Abstract
Clostridium difficile isolates from presumed community-associated infections (n = 92) were characterized by toxinotyping, pulsed-field gel electrophoresis, tcdC and cdtB PCR, and antimicrobial susceptibility. Nine toxinotypes (TOX) and 31 PFGE patterns were identified. TOX 0 (48, 52%), TOX III (18, 20%), and TOX V (9, 10%) were the most common; three isolates were nontoxigenic.
Clostridium difficile infection (CDI) is an important cause of health care-associated diarrhea, especially in patients receiving antibiotics. C. difficile also causes diarrhea among patients in community settings (1, 6, 13, 19, 20), although diarrheic outpatients are not routinely tested for CDI. Community-associated CDI (CA-CDI) appears to be increasing (3, 11, 22), but little is known about strains that cause it. We collected and characterized isolates from patients with presumed CA-CDIs.
Surveillance for CA-CDI was conducted in 2006 for 3 months at 19 clinical laboratories in nine states participating in the Food-Borne Disease Active Surveillance Network (FoodNet; California, Connecticut, Georgia, Maryland, Minnesota, New Mexico, New York, Oregon, and Tennessee). Stool specimens from patients with positive physician-ordered C. difficile toxin tests were stored at −20°C until medical record review confirmed study eligibility. Presumed CA-CDI was defined as a C. difficile toxin-positive stool specimen from an outpatient (or within 72 h of hospital admission) without a previous positive result in the same laboratory for >8 weeks or an overnight health care stay in the preceding 3 months. Diagnostic assays varied and included toxin A-only or toxin A and B enzyme immunoassays. The sensitivity and specificity of diagnostic assays were not evaluated.
C. difficile toxin-positive stool samples were submitted frozen to the state's public health laboratory or the Durham Veterans Affairs Medical Center laboratory (Durham, NC) for culture. Thawed stool samples were cultured by alcohol shock (12) on anaerobe blood agar plates (BD, Franklin Lakes, NJ), direct inoculation of cycloserine-cefoxitin fructose agar (4), or both and then incubated anaerobically at 35°C for 72 to 96 h. Plates were examined daily for characteristic colonies (4), p-cresol odor, and yellow-green fluorescence under UV light. Chopped meat broth (BD) was inoculated with a single colony and shipped to the Centers for Disease Control and Prevention (CDC) for confirmatory identification by the characteristics noted above, a negative indole reaction, and a positive l-proline-aminopeptidase (Remel, Lenexa, KS) reaction. Isolates were characterized by toxinotyping, pulsed-field gel electrophoresis (PFGE), PCR evaluation of tcdC (10) and cdtB (20), and antimicrobial susceptibility testing.
Of 175 specimens obtained from patients with presumed CA-CDIs, 162 were cultured; 103 isolates were submitted to the CDC, and 92 were confirmed as C. difficile. The C. difficile recovery rate from 162 toxin-positive stool samples was 57% (range, 29 to 100% by laboratory).
Toxinotyping was based on restriction fragment length polymorphisms in the A3 and B1 fragments of the pathogenicity locus, as described previously (17). Three isolates were nontoxigenic. We tested only one isolate per specimen, so it is unknown whether nontoxigenic isolates represent coinfection or false-positive diagnostic tests.
Nine toxinotypes were identified among 89 toxigenic C. difficile isolates. Most (n = 48; 54%) of the isolates were TOX 0; the remaining 41 (46%) had variant toxinotypes. TOX 0 strains (15) are a common cause of health care-associated CDI (HA-CDI) (6, 15, 20), whereas variant toxinotypes were rare prior to the emergence of the epidemic strain (6, 15, 20). TOX III was the most common variant (n = 18; 20%) identified, followed by TOX V (n = 9; 10%) and TOX IX (n = 7; 8%). Excepting TOX VIII and TOX XII, variant toxinotypes correlated with cdtB presence (n = 39; 42%), as expected (16, 20). The pathogenic contribution of binary toxin is unclear (5, 18), but binary toxin-positive strains are increasingly isolated from human CDIs in Europe and the United States (7, 8, 13, 18, 19).
