Abstract
The 15 proteolytic Clostridium botulinum type B strains, including 3 isolates associated with infant botulism in Japan, were genetically characterized by phylogenetic analysis of boNT/B gene sequences, genotyping, and determination of the boNT/B gene location by using pulsed-field gel electrophoresis (PFGE) for molecular epidemiological analysis of infant botulism in Japan. Strain Osaka05, isolated from a case in 2005, showed a unique boNT/B gene sequence and was considered to be a new BoNT/B subtype by phylogenetic analysis. Strain Osaka06, isolated from a case in 2006, was classified as the B2 subtype, the same as strain 111, isolated from a case in 1995. The five isolates associated with infant botulism in the United States were classified into the B1 subtype. Isolates from food samples in Japan were divided into the B1 and the B2 subtypes, although no relation with infant botulism was shown by PFGE genotyping. The results of PFGE and Southern blot hybridization with undigested DNA suggested that the boNT/B gene is located on large plasmids (approximately 150 kbp, 260 kbp, 275 kbp, or 280 kbp) in five strains belonging to three BoNT/B subtypes from various sources. The botulinum neurotoxin (BoNT) of Osaka05 was suggested to have an antigenicity different from the antigenicities of BoNT/B1 and BoNT/B2 by a sandwich enzyme-linked immunosorbent assay with the recombinant BoNT/B-C-terminal domain. We established a multiplex PCR assay for BoNT/B subtyping which will be useful for epidemiological studies of type B strains and the infectious diseases that they cause.
Infant botulism is neuromuscular paralysis caused by the botulinum neurotoxin (BoNT) produced in the intestines after the germination and outgrowth of ingested spores of Clostridium botulinum, which is an anaerobic spore-forming bacterium (7, 9). On the basis of the antigenic specificity of BoNT, C. botulinum strains are divided into seven serotypes (serotypes A to G), and the species has been separated into four groups (groups I to IV) by cultural characteristics (24). BoNT is encoded by an approximately 3.8-kb gene, which is preceded by several nontoxic component genes (17, 30). BoNT is released from the bacteria as a single polypeptide chain of 150 kDa and is cleaved by endogenous or exogenous proteases into a 50-kDa light chain and a 100-kDa heavy chain. The heavy chain contains two functional domains, the N-terminal domain (HN) and the C-terminal domain (HC). HC can be further divided into two distinct subdomains: the N-terminal domain (HCN) and the C-terminal domain (HCC) (5, 38).
Recently, the subtype classification was confirmed by the diversity of the amino acid sequences within each serotype (13, 37). BoNT serotype A (BoNT/A) has been divided into four subtypes (subtypes A1, A2, A3, and A4) (2, 10). BoNT/B has been divided into three subtypes from type B group I (subtypes B1, B2, and B3), one subtype from group I bivalent strains that express another BoNT type, in addition to BoNT/B (bivalent), and one subtype from type B group II (nonproteolytic) (13). BoNT/E has been divided into four subtypes from C. botulinum type E (subtypes E1, E2, E3, and E6) and two subtypes from BoNT/E-producing C. butyricum (subtypes E4 and E5) (6).
Since infant botulism was first recognized in the United States in 1976 (27, 31), it is now the most common disease caused by C. botulinum. This disease affects children up to 6 months old, but with rare exceptions it affects individuals of other ages. The symptoms are characterized by constipation, generalized weakness, and various neurological disorders (9). Cases represent a spectrum of disease, ranging from subclinical infection to the most fulminant form of the disease, which is unexpected sudden death (3). Almost all cases of infant botulism have been caused by proteolytic C. botulinum type A and B strains. Since the first occurrence of infant botulism in Japan caused by C. botulinum type A in 1986 (29), there have been 24 cases; 16 were caused by type A strains, 3 were caused by type B strains, 1 was caused by a type C strain, and 1 was caused by a C. butyricum strain producing BoNT/E. The types of toxin in the other three cases were not described (16). We previously indicated that the original BoNT/B2 produced by strain 111, which was isolated from the first case of type B infant botulism in Japan in 1995, showed antigenic and biological properties different from those of the authentic BoNT/B (B1) produced by strain Okra (15, 20, 22). Two additional cases of type B infant botulism with typical symptoms occurred in Osaka Prefecture in 2005 and 2006. We eventually isolated two proteolytic C. botulinum type B strains, designated Osaka05 and Osaka06, respectively.
In the study described here, to better understand the background of type B infant botulism, we determined the genetic characteristics of proteolytic C. botulinum type B isolates by comparison of the nucleotide sequences of boNT/B and nontoxic component genes, the pulsed-field gel electrophoresis (PFGE) genotypes, the boNT/B gene location by PFGE and Southern blot hybridization, and the antigenicity of the new BoNT/B subtype. We developed multiplex PCR assays for the detection and identification of the BoNT/B subtypes.
MATERIALS AND METHODS
Type B infant botulism in Japan. (i) Case 1.
The first case of type B infant botulism occurred in a female infant aged 6 months in Ishikawa Prefecture in 1995, and type B strain 111 was isolated from this case (18, 42). Strain 111 was provided by S. Nakamura (Kanazawa University School of Medicine, Ishikawa, Japan).
