-553 |
| ROTFOR1C | AANGAGTGGTTATGTAATCCNATGG | 529-553 |
| ROTFOR1D | AATGAATGGTTATGNAACCCAATGG | 529-553 |
| ROTFOR1E | AATGAGTGGCTTTGTAATCCAATGG | 529-553 |
| ROTFOR1F | AATCAATGGTTATGTAATCCGATGG | 529-553 |
| VP7rev4a | GCCACCATTTTTTCCAATTCAC | 928-907 |
| VP7rev4b | GCCACCATTTTTTCCAATTTAT | 928-907 |
| VP7rev4c | GCCACCATTTTTTCCAATTTAC | 928-907 |
| VP7rev4d | GCCACCATTTCTTCCAATTAAC | 928-907 |
| VP7rev4e | GCCACCATTTTTTCCAGTTCAC | 928-907 |
The letter N stands for inosine.
All sequence positions are given according to the sequence of the G1 strain WA (GenBank accession number M21843).
Reverse hybridization.
Genotype-specific oligonucleotide probes ( Tables 4 and 5) were provided with a 3′ poly(T) tail using terminal deoxynucleotidyltransferase and were immobilized in parallel lines on nitrocellulose membrane strips. Reverse hybridization was performed as follows. Briefly, 10 μl of the biotin-labeled RT-PCR product was denatured by addition of 10 μl of 0.1 M NaOH solution at room temperature. Hybridization solution and a reverse hybridization strip were added and incubated at 50°C for 1 h, allowing hybridization of the denatured PCR product to probes on the strip under stringent conditions. After stringent washing at 50°C, the hybrids were detected by an alkaline phosphatase-streptavidin conjugate and a substrate (5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium), resulting in a purple precipitate at the positive probe lines. After the strips were dried, the hybridization patterns were interpreted visually.
TABLE 4.
Probes for identification of rotavirus VP4 types
| Probe designation | Sequence (5′→3′)a | Positionb |
|---|---|---|
| Uniprobes | ||
| VP4-uni_1 | TAAACCATTATTAATATATTCATTACANTTAGANTCTTG | 678-640 |
| VP4-uni_2 | ACCGTTGTTAATATATTCATTACANTTAGANTCTTG | 675-640 |
| VP4-uni_3 | TTGGTGGNAANCCAGTATTTATATATTCA | 685-657 |
| VP4-uni_4 | TTGGTGGTAACCCNTTATTTATGTANTCAG | 685-656 |
| VP4-uni_5 | GGTGGTAAACCATTATTTATATATTGTGTACACATAG | 683-647 |
| VP4-uni_6 | TTGGAGGCANCCCATTATTTATATATTGCGC | 685-655 |
| Type-specific probes | ||
| P1_3_1 | CAAATAGGTGGTTAGCGA | 305-322 |
| P1_3_2 | CAAACAGATGGTTAGCGA | 305-322 |
| P 4_1_1c | AATAGACTTGTAGGAATGCTAA | 490-511 |
| P 4_2c | TTCGAAATGTTTAAAGGTAGC | 427-447 |
| P6_3 | TATAATAGTGTTTGGACTTTC | 517-537 |
| P8_VAC-str1b | CTGATACCAGACTTGTAGGA | 485-504 |
| P8_VAC-str2b | GTAGGAATATTTAAATATGGTG | 499-520 |
| P 8_1_2b | ACCACCTACTGATTACTGG | 234-252 |
| P 8_4_1a | ACCCAGTAGATAGACAATATATG | 347-369 |
| P 8_4_3a | GATCCAGTAGATAGACAATATAATG | 346-370 |
| P 9_1c | ACTTCGTGGAAATTTATATTATT | 415-437 |
| P_9_2d | AGATGGGCAAAATGTCCAAGG | 372-392 |
| P 10b | AGCCCCATTAAATGCTG | 261-277 |
The letter N stands for inosine.
All sequence positions are given according to the sequence of the P[8] strain WA (GenBank accession number L34161).
TABLE 5.
Probes for identification of rotavirus VP7 types
| Probe designation | Sequence (5′→3′)a | Positionb |
|---|---|---|
| Uniprobes | ||
| Rota-uni_2 | ATTAGACATAACAGCAGATCCAACGAC | 852-878 |
| Rota-uni_3 | TAGATATCACTGCTGATCCAACAAC | 854-878 |
| Rota-uni_4 | ATTAGATATAACGGCTGATCCCACAAC | 852-878 |
| Rota-uni_5 | TAGATATTACAGCTGATCCGACGAC | 854-878 |
| Rota-uni_6 | TTGGATATTACNGCNGATCCAACAAC | 853-878 |
| Type-specific probes | ||
| G1vac-605T1 | GAATACGCAAATGTTAGG | 642-659 |
| G1WT-605C3 | AATACGCAAACGTTAGGA | 643-661 |
| G1WT-605aC4 | ACTGAATACACAAACGTTAG | 639-658 |
| G1WT-605taC1 | CTGAATATACAAACGTTAGG | 640-659 |
| G1WT-1_2 | TTAGCTATAGTGGATGTCGGG | 718-739 |
| G1WT-1a_10 | AAATTAGCTATAGTAGATGTCGGG | 715-739 |
| G1WT-2_3 | CGTTGATGGGATAAATCAT | 735-753 |
| G1WT-2t_10 | CGTTGATGGGATAAATTAT | 735-753 |
| G2_11 | AGAAAATGTTGCTATAATTCA | 813-833 |
| G3_11 | AGCAGTTATACAGGTTGGT | 822-840 |
| G3B_1 | GGCGGTCATACAAGTT | 822-837 |
| G4_11 | CAGCTACTTTTGAAACAGTT | 683-702 |
| G4VA70-3 | AGCTACTTTTGAAATGGTG | 684-702 |
| G4_Arg1 | CAGCTACTTTTGAAACGGGG | 683-702 |
| G5_2 | TATCTATGGGTTCTTCATGG | 602-624 |
| G6_2 | CTCGGTATCGGATGTCT | 655-671 |
| G8_n10 | ACTACAACTTTTGAAGAAGTTGC | 682-704 |
| G8_n21 | ACACTACGACTTTTGAAGAA | 680-699 |
| G9_1 | ATGGGACAGTCTTGTAC | 607-623 |
N stands for inosine.