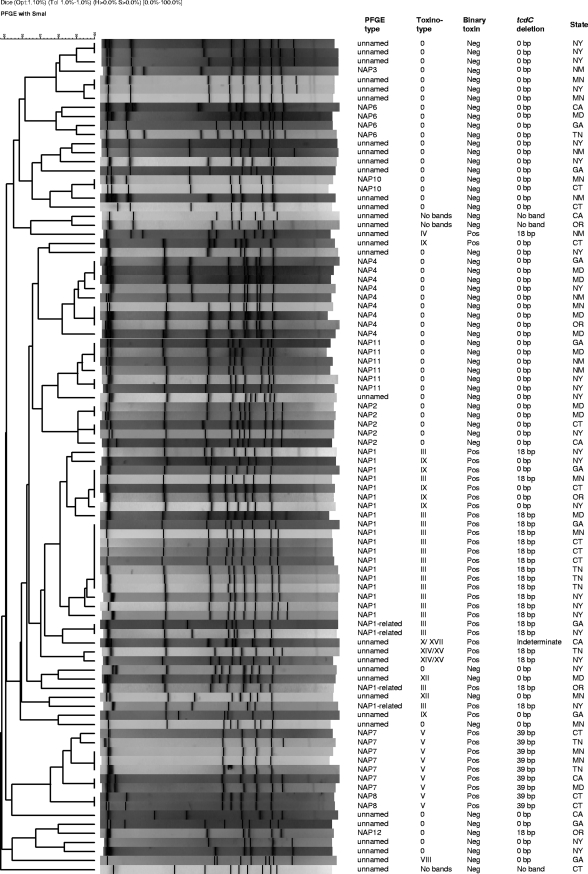
Isolates were characterized by SmaI PFGE and analyzed with BioNumerics v5.01 (Applied Maths, Austin, TX), as described previously (10), and then compared to the entire CDC C. difficile database. North American pulsed-field (NAP) types were assigned to patterns ≥80% similar to established NAP clusters. Thirty-one PFGE types were identified among 92 CA-CDI study isolates, 65% of which were assigned established NAP types. The 32 isolates not assigned NAP types belonged to 21 unnamed patterns (Fig. 1). Nontoxigenic isolates yielded unnamed patterns distinct from toxigenic isolates.
FIG. 1.
PFGE patterns and characteristics of 92 isolates from patients with CA-CDI in nine FoodNet states in 2006. Neg, negative; Pos, positive.
NAP1 was the most common PFGE type identified (n = 19; 21%) and was uniformly binary toxin positive. Fourteen isolates were typical of the epidemic strain (TOX III, 18-bp tcdC deletion); five were unusual (TOX IX, wild-type tcdC [Fig. 1]). Four isolates were typical of NAP1 (TOX III, 18-bp tcdC deletion, binary toxin positive) but <80% related to NAP1 by PFGE and thus classified as NAP1 related. Other recognized PFGE types included NAP4 (10%), NAP7 (8%), NAP11 (6%), NAP2 (5%), and others (each less than 5% of the isolates [Fig. 1]). NAP2 (restriction endonuclease type J; PCR ribotype 001; TOX 0), a previous epidemic strain in the United States and Europe (9, 21), and NAP4 (restriction endonuclease type Y; PCR ribotype 077; TOX 0) were previously common in health care settings (1) but have been uncommon in recent years (CDC, unpublished data). NAP11 has not been associated with outbreaks in the United States.
MICs of moxifloxacin, levofloxacin, ciprofloxacin, clindamycin, erythromycin, metronidazole, and vancomycin were determined for 87 isolates by broth microdilution (2). This method is not approved for C. difficile reference testing and was used for surveillance only. Five isolates received after broth microdilution panels had been exhausted were evaluated by Etest (AB Biodisk, Piscataway, NJ) as recommended (EAS 007; AB Biodisk). The frequencies at which the MICs for NAP1/TOX III isolates and non-NAP1/TOX III isolates were above the MICs for 50 and 90% of the entire isolate population tested (MIC50s and MIC90s, respectively) were compared by using Fisher's exact t test.