(ii) Case 2.
In Osaka City in October 2005, a previously healthy breast-fed female infant aged 3 months with a 2-day history of fever, pituita, and poor feeding was hospitalized for 45 days and then discharged, and she recovered fully. BoNT/B and type B strain Osaka05 were detected in stool samples on the third hospital day (28).
(iii) Case 3.
In Osaka Prefecture in May 2006, a previously healthy mainly breast-fed female infant aged 5 months with a 1-week history of constipation, poor feeding, and weakness was hospitalized for 32 days and then discharged, and she recovered fully. BoNT/B and type B strain Osaka06 were detected in the stool on the fifth hospital day (1). Strain Osaka06 was provided by T. Asao (Osaka Prefectural Institute of Public Health, Osaka, Japan).
C. botulinum was not detected in food, drink, or house dust samples obtained from the patients' homes for cases 2 and 3. In all cases, the patients had no history of honey consumption.
Bacterial strains and DNA extraction.
Fifteen proteolytic C. botulinum type B strains, including 3 strains associated with infant botulism in Japan, were used in this study (Table 1). The biochemical characteristics, including the proteolytic activities, of all strains were tested by use of an API 20A kit (Biomerieux, Marcy l'Etoile, France). No strains with any unexpressed boNT genes were detected by PCR assay (40). Individual strains were cultured in 10 ml cooked meat medium (Difco, Becton Dickinson and Co., Franklin Lakes, NJ) supplemented with 0.3% glucose and 0.2% soluble starch (Difco, Becton Dickinson and Co.) under anaerobic conditions at 30°C for 18 h. Bacterial DNA was extracted from 1 ml culture with a DNeasy tissue kit (Qiagen Inc., Valencia, CA), according to the manufacturer's instructions. Finally, DNA was eluted with 100 μl elution buffer and stored at 4°C until use.
TABLE 1.
C. botulinum type B strains and sequences used in this studya
| Strain identifier | BoNT/B subtypeb | GenBank accession no. | Origin (country, yr) | Reference or source |
|---|---|---|---|---|
| Bacterial strain | ||||
| 111 | B2 | AB302854, AB084152 | Infant botulism (Japan, 1995) | 15 |
| Osaka05 | Osaka05 | AB302852 | Infant botulism (Japan, 2005) | In this study |
| Osaka06 | B2 | AB302853 | Infant botulism (Japan, 2006) | In this study |
| 89E00061-2 | B1 | AB302861 | Infant botulism (United States, 1989) | In this study |
| 89E00067-4 | B1 | AB302862 | Infant botulism (United States, 1989) | In this study |
| 89E00123-1 | B1 | AB302963 | Infant botulism (United States, 1989) | In this study |
| 90E00001-3 | B1 | AB302864 | Infant botulism (United States, 1990) | In this study |
| 3129-2-77 | B1 | AB302865 | Infant botulism (United States, 1990) | In this study |
| Okra | B1 | AB232927 | Food borne (unknown) | 23 |
| 326 | B1 | AB302858 | Pork meat (Japan) | In this study |
| 407 | B1 | AB302856 | Pork meat (Japan) | In this study |
| Ginger | B2 | AB302857 | Ginger (Japan) | In this study |
| 7H215S | B2 | AB302860 | Honey (Japan) | In this study |
| 9B | B1 | AB302859 | Stocked strain (United States) | In this study |
| 67B | B1 | AB302855 | Stocked strain (United States) | In this study |
| GenBank | ||||
| CDC1758 | B1 | EF033127 | Unknown | 13 |
| Danish | B1 | M81186 | Unknown | 41 |
| Hall6517(B) | B1 | EF028399 | Unknown | 13 |
| CDC1656 | B1 | EF028396 | Unknown | 13 |
| Prevot25 NCASE | B2 | EF033129 | Unknown | 13 |
| 213B | B2 | EF028395 | Unknown | 13 |
| Smith L-590 | B2 | EF028398 | Unknown | 13 |
| Prevot59 | B2 | EF033128 | Unknown | 13 |
| CDC1828 | B2 | EF051571 | Unknown | 13 |
| CDC6291 | B2 | EF028401 | Unknown | 13 |
| Korean soil 1 | B2 | DQ417353 | Korean soil | GenBank |
| Korean soil 2 | B2 | DQ417354 | Korean soil | GenBank |
| CDC795 | B3 | EF028400 | Unknown | 13 |
| CDC593 | Bivalent, A(B)c | AF300466 | Dog feces (United States, 1976) | 19 |
| CDC1436 | Bivalent, ABd | AF295926 | Stool sample (United States, 1977) | 19 |
| 657Ba | Bivalent, Bad | EF033130 | Unknown | 13, 36 |
| CDC588 | Bivalent, Abd | AF300465 | Food borne (United States, 1976) | 19 |
| CDC3281 | Bivalent, Bfd | Y13630 | Infant botulism (United States, 1980) | 33 |
| ATCC 17844 | Nonproteolytic | EF028394 | Unknown | 13 |
| Eklund17B (B257) | Nonproteolytic | EF051570 | Unknown | 13 |
| 10068 | Nonproteolytic | EF028402 | Unknown | 13 |
| Eklund17B | Nonproteolytic | X71343 | Unknown | 14 |
The subtypes and GenBank accession numbers for strains determined in this study are indicated in boldface.