All sequence positions are given according to the sequence of the G1 strain WA (GenBank accession number M21843).
Two reverse hybridization strip assays (RHA), designated the Rota VP7 and Rota VP4 strips, were developed for identification of G and P genotypes, respectively. Both the VP7 and VP4 strips contain universal probes for the detection of VP7 and VP4 sequences, irrespective of the G or P genotype. These would permit the detection of rotavirus genotypes for which the strip does not contain type-specific probes.
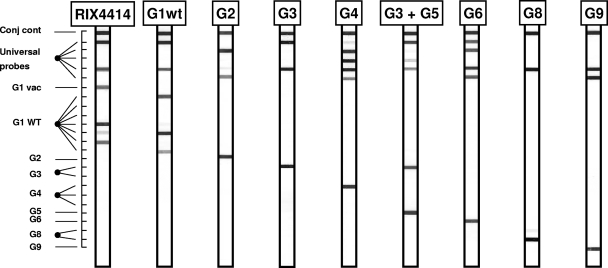
The VP7 strip (Fig. 1) contained type-specific probes for G1, G2, G3, G4, G5, G6, G8, and G9. In addition, probes to distinguish between wild-type and vaccine strain G1 sequences (G1WT and G1vac sequences, respectively) were present. The live attenuated HRV G1P[8] vaccine strain RIX4414 can be identified by a specific probe (G1vac) that targets a unique single-nucleotide difference from G1WT sequences at position 605 (605C in the wild-type strains; 605T in the vaccine strain).
FIG. 1.
Outline and representative examples of the VP7 RHA. The top line (Conj cont) is a control line for checking the detection reaction on the strip. The positions of the universal probe lines as well as those for type-specific probes are indicated. The VP7 genotype can be deduced from the hybridization pattern and is given at the top of each strip.
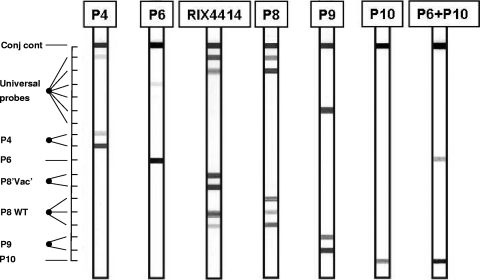
The VP4 strip (Fig. 2) contained type-specific probes for P[4], P[6], P[8], P[9], and P[10]. Two probes were added to recognize the P[8] sequence present in the vaccine strain. It should be noted that this sequence is not completely unique to the vaccine strain, since it also occurs in some nonvaccine strains.
FIG. 2.
Outline and representative examples of the VP4 RHA. The top line (Conj cont) is a control line for checking the detection reaction on the strip. The positions of the universal probe lines as well as those for type-specific probes are indicated. The VP4 genotype can be deduced from the hybridization pattern and is given at the top of each strip.
If the strip yields positive results only for the universal VP4 or VP7 probes, the sample is rotavirus RNA positive, but the probes on the strip do not permit identification of the correct VP4 or VP7 genotype. Therefore, such amplimers are subjected to sequence analysis.
Sequence analysis.
PCR amplimers were analyzed by direct sequencing using the ABI BigDye Terminator cycle sequencing kit (version 1.1; Applied Biosystems). Sequences were analyzed by comparison to sequences from rotavirus genotype reference strains. Sequences were also compared to rotavirus sequences in GenBank by using BLAST software (1).
Genotype-specific PCR.
As a reference method, genotype-specific RT-PCR was used, based on the RT-PCR methods for G typing of Gault et al. and Gouvea et al. (19, 22) and on the RT-PCR technique for P typing of Khetawat et al. (29).
Briefly, RNA was subjected to individual RT-PCRs with combinations of universal reverse primers and type-specific forward primers. Amplimers were analyzed by gel electrophoresis, and genotypes could be deduced based on amplimer length.
RESULTS
Design of the Rota VP7 and VP4 assays.
Sequences from nine different HRV VP7 genotypes (including G1, G2, G3, G4, G5, G6, G8, and G9) were aligned in order to identify conserved genomic regions suitable for the design of broad-spectrum RT-PCR primers. Based on this sequence alignment, target regions for forward and reverse primers for the broad-spectrum VP7 RT-PCR were selected. Six forward primers were aimed at the conserved region of VP7 between positions 529 and 553 (all positions for VP7 are given according to those of the G1 strain WA [GenBank accession number M21843]). Five reverse primers were aimed at the conserved region between positions 907 and 927. Forward and reverse primers were designed to cover all relevant genotypes, and some primers contain one or two inosines (primer degeneracy). This cocktail of 11 primers is called the VP7 primer set. The expected amplimer size is 400 bp. Subsequently, universal probes, aimed at the region between positions 852 and 878, were designed. Each of the nine HRV VP7 genotypes should hybridize to at least one of the universal VP7 probes. Type-specific probes, aimed at target sequences located in the interprimer region, were selected and used on the reverse hybridization strip. Examples of the VP7 assay are shown in Fig. 1.