NAP1/TOX III isolates displayed different MIC distributions than others and thus are presented together and separately (Table 1). MICs for NAP1/TOX III isolates were more likely than those for non-NAP1/TOX III isolates to be above the aggregate MIC50 of all of the antimicrobials tested (P < 0.05), except clindamycin and vancomycin, and above the aggregate MIC90 of ciprofloxacin (P < 0.05). All of the isolates were susceptible to metronidazole and vancomycin (Table 1).
TABLE 1.
Ranges of MICs for NAP1/TOX III and non-NAP1/TOX III C. difficile isolates from patients with presumed CA-CDIs in nine FoodNet states in 2006
| Drug | All isolates (n = 92)
|
NAP1/TOX III (n = 14)
|
Others (n = 78)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | MIC range | MIC50 | MIC90 | Range | MIC50 | MIC90 | MIC range | |
| Moxifloxacin | 1a | 16 | ≤0.5-256 | 16 | 128 | 1-256 | 1 | 16 | ≤0.5-128 |
| Levofloxacin | 4 | 128 | ≤0.5-256 | 64 | 128 | 2-256 | 4 | 64 | ≤0.5-256 |
| Ciprofloxacin | 8 | 64 | 2-256 | 128 | 256 | 4-256 | 8 | 32 | 2-64 |
| Clindamycin | 4 | 256 | ≤0.5->256 | 2 | 128 | 1-128 | 4 | 256 | ≤0.5->256 |
| Erythromycin | 1 | >256 | ≤0.5->256 | >256 | >256 | ≤0.5->256 | 1 | >256 | ≤0.5->256 |
| Metronidazole | 0.5 | 2 | ≤0.25-4 | 2 | 4 | 0.5-4 | 0.5 | 1 | ≤0.25-4 |
| Vancomycin | 0.5 | 1 | ≤0.25-2 | 0.5 | 2 | 0.5-2 | 0.5 | 1 | ≤0.25-2 |
Values are in micrograms per milliliter.
Our findings suggest diverse molecular epidemiology of CA-CDI strains, with similarities to and differences from HA-CDI strains. Two recently described strains with importance in the epidemiology of human CDI were prevalent in this study, the NAP1/TOX III and TOX V strains. NAP1/TOX III was the most common strain type identified. This may be due to health care exposures undocumented in medical records or lacking in the case definition of CA-CDI or to a reservoir for the epidemic strain outside of health care settings. TOX V strains are a frequent cause of CDI in food-producing animals in Europe (14) and the United States. The presence of TOX V strains causing CA-CDI at a time when such strains are uncommon in health care settings may provide ecological evidence of the transmission of C. difficile from animals to humans (8). Further research into the molecular epidemiology and the sources of strains causing CA-CDI will advance understanding of the evolving epidemiology of human CDI and could inform additional strategies to prevent CDI.
Acknowledgments
We thank Chris Woods and Brad Nicholson, Durham Veterans Affairs Medical Center, Durham, NC, for assistance with primary culture of C. difficile.
The findings and conclusions in this report are ours and do not necessarily represent the official position of the CDC.
Footnotes
Published ahead of print on 1 July 2009.