Subtyped by Hill et al. (13); the other strains were subtyped in this study.
BoNT/A producing and unexpressed boNT/B gene possessing.
Dual toxin-producing strains; the major toxin type is indicated in uppercase letters and the minor type is indicated in lowercase letters.
Nucleotide sequencing of boNT/B and nontoxic component genes.
The boNT/B gene and nontoxic component genes (ha70, ha17, ha33, p21, and ntnh) were amplified by using the overlapping primer pairs listed in Table 2, which were designed on the basis of the nucleotide sequences available from GenBank. PCR was performed with a 50-μl reaction mixture containing 10 ng extracted DNA, 0.5 μM of each primer, 2.5 U LA Taq (TaKaRa Shuzo, Kyoto, Japan), 5 μl LA Taq buffer, 2.5 mM MgCl2, and 400 μM of each deoxynucleoside triphosphate (dNTP). Each PCR cycle consisted of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 65°C for 3 min and was repeated 25 times. Final extension was carried out at 65°C for 5 min. The amplicons were directly sequenced by primer walking, and the sequence in each direction was confirmed with an ABI Prism BigDye cycle sequencing kit (Applied Biosystems Inc., Foster City, CA).
TABLE 2.
Primers used for amplification of boNT/B and nontoxic component genes
| Gene(s) included in amplified fragment | Primer pair sequences (5′-3′)a | Location (positions)a,b |
|---|---|---|
| ha70, ha17, and first half of ha33 | CAAAATATGATTTCCTTGT/AGCAGCATACCAGTTTT | 2725-2743/5578-5562 |
| Latter half of ha33, botR, and first half of ntnh | CGCGTAGATTAGTAATTG/AAGTGCATTATTAAATCTATCT | 5406-5423/8166-8145 |
| Latter half of ntnh | AGGAAATAATGCCATTG/CTTTATAATATCTCCCCGT | 8022-8038/10826-10808 |
| boNT/B light chain | TTTATGGGCATTAAAAG/CATCTGAAAAACTATTTTTAT | 10671-10687/12116-12096 |
| boNT/B heavy chain | AGAGGTCAGAATAAAGCTA/CAAAATTTAGCTACATCCT | 11948-11966/14682-14664 |
Sequences and positions are for forward primer/reverse primer.
Location of primer sequence of boNT/B and nontoxic component genes in the sequence reported under DDBJ accession no. AB232927.
Nucleotide sequencing of the C-terminal region of BoNT/B.
The nucleotide sequences of the C-terminal region (400 bp) of BoNT/B were determined by direct sequencing with the following primers: forward primer 5′-GAAAGTCAAATTCTCAATC-3′ (positions in the coding region, bases 3445 to 3463) and reverse primer 5′-CAAAATTTAGCTACATCCT-3′ (positions in the coding region, bases 3961 to 3943). PCR was performed with a 50-μl reaction mixture containing 1 ng extracted DNA, 0.5 μM of each primer, 2.5 U LA Taq (TaKaRa Shuzo), 5 μl LA Taq buffer, 2.5 mM MgCl2, and 200 μM of each dNTP. Each PCR cycle consisted of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 65°C for 2 min and was repeated 25 times. Final extension was carried out at 65°C for 5 min.
Phylogenetic analysis.
The nucleotide sequences were aligned by use of the Clustal X program (version 1.83) with the parameters provided in Clustal W, version 1.6. A phylogenetic tree with 1,000 bootstrap replications was constructed by the neighbor-joining method, and genetic distances were calculated by the Kimura two-parameter method (8). The resulting tree was drawn with NJplot software.
PFGE analysis.
PFGE plugs were prepared as described by Hielm et al. (12). DNA was left undigested or was digested with 20 U of SmaI (GE Healthcare UK Ltd., Little Chalfont, Buckinghamshire, United Kingdom) in 200 μl of optimal buffer at 25°C for 18 h. The plugs were electrophoresed in a CHEF-DRIII apparatus (Bio-Rad Laboratories, Hercules, CA) through a 1% pulse-field-certified agarose gel (Bio-Rad Laboratories) in 0.5× Tris-borate-EDTA buffer at 14°C and 6 V/cm. The switching times were ramped from 0.5 to 40 s, and the bands were visualized by ethidium bromide staining. Dendrogram analysis of the band patterns was performed with Fingerprinting II software (Bio-Rad Laboratories). Similarity analysis was performed by using the Dice coefficient, and clustering was examined by the unweighted pair group method with arithmetic averages. Approximate fragment sizes (kbp) were measured by use of a molecular size marker.
Southern blot hybridization.