Sequences from six different HRV VP4 genotypes (P[1], P[4], P[6], P[8], P[9], and P[10]) were aligned to identify conserved genomic regions suitable for the design of broad-spectrum RT-PCR primers. Based on this sequence alignment, target regions for forward and reverse primers were chosen for the broad-spectrum VP4 RT-PCR. Six forward primers were aimed at the conserved region of VP4 between positions 204 and 224 (all positions for VP4 are given according to those of the P[8] strain WA [GenBank accession number L34161]). Three reverse primers were aimed at the conserved region between positions 680 and 703. Forward and reverse primers were designed to cover all relevant genotypes, and some contain one or two inosines (primer degeneracy). This cocktail of nine primers is called the VP4 primer set. The expected amplimer size is 500 bp. Subsequently, universal probes, aimed at the region between positions 640 and 685, were designed. Each of the six HRV VP7 genotypes should hybridize to at least one of the universal VP4 probes. Type-specific probes, aimed at target sequences located in the interprimer region, were selected and used on the reverse hybridization strip. The results of the VP4 assay are shown in Fig. 2.
In order to achieve high specificity for all probes on the strips under the uniform RHA conditions, the length and composition of each probe were optimized.
Specificities of the VP4 and VP7 assays.
The specificities of the VP4 and VP7 assays were first assessed by comparing the primer and probe sequences to the GenBank database (performed in April 2004) using BLAST software (1). This in silico analysis confirmed the specificities of all primers and probes, Probe G1vac was specifically aimed at recognizing the VP7 sequence of the vaccine strain. The C605T variant was not found in GenBank; therefore, this probe sequence appears completely specific for the identification of the vaccine strain.
Probes P8vac-WT1 and P8vac-WT2 were aimed at recognizing specifically the VP4 P[8] sequence of the vaccine strain. However, GenBank contained several VP4 P[8] wild-type sequences fully matching the VP4 vaccine strain sequence. These strains were first isolated in Finland (35). Thus, the VP4 P[8] sequence of the vaccine strain is not completely unique, since it also occurs in some natural strains. In conclusion, the vaccine strain can be reliably identified only by the VP7 G1vac sequence, and the VP4 P[8] sequence should be used for confirmation only when the VP7 result indicates the presence of the vaccine strain.
The specificities of both the VP4 and VP7 assays were experimentally tested by analysis of reference strains representing established G and P genotypes. All reference strains (listed in Table 1) were correctly identified on the strips, confirming the specificity of the genotyping probes on the strips (Fig. 1 and 2). Amplimers from each reference strain were also sequenced and confirmed the presence of the expected VP4 or VP7 genotypes.
To assess potential cross-reactivity of the VP4 and VP7 RT-PCR assays with other gastrointestinal pathogens, the rotavirus vaccine strain (G1vacP[8]vac; 50 focus-forming units [FFU] per RT-PCR) was mixed with various enteroviruses included in the third Quality Control for Molecular Diagnostics enterovirus panel (www.qcmd.org) or with poliovirus (Sabin strain, produced at GlaxoSmithKline Biologicals, Rixensart, Belgium). The results (data not shown) indicate that these organisms do not interfere with the detection and genotyping of the HRV G1vacP[8]vac strain by the VP4 and VP7 RT-PCR RHAs.
For assessment of the specificity of identification of the G1 vaccine strain, high concentrations of PCR amplimers from two different cloned G1WT sequences were used for reverse hybridization in order to determine whether this could cause cross-reactivity on the G1vac probe. Only specific hybridization patterns were observed, without any visible cross-reaction (data not shown).
Furthermore, experimental mixtures of G1vac and G1WT amplimers were tested. PCR products were generated from plasmid DNA containing VP7 sequences from either the G1WT or the G1vac strain by using the VP7 primer set. Mixtures with different relative concentrations were prepared, leaving the total amount of amplimer constant, and these mixtures were used for reverse hybridization and direct sequence analysis.
RHA allowed the detection of the G1vac strain in samples with G1vac-G1WT mixtures in 50:50, 25:75, and 10:90 ratios. In mixtures containing less G1vac (5:95 and 1:99 ratios), only G1WT was observed on the strip. Direct sequencing detected only G1vac in samples with G1vac-G1WT mixtures at 50:50 and 25:75 ratios, but in the 10:90, 5:95, and 1:99 mixtures, only G1WT could be identified.
Sensitivities of the assays.
In order to determine the sensitivities of the assays, dilution series of the following reference strains (genotypes) were used: RIX4414 (G1P[8]vac), WA (G1P[8]WT), WI61 (G9P[6]), and ST3 (G4P[6]). The choice of these strains was based on the high prevalence of these genotypes around the world (21). These strains were quantified by virus titration after culture, and virus levels were expressed in focus-forming units.
The limit of detection (LOD) was determined by testing 10-fold dilutions of these strains in EBSS in four different runs, using six replicates per dilution, yielding 24 test results per dilution. From these data, the smallest amount of virus detected in 95% of the assays was calculated as the LOD, and values for each genotype are shown in Table 6. The sensitivity of the RT-PCR assay ranged from 1.3 FFU per PCR to 3.5 × 10−3 FFU per PCR, depending on the genotype of the virus. In general, the VP4 PCR was less sensitive than the VP7 PCR.