REFERENCES
- 1.Clabots, C. R., S. Johnson, K. M. Bettin, P. A. Mathie, M. E. Mulligan, D. R. Schaberg, L. R. Peterson, and D. N. Gerding. 1993. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J. Clin. Microbiol. 311870-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard—7th edition. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Dial, S., J. A. Delaney, A. N. Barkun, and S. Suissa. 2005. Use of gastric acid-suppressive agents and the risk of community-acquired Clostridium difficile-associated disease. JAMA 2942989-2995. [DOI] [PubMed] [Google Scholar]
- 4.George, W. L., V. L. Sutter, D. Citron, and S. M. Finegold. 1979. Selective and differential medium for isolation of Clostridium difficile. J. Clin. Microbiol. 9214-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geric, B., R. J. Carman, M. Rupnik, C. W. Genheimer, S. P. Sambol, D. M. Lyerly, D. N. Gerding, and S. Johnson. 2006. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J. Infect. Dis. 1931143-1150. [DOI] [PubMed] [Google Scholar]
- 6.Geric, B., M. Rupnik, D. N. Gerding, M. Grabnar, and S. Johnson. 2004. Distribution of Clostridium difficile variant toxinotypes and strains with binary toxin genes among clinical isolates in an American hospital. J. Med. Microbiol. 53887-894. [DOI] [PubMed] [Google Scholar]
- 7.Goorhuis, A., S. B. Debast, L. A. van Leengoed, C. Harmanus, D. W. Notermans, A. A. Bergwerff, and E. J. Kuijper. 2008. Clostridium difficile PCR ribotype 078: an emerging strain in humans and in pigs? J. Clin. Microbiol. 461157-1158. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jhung, M. A., A. D. Thompson, G. E. Killgore, W. E. Zukowski, G. Songer, M. Warny, S. Johnson, D. N. Gerding, L. C. McDonald, and B. M. Limbago. 2008. Toxinotype V Clostridium difficile in humans and food animals. Emerg. Infect. Dis. 141039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, S., M. H. Samore, K. A. Farrow, G. E. Killgore, F. C. Tenover, D. Lyras, J. I. Rood, P. DeGirolami, A. L. Baltch, M. E. Rafferty, S. M. Pear, and D. N. Gerding. 1999. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N. Engl. J. Med. 3411645-1651. [DOI] [PubMed] [Google Scholar]
- 10.Killgore, G. E., A. D. Thompson, S. Johnson, J. Brazier, E. Kuijper, J. Pepin, E. H. Frost, P. Savelkoul, B. Nicholson, R. J. van den Berg, H. Kato, S. P. Sambol, W. Zukowski, C. Woods, B. M. Limbago, D. N. Gerding, and L. C. McDonald. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyne, L., C. Merry, B. O'Connell, C. Keane, and D. O'Neill. 1998. Community-acquired Clostridium difficile infection. J. Infect. 36287-288. [DOI] [PubMed] [Google Scholar]
- 12.Marler, L. M., J. A. Siders, L. C. Wolters, Y. Pettigrew, B. L. Skitt, and S. D. Allen. 1992. Comparison of five cultural procedures for isolation of Clostridium difficile from stools. J. Clin. Microbiol. 30514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald, L. C., G. E. Killgore, A. D. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 3532433-2441. [DOI] [PubMed] [Google Scholar]
- 14.Pirs, T., M. Ocepek, and M. Rupnik. 2008. Isolation of Clostridium difficile from food animals in Slovenia. J. Med. Microbiol. 57790-792. [DOI] [PubMed] [Google Scholar]
- 15.Rupnik, M. 2008. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol. Rev. 32541-555. [DOI] [PubMed] [Google Scholar]
- 16.Rupnik, M. 2001. How to detect Clostridium difficile variant strains in a routine laboratory. Clin. Microbiol. Infect. 7417-420. [DOI] [PubMed] [Google Scholar]
- 17.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmee. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 362240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rupnik, M., M. Grabnar, and B. Geric. 2003. Binary toxin producing Clostridium difficile strains. Anaerobe 9289-294. [DOI] [PubMed] [Google Scholar]
- 19.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 403470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186307-312. [DOI] [PubMed] [Google Scholar]
- 21.Stubbs, S. L., J. S. Brazier, G. L. O'Neill, and B. I. Duerden. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37461-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilcox, M. H., L. Mooney, R. Bendall, C. D. Settle, and W. N. Fawley. 2008. A case-control study of community-associated Clostridium difficile infection. J. Antimicrob. Chemother. 62388-396. [DOI] [PubMed] [Google Scholar]