After PFGE, the DNA fragments in the gel were transferred onto Hybond N+ nylon membranes (GE Healthcare) with a capillary transfer system and 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 18 h. The DNA was fixed to the membrane by UV irradiation. The boNT/B and 16S rRNA gene probes were prepared by using the primers reported by Szabo et al. (39) and Marshall et al. (25), and DNA extracted from strain Okra was labeled by use of a PCR DIG probe synthesis kit (Roche Diagnostics, Mannheim, Germany). The membranes were incubated at 37°C for 2 h with hybridization solution (DIG-Easy Hyb; Roche Diagnostics), and hybridizations were performed at 42°C for 18 h with fresh DIG-Easy Hyb containing 20 ng/ml of each probe. The membranes were then washed three times for 15 min each time at room temperature with 1× SSC containing 0.1% sodium dodecyl sulfate and twice for 15 min each time with 0.1× SSC containing 0.1% sodium dodecyl sulfate at 60°C for the boNT/B-specific probe and at 65°C for the 16S rRNA-specific probe. Hybridization signals were detected with a DIG luminescent detection kit (Roche Diagnostics).
PCR assays for BoNT/B subtyping.
PCR was performed with four primer mixtures, as follows: primer B-forward (5′-GATTTTTGGGGAAATCTTT-3′; positions in the coding region, bases 3256 to 3275), primer B1-reverse (5′-CCAATTACATCCCAATTTTAAA-3′; positions, bases 3840 to 3819), primer B2-reverse (5′-GTATAGTTTTGTAAAATTCATTAGAATCATA-3′; positions, bases 3625 to 3595), and primer Osaka05-reverse (5′-TCTTCCTTTTTCTTAAAATTTTTAAG-3′; positions, bases 3572 to 3547). These primers were designed on the basis of the boNT sequences published by GenBank and determined in this study. PCR was performed with a 25-μl reaction mixture containing 0.3 to 1 ng template DNA, 0.25 μM of each primer, 1.25 U Ex Taq (TaKaRa Shuzo), 2.5 μl Ex Taq buffer (TaKaRa Shuzo), and 200 μM of each dNTP. Each PCR cycle consisted of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min and was repeated 25 to 35 times. Final extension was carried out at 72°C for 5 min. The amplicons were visualized in 3% Nu-Sieve agarose gels (Camblex Bio Science, Rockland, ME) stained with ethidium bromide.
Sandwich enzyme-linked immunosorbent assay (ELISA) with rHC.
Recombinant BoNT/B HC (rHC) from strain Osaka05 (rHC/Osaka05), rHC/Okra, and rHC/111 were expressed as described in our previous report (15), except that the primers used for the amplification of the DNA fragment encoding BoNT/Osaka05 HC were as follows: forward primer 5′-CACGGATCCAAAAAATATAATAGCGAAATTTTAAA-3′ and reverse primer 5′-ATTAAGCTTTTATTCAATCCATCCTTCATCTTT-3′. The rHC that was expressed was purified with nickel-nitrilotriacetic acid agarose (Qiagen) and by CM-Sephadex C-25 (GE Healthcare) chromatography. Polyclonal antibody against each rHC was prepared with rabbits. The first subcutaneous injection was performed with purified 50 μg rHC in Freund's complete adjuvant. After 2 weeks, the animals received three booster injections of the same amount of rHC in Freund's incomplete adjuvant at 2-week intervals. The animals were bled 2 weeks after the last booster injection. The immunoglobulin G fraction was purified by ammonium sulfate precipitation and was subsequently purified on an rHC-coupled HiTrap N-hydroxysuccinimide-activated high-performance column (GE Healthcare), according to the manufacturer's instructions. Biotinylated antibodies were prepared with EZ-Link sulfosuccinimidyl-6-[biotinamido]hexanoate (Pierce, Rockford, IL).
For the sandwich ELISA, 96-well microtiter plates (Iwaki, Japan) were coated with 0.1 ml polyclonal antibody against rHC (3 μg/ml) at 37°C for 2 h. After the plates were blocked at 4°C for 18 h with 1.0% Blockace blocking reagent (Dainippon Sumitomo Pharma Co., Japan), rHC was applied at dilutions ranging from 10 ng/ml to 5,000 ng/ml in duplicate and the plates were incubated at 37°C for 1 h. The plates were washed and incubated with 1 μg/ml biotinylated antibody against rHC at 37°C for 1 h, followed by washing and incubation with 0.2 μg/ml streptavidin-conjugated horseradish peroxidase (Pierce) at 37°C for 1 h. The plates were developed with 2,2-azinobis-(3-ethylbenzthiazoline sulfonic acid) (Roche) at 37°C for 1 h. The color was monitored by measuring the absorbance at 415 nm on a Labsystem Multiscan mass spectrometer (Labsystems, Finland).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this work were submitted to the DDBJ database and may be found under accession numbers AB302852 to AB302865 (Table 1).
RESULTS
Comparison of boNT/B and nontoxic component gene sequences among strains Osaka05, Okra (subtype B1), and 111 (subtype B2).