TABLE 6.
LODs of the VP7 and VP4 RT-PCR RHAs
| Genotype | LOD (FFU of HRV)/RT-PCR RHA for:
|
|
|---|---|---|
| VP7 | VP4 | |
| G1vacP8vac | ∼ 1.3 × 10−2 | ∼ 1.2 × 10−1 |
| G1WTP8WT | ∼ 3.5 × 10−3 | ∼ 2 × 10−2 |
| G9P6 | ∼ 6.4 × 10−2 | ∼1.3 |
| G4P6 | ∼ 10−1 | NDa |
ND, not determined.
Accuracy of the assay.
In order to assess the accuracy of the VP7 and VP4 RT-PCR RHAs, a panel of human stool samples was tested. This panel consists of 149 stools that tested positive for rotavirus by an enzyme-linked immunosorbent assay (ELISA) for HRV antigen. These samples were analyzed under code by the novel VP4 and VP7 RT-PCRs and reverse hybridization as well as by conventional type-specific PCR methods (20, 23, 30). It should be noted that the conventional type-specific PCRs do not allow discrimination between the G1 vaccine strain and G1 WT strains. The results are summarized in Tables 7 to 9. Comparison of the results obtained by the two methods showed that for G typing, 88% of the results were identical (exactly the same genotype by both methods), 9% were compatible (at least one HRV genotype in common), and 3% were discordant. All discordant cases were due to negative results obtained by the conventional type-specific RT-PCR and positive typing results found by the novel RHA.
TABLE 7.
Rotavirus VP7 genotyping results for 149 stool samples, obtained by conventional type-specific PCR and the novel PCR-reverse hybridization method
| Genotype by TS PCRa | No. of isolates with the following VP7 genotype by broad-spectrum RT-PCR and reverse hybridization:
|
Total | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1vac | G1WT | G1WT + G2 | G1WT + G2 + G3 | G1WT + G4 | G1WT + G9 | G2 | G2 + G3 | G2 + G4 | G2 + G4 + G9 | G3 | G1 + G2 + G3 | G4 | G9 | ||
| G1vac | 20 | 20 | |||||||||||||
| G1WT | 71 | 2 | 1 | 1 | 75 | ||||||||||
| G1WT + G2 | 1 | 1 | 2 | ||||||||||||
| G1WT + G9 | 2 | 2 | |||||||||||||
| G1 + G9 | 1 | 1 | |||||||||||||
| G2 | 1 | 7 | 8 | ||||||||||||
| G3 | 1 | 1 | 6 | 8 | |||||||||||
| G4 | 1 | 1 | 11 | 13 | |||||||||||
| G9 | 16 | 16 | |||||||||||||
| Negative | 1 | 1 | 1 | 1 | 4 | ||||||||||
| Total | 21 | 72 | 4 | 1 | 1 | 1 | 8 | 1 | 1 | 1 | 6 | 1 | 11 | 20 | 149 |
TS PCR, type-specific PCR.
TABLE 9.
Combined genotyping results for 149 stool samples, obtained by conventional type-specific PCR and the novel PCR-reverse hybridization method
| G type | No. of isolates with the following P type:
|
Total | ||||||
|---|---|---|---|---|---|---|---|---|
| P[4] | P[6] | P[8] | P[8]vac | P[4]/P[8] | P[8]/P[4]/P[6] | P[8]/P[6] | ||
| G1vac | 21 | 21 | ||||||
| G1WT | 68 | 1 | 1 | 2 | 72 | |||
| G1WT/G2 | 1 | 2 | 1 | 4 | ||||
| G1WT/G2/G3 | 1 | 1 | ||||||
| G1WT/G4 | 1 | 1 | ||||||
| G1WT/G9 | 1 | 1 | ||||||
| G1/G2/G3 | 1 | 1 | ||||||
| G2 | 8 | 8 | ||||||
| G2/G3 | 1 | 1 | ||||||
| G2/G4 | 1 | 1 | ||||||
| G2/G4/G9 | 1 | 1 | ||||||
| G3 | 2 | 4 | 6 | |||||
| G4 | 8 | 3 | 11 | |||||
| G9 | 6 | 14 | 20 | |||||
| Total | 9 | 17 | 95 | 22 | 2 | 1 | 3 | 149 |
For P typing, 90% of the results were identical, 4% were compatible, and 6% were discordant. As with G typing, discordance was completely due to negative results obtained by the type-specific RT-PCR methods. Most compatible results were due to the fact that the novel method found additional genotypes besides those identified by the conventional method.
These results show that genotyping agreement between the two methods was very high and that the novel method was slightly more sensitive, mainly in cases of mixed rotavirus infections.
As shown in Table 9, there is a very close correlation between the identification of G1vac and that of P[8]vac: all 21 samples in which G1vac was detected also contained P[8]vac. However, one sample contained P[8]vac in combination with G1WT, as determined by the RHA method. This confirmed the fact that the sequence identified by the P[8]vac probes is not completely specific to the vaccine strain alone but also occurs in some natural strains. Interestingly, the G1WT/P[8]vac sample was obtained from a patient in Finland, and natural strains containing this P[8]vac sequence have also been reported previously in Finland (35).