The nucleotide sequences of the boNT/B gene (3,876 bp) from two isolates associated with infant botulism in Japan (strains Osaka05 and Osaka06) and nontoxic component genes (ha70, 1,881 bp; ha17, 441 bp; ha33, 879 bp; botR, 537 bp; ntnh, 3,594 bp) from strains Osaka05 and 111 were determined. The boNT/B gene nucleotide sequences of strains Osaka05 and Osaka06 shared 98.8% identity. These sequences were further compared with 24 full-length boNT/B gene sequences available in the GenBank database (Table 1). The boNT/B gene sequence of strain Osaka06 was identical to that of strain CDC6291, which was classified as subtype B2. Strain Osaka05 was shown to possess a unique sequence in comparison with the sequences of the other boNT/B genes.
The nucleotide and amino acid identities of boNT/B and nontoxic components among strains Osaka05, Okra (subtype B1), and 111 (subtype B2) are summarized in Table 3. At the nucleotide and amino acid levels in boNT/B, Osaka05 had identities of 97.9% and 96.2%, respectively, with Okra and 99.0% and 98.5%, respectively, with 111, while Okra and 111 possessed identities of 97.6% and 95.7% at the nucleotide and amino acid levels in boNT/B, respectively. The organization of nontoxic component genes (composed of ha70, ha17, ha33, botR, and ntnh) and the boNT/B gene was shown to be common among the three strains. For all nontoxic components except ha33, Osaka05 showed 97.9% to 99.6% nucleotide sequence identities and 96.7% to 99.0% amino acid sequence identities with Okra and 98.6% to 99.6% nucleotide sequence identities and 98.4% to 100.0% amino acid sequence identities with 111, while Okra had 97.8% to 98.9% nucleotide sequence identities and 96.4% to 98.1% amino acid sequence identities with 111. For ha33, the three strains shared 92.3% to 95.2% nucleotide sequence identities and 83.6% to 90.1% amino acid sequence identities, which were remarkably lower than the levels of identity for the other nontoxic component genes.
TABLE 3.
Identities among strains Osaka05, Okra (subtype B1), and 111 (subtype B2) in the boNT/B and nontoxic componentsa
| Strains compared | % Nucleotide/% amino acid identity
|
|||||
|---|---|---|---|---|---|---|
| HA70 (1,881/626)a | HA17 (441/146) | HA33 (885[879]/294[292]b) | BotR (537/178) | NTNH (3,594/1,197) | BoNT/B (3,876/1,291) | |
| Osaka05 and Okra (B1) | 99.6/99.0 | 99.3/97.9 | 95.2/90.1 | 98.5/97.2 | 97.9/96.7 | 97.9/96.2 |
| Osaka05 and 111 (B2) | 99.1/98.4 | 98.6/100.0 | 92.3/83.6 | 99.4/98.9 | 99.6/99.3 | 99.0/98.5 |
| Okra and 111 | 98.9/98.1 | 98.0/97.9 | 93.1/84.9 | 98.3/96.6 | 97.8/96.4 | 97.6/95.7 |
Data in parentheses are number of nucleotide base pairs/number of amino acid residues.
Data in brackets are for strains Osaka05 and 111.
The amino acid substitutions in BoNT/B among strains Okra, 111, and Osaka05 involved 61 residues (2 residues in the light chain, which was 441 amino acids [aa], and 59 residues in the heavy chain [which was 850 aa]), and the substitutions spread throughout the domains or the subdomains within the heavy chain were as follows: 22 in HN (420 aa), 37 in HC (430 aa), 12 in HCN (167 aa), and 25 in HCC (263 aa). These substitutions were mostly concentrated in the heavy chain, especially in the HCC subdomain. Osaka05 showed identities of 99.5% (light chain), 94.5% (heavy chain), 95.2% (HN), 92.8% (HCN), and 94.7% (HCC) with Okra at the amino acid level and identities of 100.0% (light chain), 97.6% (heavy chain), 98.6% (HN), 100.0% (HCN), and 94.3% (HCC) with 111 at the amino acid level. Okra had identities of 99.5% (light chain), 93.6% (heavy chain), 95.5% (HN), 92.8% (HCN), and 91.3% (HCC) with 111 at the amino acid level (Fig. 1).
FIG. 1.
Summary of BoNT/B amino acid substitutions between strains Osaka05, Okra (subtype B1), and 111 (subtype B2) in each domain (light chain and HN) or subdomain (HCN and HCC in HC). Hyphens indicate residues identical to those in strain Okra.
Phylogenetic analysis of full-length boNT/B gene.
The phylogenetic tree was constructed on the basis of alignment of the full length of the nucleotide sequences of the boNT/B genes from strains Osaka05 and Osaka06 with 24 boNT/B sequences, including the sequences of the five BoNT/B subtypes, published in GenBank (Fig. 2A). Overall, among the 26 strains producing BoNT/B, 25 (the exception was Osaka05) were classified into five clusters, which was the same classification as the BoNT/B subtypes. Strain Osaka06 was classified into the subtype B2 cluster and showed 99.3% to 100% identity with the subtype B2 strains. The new isolate, strain Osaka05, was different from the strains in the other five clusters and shared 98.8% to 99.1% nucleotide sequence identity with the subtype B2 strains, 98.9% identity with the subtype B3 strains, identities of 97.7% to 98.0% with the subtype B1 strains, identities of 97.4% with the bivalent strains, and identities of 95.7% to 96.2% with the nonproteolytic strains.
FIG. 2.
Phylogenetic trees based on boNT/B gene sequences. The full-length boNT/B gene nucleotide sequences (3,876 bp) (A) and the nucleotide sequences at the C-terminal region (400 bp) of BoNT/B (B) were constructed by use of the reference sequences listed in Table 1. The sequences determined in this study are indicated in boldface. The outgroup was the neurotoxin sequence of C. tetani CN3911 (GenBank accession no. X06214) and was removed from the final figure (A), and nonproteolytic C. botulinum Eklund17B type B was the outgroup for the BoNT/B sequences (B). The five BoNT/B subtypes (subtypes B1, B2, B3, bivalent, and nonproteolytic) were described by Hill et al. (13). The BoNT/B subtype newly identified in this study is boxed. Numbers on each branch indicate bootstrap values (>950) for the cluster supported by that branch.
Phylogenetic analysis of the C-terminal region of BoNT/B.
The phylogenetic tree was also constructed on the basis of the alignment of the nucleotide sequences at the C-terminal region (400 bp) of BoNT/B, which partially encoded the HCC subdomain (Fig. 2B). Overall, except for Osaka05, 28 proteolytic type B strains (the 15 bacterial strains and the 13 reference strains; Table 1) were classified into three subtypes (subtypes B1, B2, and B3), which was the same as the classification for the full-length boNT/B genes (Fig. 2A). Subtype B1 included five isolates associated with U.S. infant botulism, two isolates from pork meat, two stocked strains, and five reference strains. Subtype B2 included two isolates associated with Japanese infant botulism, isolates from ginger and honey, and eight reference strains. Strains CDC795 (B3 subtype) and Osaka05 were also different from the strains in the other clusters and were not classified in the same cluster.
PFGE and Southern blot hybridization.
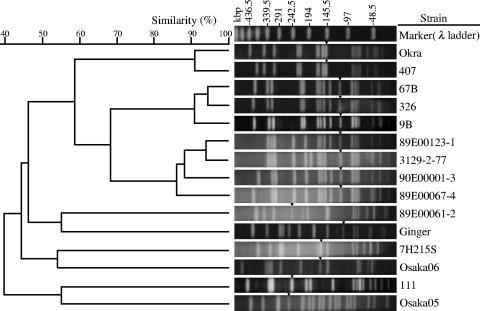
The PFGE patterns of SmaI-digested DNA from 15 C. botulinum type B strains and a dendrogram based on the similarities between normalized PFGE patterns are presented in Fig. 3. All strains were distinguishable, and their similarities ranged from 25.8% to 94.1%. The BoNT/B subtypes were clearly different on PFGE analysis. Strain Osaka05 exhibited a pattern entirely different from the patterns of the other 14 strains, having similarities of 28.6% to 45.7% with subtype B1 strains and similarities of 25.8% to 54.6% with subtype B2 strains. The similarities between subtype B1 and subtype B2 strains ranged from 29.4% to 60.0%. Homologous subtypes of subtype B1 strains showed similarities of 41.2% to 94.1%. Nine of the 10 subtype B1 strains (the exception was strain 89E00061-2) were separated into three clusters (Okra and 407; 326, 9B, and 67B; and 89E00067-4, 89E00123-1, 90E00001-3, and 3129-2-77) with more than 80% similarity. The four subtype B2 strains (strains 111, Osaka06, Ginger, and 7H215S) showed more diversity and had 35.7% to 53.9% similarities.
FIG. 3.
PFGE genotyping. The dendrogram and PFGE patterns of SmaI-digested DNA from 15 C. botulinum type B strains are shown. Arrowheads indicate the DNA fragment containing the boNT/B gene detected by Southern blot hybridization.
Fragments containing boNT/B genes were detected by Southern blot hybridization and are indicated by arrowheads in the gel in Fig. 3. An approximately 100-kbp fragment was detected in strain Ginger; a 110-kbp fragment was detected in strains 89E0067-4, 89E123-1, 90E00001-3, 3129-2-77, 326, 9B, and 67B; a 150-kbp fragment was detected in strains Okra and 407; a 170-kbp fragment was detected in strain 7H215S; a 175-kbp fragment was detected in strain Osaka06; a 260-kbp fragment was detected in strain 89E00061-2; a 275-kbp fragment was detected in strain 111; and a 280-kbp fragment was detected in strain Osaka05.
DNA fragments smaller than the chromosomal DNA (>970 kbp) were detected from strains 111, Osaka05, 89E00061-2, Okra, and 407 by PFGE of undigested DNA (Fig. 4A). The location of the boNT/B gene was confirmed by subsequent Southern blot hybridization with a boNT/B probe. The boNT/B genes of strains Okra and 407 were on the approximately 150-kbp fragment, the boNT/B gene of strain 89E00061-2 was located on the 260-kbp fragment, the boNT/B gene of strain 111 was located on the 275-kbp fragment, and the boNT/B gene of strain Osaka05 was located on the 280-kbp fragment (Fig. 4B), while the 16S rRNA genes were located on the chromosomal DNA (Fig. 4C). For the remaining 10 strains, the chromosomal DNA fragment was detected only by PFGE of undigested DNA, and both the boNT/B and the 16S rRNA genes were located on the chromosomal DNA.