To illustrate the effectiveness of the novel VP7 and VP4 assays, genotyping results from more than 2,000 clinical stool samples positive for HRV antigen by ELISA were analyzed. These samples were of various geographic origins. The VP7 PCR was positive for more than 97% of the antigen-positive samples. Reverse hybridization yielded clear VP7 genotypes for approximately 85% of the PCR-positive samples, but all samples were positive for the VP7 universal probes. Sequence analysis of the remaining samples revealed the presence of G12 (for which there is no probe on the strip) and a G9 variant (which showed weak results on the G9 probe on the strip but was confirmed by sequencing). In less than 1% of the PCR positive samples, novel variants of G2, G3, G4, G6, or G8 were observed (data not shown).
Similarly, the VP4 PCR was positive for more than 97% of the rotavirus antigen-positive samples. Reverse hybridization yielded clear VP4 genotypes for approximately 98% of the PCR-positive samples, but all samples were positive for the VP4 universal probes. For the remaining 2%, sequence analysis revealed the presence of P[14] (for which there is no probe on the strip) or of P[4], P[6], or P[8] variants (data not shown).
Robustness of the assays.
To assess the robustness of the assays, both RT-PCR and reverse hybridization were performed at temperatures deviating from the standard values. A dilution series of samples was tested by RT-PCR where the incubation temperatures during PCR were either 1.5°C lower or 1.5°C higher than the standard temperatures. Similarly, reverse hybridization was performed at temperatures 0.5°C higher or lower than the standard temperature. The results showed that small variations in temperature did not significantly impact the outcome of the assay (data not shown).
Storage of samples.
In order to determine the impact of long term-storage and repeated freeze-thawing of samples, dilutions of stool samples were subjected to as many as five freeze-thaw cycles (−20°C to room temperature). There was no impact on the HRV detection and genotyping results by the VP4 and VP7 assays. Similarly, RNAs isolated from stool samples were subjected to repeated freeze-thawing cycles (−80°C to room temperature), and again, there was no impact on the test outcome (data not shown).
Stools were stored at 4°C for 6 weeks and were tested again by the VP4 and VP7 assays. The results were identical to the original results, indicating that stool samples could be stored at 4°C for a prolonged period. Isolated RNA was stored for 6 weeks at −80°C and retested, also yielding identical results. Finally, RT-PCR products were stored at −20°C for 6 weeks; again, no impact on test results was observed.
DISCUSSION
Accurate detection and genotyping of HRV is important for several reasons. The epidemiology of rotavirus infections shows significant geographic differences, and global surveillance is crucial to determine the prevalence and incidence of different rotavirus genotypes.
In general, all recent studies confirm that the diversity of rotavirus strains is much greater than previously recognized (8, 14, 15, 21, 42).
The epidemiology of rotavirus is changing rapidly. Specific genotypes, such as G9 and G12, are emerging in various parts of the world (9, 28, 33, 39, 40, 45, 47). Outbreaks of rotavirus infections have been described, and these also require adequate genotype identification tools (18, 46, 50). Analysis of the epidemiology, molecular complexity, and evolution of the HRV is crucial to optimize the prevention of the disease. Most importantly, highly effective rotavirus vaccines have been developed and introduced for childhood immunization (41, 49). The impact of a rotavirus vaccine on local and global epidemiology should be carefully monitored.
Molecular methods to detect and genotype rotavirus isolates are hampered by the very high level of molecular heterogeneity among rotaviruses. The virus has a segmented, double-stranded RNA genome that is prone to genetic variation due to reassortment of segments and mutation during RNA replication.
Phylogenetic studies have revealed that all genomic segments show considerable heterogeneity but that some G types, such as G4, may be even more heterogeneous (6). Similarly, P[4], P[6], and P[8] show high levels of sequence variation (4, 25).
The sensitivity of the VP4 and VP7 RT-PCR RHA depends on the rotavirus genotype, which can be explained by the considerable sequence variation of the rotavirus genome. At present, there is no established correlation between focus-forming units and genome copies. However, all ELISA-positive stool samples were found positive by both the VP4 and VP7 RT-PCRs, indicating the effectiveness of these RT-PCRs in a clinical setting.
Genotyping methods should not only adequately identify known genotypes but also permit recognition of aberrant or novel genotypes. Type-specific PCR primers are aimed at highly specific amplification of known genotypes and have two major disadvantages. First, aberrant sequence will yield no amplimers at all, preventing downstream analysis. Second, and conversely, sequence heterogeneity at the primer sites may result in false-positive results, due to nonspecific annealing at modified priming sites. The method presented here employs broad-spectrum PCR primers, which allow the amplification of VP4 or VP7 sequences from a large variety of strains. After this broad-spectrum amplification, the amplimers can be analyzed in a stepwise manner to determine the precise genotype. First, hybridization to universal VP4 or VP7 probes is assessed. Subsequently, hybridization to type-specific probes is determined. For several genotypes, more than one probe was used on the strip. The aim was to prevent false-negative results and to match the different variants of certain genotypes, as described in the literature. Even if the type-specific probes yielded no signal, there was still the opportunity to perform sequence analysis on the generated amplimer.
The discrimination between the G1WT and G1vac strains in this novel assay is based on a single-nucleotide change at position 605 in VP7. This is the only specific signature sequence that can be exploited, since VP4 P[8] sequences also occur in some natural rotavirus strains (35). Therefore, VP4 sequences can be used only to confirm the specific identification of the vaccine strain by the VP7 assay.
The present method is highly effective at detecting mixed infections, which are highly prevalent (15, 16, 26, 36, 39). Mixtures of G1vac and G1WT sequences could be identified accurately, even if the two strains were not present in equimolar quantities.
The novel assay is robust, and the stability of rotavirus particles and of isolated RNA observed during the present study is in accordance with earlier observations (17). Taken together, these are important issues permitting adequate rotavirus diagnostics.