FIG. 4.
PFGE of undigested DNA and Southern blot hybridization. The PFGE patterns of undigested DNA (A) and Southern blot hybridization detection of the boNT/B genes (B) and the 16S rRNA genes (C) of strains Okra (lane 1), 407 (lane 2), 89E00061-2 (lane 3), 111 (lane 4), and Osaka05 (lane 5) are shown. Lanes M, bacteriophage λ ladder.
PCR assays for identification of BoNT/B subtypes.
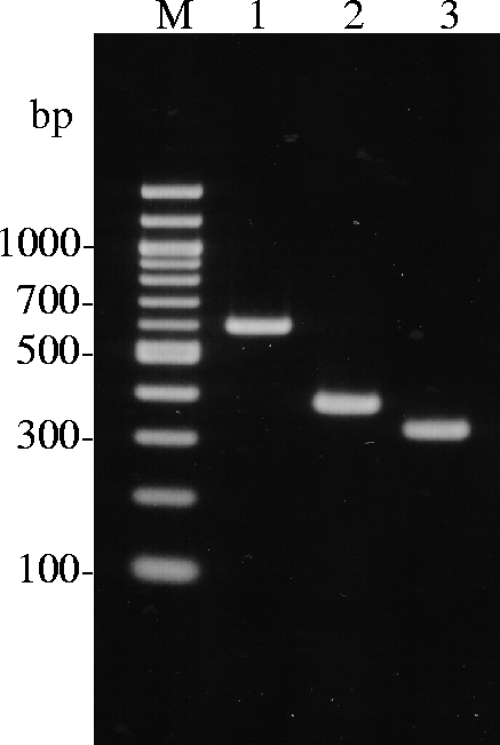
A multiplex PCR assay for the detection of the boNT/B1, boNT/B2, and boNT/Osaka05 genes was developed. The results of the PCR assay with strains Okra (subtype B1), 111 (subtype B2), and Osaka05 are shown in Fig. 5.
FIG. 5.
Multiplex PCR assay for BoNT/B subtyping. The results of the multiplex PCR assay with strains Okra (lane 1), 111 (lane 2), and Osaka05 (lane 3) were visualized. Lanes M, 100-bp ladder.
The boNT/B1 amplicon (585 bp) from strain Okra was detected in lane 1, the boNT/B2 amplicon (370 bp) from strain 111 was detected in lane 2, and the boNT/Osaka05 amplicon (317 bp) from strain Osaka05 was detected in lane 3. When the other bacterial strains listed in Table 1 were assayed, the boNT/B1 amplicon was detected in nine strains (strains 89E00061-2, 89E00067-4, 89E00123-1, 90E00001-3, 3129-2-77, 326, 407, 9B, and 67B), and the boNT/B2 amplicon was detected in three strains (strains Osaka06, Ginger, and 7H215S). The detection limit of the multiplex PCR assay was from 9.5 × 10 to 2.5 × 102 cells/ml of culture dilution (data not shown). No amplicons were detected from the 11 control strains, as follows: 2 type A strains (strains 62A and Kyoto-F), 1 type C strain (strain CB-19), 1 type D strain (strain 1873), 1 type E strain (strain Iwanai), 1 type F strain (strain Langeland), 1 BoNT/E-producing C. butylicum strain (strain 5262), 1 C. sporogenes strain (strain ATCC 19404), 1 C. bifementas strain (strain ATCC 638), 1 C. perfringens strain (strain ATCC 13124), and 1 C. difficile strain (strain ATCC 43593) (data not shown).
Comparison of antigenicities of BoNT/B HC among strains Osaka05, Okra, and 111.
In order to characterize BoNT/Osaka05 antigenically, the levels of binding of rHC/Osaka05, rHC/Okra, and rHC/111 to their specific antibodies were measured by a sandwich ELISA (Fig. 6). When the levels of binding of rHC to the antibody against rHC/Osaka05 were assayed, rHC/111 and rHC/Okra showed low binding affinities, and the binding affinity of rHC/Okra was lower than that of rHC/111 (Fig. 6A). rHC/Osaka05 and rHC/111 did not react to the antibody against rHC/Okra (Fig. 6B). rHC/Osaka05 and rHC/Okra exhibited a low binding affinity to the antibody against rHC/111, and the binding affinity of rHC/Okra was lower than that of rHC/Osaka05 (Fig. 6C).
FIG. 6.
Sandwich ELISA for determination of the binding affinity of rHC to specific antibodies. The results are expressed as the means of the optical density (OD) at 415 nm after subtraction of the optical density for the background control in three experiments. Error bars indicate standard errors. Antibodies against rHC/Osaka05 (A), rHC/Okra (B), and rHC/111 (C) were used as capture antibodies; and rHC/Osaka05 (closed circles), rHC/Okra (open triangles), and rHC/111 (open squares) were applied as antigens at various concentrations.