The novel assay has several interesting features. The strip format permits extension with additional probes, to include the identification of other genotypes. The reverse hybridization technology is known to be very specific and permits the recognition of single-nucleotide mismatches. Furthermore, reverse hybridization is very sensitive in detecting the presence of mixed genotypes. The strip assay allows visual interpretation and storage of the raw data in an easy format.
In conclusion, the novel VP4 and VP7 RT-PCR RHA is a useful tool for rotavirus diagnostics for epidemiological, natural history, and vaccine effectiveness studies.
TABLE 8.
Rotavirus VP4 genotyping results for 149 stool samples, obtained by conventional type-specific PCR and the novel PCR-reverse hybridization method
| Genotype by TS PCRa | No. of isolates with the following VP4 genotype by broad-spectrum RT-PCR and reverse hybridization:
|
Total | ||||||
|---|---|---|---|---|---|---|---|---|
| P[4] | P[6] | P[8] | P[8]vac | P[8]/P[4] | P[8]/P[6] | P[8]/P[4]/P[6] | ||
| P[4] | 9 | 1 | 1 | 1 | 12 | |||
| P[6] | 15 | 1 | 2 | 18 | ||||
| P[8] | 90 | 19 | 1 | 1 | 111 | |||
| Negative | 1 | 5 | 2 | 8 | ||||
| Total | 9 | 17 | 95 | 22 | 2 | 3 | 1 | 149 |
TS PCR, type-specific PCR.
Acknowledgments
We thank Carine Hastir and Valérie Wansard for technical support.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, E. J., and S. G. Weber. 2004. Rotavirus infection in adults. Lancet Infect. Dis. 491-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baggi, F., and R. Peduzzi. 2000. Genotyping of rotaviruses in environmental water and stool samples in southern Switzerland by nucleotide sequence analysis of 189 base pairs at the 5′ end of the VP7 gene. J. Clin. Microbiol. 383681-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bányai, K., V. Martella, F. Jakab, B. Melegh, and G. Szucs. 2004. Sequencing and phylogenetic analysis of human genotype P[6] rotavirus strains detected in Hungary provides evidence for genetic heterogeneity within the P[6] VP4 gene. J. Clin. Microbiol. 424338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barman, P., S. Ghosh, S. Das, V. Varghese, S. Chaudhuri, S. Sarkar, T. Krishnan, S. K. Bhattacharya, A. Chakrabarti, N. Kobayashi, and T. N. Naik. 2004. Sequencing and sequence analysis of VP7 and NSP5 genes reveal emergence of a new genotype of bovine group B rotaviruses in India. J. Clin. Microbiol. 422816-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bok, K., D. O. Matson, and J. A. Gomez. 2002. Genetic variation of capsid protein VP7 in genotype G4 human rotavirus strains: simultaneous emergence and spread of different lineages in Argentina. J. Clin. Microbiol. 402016-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 402398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, S., A. Sen, G. Uma, V. Varghese, S. Chaudhuri, S. K. Bhattacharya, T. Krishnan, P. Dutta, D. Dutta, M. K. Bhattacharya, U. Mitra, N. Kobayashi, and T. N. Naik. 2002. Genomic diversity of group A rotavirus strains infecting humans in eastern India. J. Clin. Microbiol. 40146-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, S., V. Varghese, S. Chaudhury, P. Barman, S. Mahapatra, K. Kojima, S. K. Bhattacharya, T. Krishnan, R. K. Ratho, G. P. Chhotray, A. C. Phukan, N. Kobayashi, and T. N. Naik. 2003. Emergence of novel human group A rotavirus G12 strains in India. J. Clin. Microbiol. 412760-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiStefano, D. J., N. Kraiouchkine, L. Mallette, M. Maliga, G. Kulnis, P. M. Keller, H. F. Clark, and A. R. Shaw. 2005. Novel rotavirus VP7 typing assay using a one-step reverse transcriptase PCR protocol and product sequencing and utility of the assay for epidemiological studies and strain characterization, including serotype subgroup analysis. J. Clin. Microbiol. 435876-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eing, B. R., G. May, H. G. Baumeister, and J. E. Kuhn. 2001. Evaluation of two enzyme immunoassays for detection of human rotaviruses in fecal specimens. J. Clin. Microbiol. 394532-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estes, M. K., and J. Cohen. 1989. Rotavirus gene structure and function. Microbiol. Rev. 53410-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estes, M. K., and A. Z. Kapikian. 2007. Rotaviruses, p. 1917-1974. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 14.Fang, Z. Y., H. Yang, J. Qi, J. Zhang, L. W. Sun, J. Y. Tang, L. Ma, Z. Q. Du, A. H. He, J. P. Xie, Y. Y. Lu, Z. Z. Ji, B. Q. Zhu, H. Y. Wu, S. E. Lin, H. P. Xie, D. D. Griffin, B. Ivanoff, R. I. Glass, and J. R. Gentsch. 2002. Diversity of rotavirus strains among children with acute diarrhea in China: 1998-2000 surveillance study. J. Clin. Microbiol. 401875-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, T. K., J. Eugen-Olsen, A. G. Pedersen, K. Molbak, B. Bottiger, K. Rostgaard, and N. M. Nielsen. 2005. Characterization of rotavirus strains in a Danish population: high frequency of mixed infections and diversity within the VP4 gene of P[8] strains. J. Clin. Microbiol. 431099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, T. K., N. A. Page, D. D. Griffin, J. Eugen-Olsen, A. G. Pedersen, P. Valentiner-Branth, K. Molbak, H. Sommerfelt, and N. M. Nielsen. 2003. Characterization of incompletely typed rotavirus strains from Guinea-Bissau: identification of G8 and G9 types and a high frequency of mixed infections. Virology 311125-133. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, T. K., H. Steinsland, and P. Valentiner-Branth. 2002. Rotavirus particles can survive storage in ambient tropical temperatures for more than 2 months. J. Clin. Microbiol. 404763-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallimore, C. I., J. S. Cheesbrough, K. Lamden, C. Bingham, and J. J. Gray. 2005. Multiple norovirus genotypes characterised from an oyster-associated outbreak of gastroenteritis. Int. J. Food Microbiol. 103323-330. [DOI] [PubMed] [Google Scholar]
- 19.Gault, E., R. Chikhi-Brachet, S. Delon, N. Schnepf, L. Albiges, E. Grimprel, J. P. Girardet, P. Begue, and A. Garbarg-Chenon. 1999. Distribution of human rotavirus G types circulating in Paris, France, during the 1997-1998 epidemic: high prevalence of type G4. J. Clin. Microbiol. 372373-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 301365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentsch, J. R., A. R. Laird, B. Bielfelt, D. D. Griffin, K. Banyai, M. Ramachandran, V. Jain, N. A. Cunliffe, O. Nakagomi, C. D. Kirkwood, T. K. Fischer, U. D. Parashar, J. S. Bresee, B. Jiang, and R. I. Glass. 2005. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J. Infect. Dis. 192(Suppl. 1)S146-S159. [DOI] [PubMed] [Google Scholar]
- 22.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouvea, V., M. S. Ho, R. Glass, P. Woods, B. Forrester, C. Robinson, R. Ashley, M. Riepenhoff-Talty, H. F. Clark, K. Taniguchi, et al. 1990. Serotypes and electropherotypes of human rotavirus in the USA: 1987-1989. J. Infect. Dis. 162362-367. [DOI] [PubMed] [Google Scholar]
- 24.Iturriza Gómara, M., C. Wong, S. Blome, U. Desselberger, and J. Gray. 2002. Rotavirus subgroup characterisation by restriction endonuclease digestion of a cDNA fragment of the VP6 gene. J. Virol. Methods 10599-103. [DOI] [PubMed] [Google Scholar]
- 25.Iturriza-Gómara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J. Clin. Microbiol. 38898-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain, V., B. K. Das, M. K. Bhan, R. I. Glass, and J. R. Gentsch. 2001. Great diversity of group A rotavirus strains and high prevalence of mixed rotavirus infections in India. J. Clin. Microbiol. 393524-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiraphongsa, C., J. S. Bresee, Y. Pongsuwanna, P. Kluabwang, U. Poonawagul, P. Arporntip, M. Kanoksil, N. Premsri, and U. Intusoma. 2005. Epidemiology and burden of rotavirus diarrhea in Thailand: results of sentinel surveillance. J. Infect. Dis. 192(Suppl. 1)S87-S93. [DOI] [PubMed] [Google Scholar]
- 28.Khamrin, P., S. Peerakome, L. Wongsawasdi, S. Tonusin, P. Sornchai, V. Maneerat, C. Khamwan, F. Yagyu, S. Okitsu, H. Ushijima, and N. Maneekarn. 2006. Emergence of human G9 rotavirus with an exceptionally high frequency in children admitted to hospital with diarrhea in Chiang Mai, Thailand. J. Med. Virol. 78273-280. [DOI] [PubMed] [Google Scholar]
- 29.Khetawat, D., T. Ghosh, M. K. Bhattacharya, S. K. Bhattacharya, and S. Chakrabarti. 2001. Molecular characterization of the VP7 gene of rotavirus isolated from a clinical sample of Calcutta, India. Virus Res. 7453-58. [DOI] [PubMed] [Google Scholar]
- 30.Laird, A. R., J. R. Gentsch, T. Nakagomi, O. Nakagomi, and R. I. Glass. 2003. Characterization of serotype G9 rotavirus strains isolated in the United States and India from 1993 to 2001. J. Clin. Microbiol. 413100-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laird, A. R., V. Ibarra, G. Ruiz-Palacios, M. L. Guerrero, R. I. Glass, and J. R. Gentsch. 2003. Unexpected detection of animal VP7 genes among common rotavirus strains isolated from children in Mexico. J. Clin. Microbiol. 414400-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovmar, L., C. Fock, F. Espinoza, F. Bucardo, A. C. Syvanen, and K. Bondeson. 2003. Microarrays for genotyping human group A rotavirus by multiplex capture and type-specific primer extension. J. Clin. Microbiol. 415153-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martella, V., V. Terio, G. Del Gaudio, M. Gentile, P. Fiorente, S. Barbuti, and C. Buonavoglia. 2003. Detection of the emerging rotavirus G9 serotype at high frequency in Italy. J. Clin. Microbiol. 413960-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthijnssens, J., M. Ciarlet, M. Rahman, H. Attoui, K. Bányai, M. K. Estes, J. R. Gentsch, M. Iturriza-Gómara, C. D. Kirkwood, V. Martella, P. P. Mertens, O. Nakagomi, J. T. Patton, F. M. Ruggeri, L. J. Saif, N. Santos, A. Steyer, K. Taniguchi, U. Desselberger, and M. Van Ranst. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 1531621-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maunula, L., and C. H. Von Bonsdorff. 2002. Frequent reassortments may explain the genetic heterogeneity of rotaviruses: analysis of Finnish rotavirus strains. J. Virol. 7611793-11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen, N. M., J. Eugen-Olsen, P. Aaby, K. Molbak, A. Rodrigues, and T. K. Fischer. 2005. Characterisation of rotavirus strains among hospitalised and non-hospitalised children in Guinea-Bissau, 2002. A high frequency of mixed infections with serotype G8. J. Clin. Virol. 3413-21. [DOI] [PubMed] [Google Scholar]
- 37.Parashar, U. D., C. J. Gibson, J. S. Bresse, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pongsuwanna, Y., R. Guntapong, M. Chiwakul, R. Tacharoenmuang, N. Onvimala, M. Wakuda, N. Kobayashi, and K. Taniguchi. 2002. Detection of a human rotavirus with G12 and P[9] specificity in Thailand. J. Clin. Microbiol. 401390-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reidy, N., F. O'Halloran, S. Fanning, B. Cryan, and H. O'Shea. 2005. Emergence of G3 and G9 rotavirus and increased incidence of mixed infections in the southern region of Ireland 2001-2004. J. Med. Virol. 77571-578. [DOI] [PubMed] [Google Scholar]
- 40.Rubilar-Abreu, E., K. O. Hedlund, L. Svensson, and C. Mittelholzer. 2005. Serotype G9 rotavirus infections in adults in Sweden. J. Clin. Microbiol. 431374-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz-Palacios, G. M., I. Perez-Schael, F. R. Velázquez, H. Abate, T. Breuer, S. C. Clemens, B. Cheuvart, F. Espinoza, P. Gillard, B. L. Innis, Y. Cervantes, A. C. Linhares, P. López, M. Macías-Parra, E. Ortega-Barría, V. Richardson, D. M. Rivera-Medina, L. Rivera, B. Salinas, N. Pavía-Ruz, J. Salmerón, R. Rüttimann, J. C. Tinoco, P. Rubio, E. Nuñez, M. L. Guerrero, J. P. Yarzábal, S. Damaso, N. Tornieporth, X. Sáez-Llorens, R. F. Vergara, T. Vesikari, A. Bouckenooghe, R. Clemens, B. De Vos, and M. O'Ryan for the Human Rotavirus Vaccine Study Group. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 35411-22. [DOI] [PubMed] [Google Scholar]
- 42.Sánchez-Fauquier, A., I. Wilhelmi, J. Colomina, E. Cubero, and E. Roman. 2004. Diversity of group A human rotavirus types circulating over a 4-year period in Madrid, Spain. J. Clin. Microbiol. 421609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santos, N., S. Honma, M. C. Timenetsky, A. C. Linhares, H. Ushijima, G. E. Armah, J. R. Gentsch, and Y. Hoshino. 2008. Development of a microtiter plate hybridization-based PCR-enzyme-linked immunosorbent assay for identification of clinically relevant human group A rotavirus G and P genotypes. J. Clin. Microbiol. 46462-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 1529-56. [DOI] [PubMed] [Google Scholar]
- 45.Shinozaki, K., M. Okada, S. Nagashima, I. Kaiho, and K. Taniguchi. 2004. Characterization of human rotavirus strains with G12 and P[9] detected in Japan. J. Med. Virol. 73612-616. [DOI] [PubMed] [Google Scholar]
- 46.Stelzmueller, I., K. M. Dunst, P. Hengster, H. Wykypiel, W. Steurer, S. Wiesmayr, R. Margreiter, and H. Bonatti. 2005. A cluster of rotavirus enteritis in adult transplant recipients. Transpl. Int. 18470-474. [DOI] [PubMed] [Google Scholar]
- 47.Steyer, A., M. Poljsak-Prijatelj, D. Barlic-Maganja, T. Bufon, and J. Marin. 2005. The emergence of rotavirus genotype G9 in hospitalised children in Slovenia. J. Clin. Virol. 337-11. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki, H. 1996. A hypothesis about the mechanism of assembly of double-shelled rotavirus particles. Arch. Virol. Suppl. 1279-85. [DOI] [PubMed] [Google Scholar]
- 49.Vesikari, T., D. O. Matson, P. Dennehy, P. Van Damme, M. Santosham, Z. Rodriguez, M. J. Dallas, J. F. Heyse, M. G. Goveia, S. B. Black, H. R. Shinefield, C. D. Christie, S. Ylitalo, R. F. Itzler, M. L. Coia, M. T. Onorato, B. A. Adeyi, G. S. Marshall, L. Gothefors, D. Campens, A. Karvonen, J. P. Watt, K. L. O'Brien, M. J. DiNubile, H. F. Clark, J. W. Boslego, P. A. Offit, and P. M. Heaton for the Rotavirus Efficacy and Safety Trial (REST) Study Team. 2006. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 35423-33. [DOI] [PubMed] [Google Scholar]
- 50.Yan, H., T. Abe, T. G. Phan, T. A. Nguyen, T. Iso, Y. Ikezawa, K. Ishii, S. Okitsu, and H. Ushijima. 2005. Outbreak of acute gastroenteritis associated with group A rotavirus and genogroup I sapovirus among adults in a mental health care facility in Japan. J. Med. Virol. 75475-481. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, M., C. Q. Zeng, Y. Dong, J. M. Ball, L. J. Saif, A. P. Morris, and M. K. Estes. 1998. Mutations in rotavirus nonstructural glycoprotein NSP4 are associated with altered virus virulence. J. Virol. 723666-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]