DISCUSSION
Two cases of infant botulism occurred in Osaka, Japan, in 2005 and 2006, and type B strains (strains Osaka05 and Osaka06) were successfully isolated from both cases. The full-length boNT/B gene sequences of the two isolates were determined and compared with the sequences of the boNT/B genes in the GenBank database. Strain Osaka05 possessed a unique boNT/B gene. The BoNT subtypes within a serotype were defined as differing by at least 2.6% at the amino acid level (37). Currently, it is usual to determine BoNT subtypes by phylogenetic analysis of full-length boNT gene sequences (6, 13). Phylogenetic analysis of the boNT/B genes indicated that strain Osaka05 should be classified into a group other than the five BoNT/B subtypes (13), and strain Osaka05 was shown to be a new BoNT/B subtype. On the other hand, the boNT/B gene sequence of strain Osaka06 was identical to that of strain CDC6291 and was classified in the B2 subtype (Fig. 2A).
The amino acid substitutions in the BoNT/B subtypes were concentrated in the heavy chain, especially in HCC, the same as in Fig. 1. The sequences in the C-terminal region of BoNT/B, which encoded HCC, were available for BoNT/B subtyping by phylogenetic analysis, as was the sequence of the full-length of boNT/B genes (Fig. 2B). We also established a multiplex PCR assay to detect BoNT/B subtypes B1, B2, and Osaka05 (Fig. 5). The PCR results were well correlated with the subtypes identified by phylogenetic analysis of boNT/B genes.
Genotyping by PFGE with SmaI-digested DNA revealed genetic diversity among subtypes B1, B2, and Osaka05 (Fig. 3). The diversity within subtype B2 strains was greater than that within B1 strains. The seven isolates from Japan (associated with infant botulism and food samples) were clearly distinct from each other, in contrast to the isolates associated with infant botulism in the United States, which showed high degrees of similarity.
The location of the boNT gene and its associated nontoxic component gene cluster varied among serotypes and strains; the boNT/A, boNT/B, boNT/E, and boNT/F genes were considered to be located on the chromosome, while the boNT/C and boNT/D genes were carried on bacteriophages, and the boNT/G gene was located on plasmids (4, 30, 41, 43). Recently, sequencing of the complete genome of strain Okra revealed that its boNT/B gene was present within the 149-kbp plasmid (36). The boNT genes of strain Loch Maree (subtype A3) and strain 657Ba (type B and subtype A4) were also found to be located on the approximately 270-kbp plasmid (25, 36). In this study, we found that the boNT/B genes were located on extrachromosomal DNA, assumed to be plasmids, in five strains belonging to distinct BoNT/B subtypes (Fig. 4). The plasmids were approximately 150 kbp, 260 kbp, 275 kbp, and 280 kbp. These findings indicate that the boNT/B gene location is not correlated with the BoNT/B subtype. Detailed characterization of the boNT/B gene-encoding plasmids is required to understand the mechanisms of boNT/B expression and evaluation of the boNT/B gene within C. botulinum.
The nontoxic component genes encode the proteins that protect the neurotoxin from the acids and proteases in the stomach and assist with transportation of the neurotoxin from the intestine to the bloodstream (30). This study and previous reports (23, 30, 36) indicated that the hemagglutinin genes (ha70, ha33, and ha17), the regulator gene (botR), and the nontoxic-nonhemagglutinin gene (ntnh) exist upstream of the boNT/B gene. The amino acid identities of HA33 were significantly lower than those of the other nontoxic components and BoNT/B (Table 3). It was suggested that HA33 acts as an adhesin, allowing the complex of the neurotoxin and nontoxic components to bind to intestinal epithelial cells and erythrocytes (11, 26); however, the influence of amino acid substitutions in HA33 on symptoms of infant botulism is unknown.
The variation in the BoNT amino acid sequence within serotypes is capable of causing significant differences in the immunological and biological properties of the neurotoxin. We previously indicated immunological differences between BoNT/B1 and BoNT/B2 (15, 22). Briefly, most monoclonal antibodies against the HC of BoNT/Okra did not react with BoNT/111, while monoclonal antibodies against the light chain and the HN of BoNT/Okra could react with BoNT/111 by immunoblotting and ELISA. In this study, the antigenicity of BoNT/Osaka05 was suggested to be different from the antigenicities of BoNT/Okra and BoNT/111 by sandwich ELISA with rHC and their specific antibodies (Fig. 6). Strain Osaka05 was confirmed to be a new BoNT/B subtype by its antigenic specificity, in addition to by subtype classification by phylogenetic analysis of the boNT/B gene. The antigenic difference between BoNT/Osaka05 and BoNT/Okra was greater than that between BoNT/Osaka05 and BoNT/111, depending upon the difference in BoNT amino acid similarities. We also previously demonstrated that two different subtypes in BoNT/A (subtypes A1 and A2), which differ by 10% at the amino acid level, had different antigenicities by ELISA with monoclonal antibodies against BoNT/A (21). Similar findings were presented by Smith et al. (37). This information will be important for the development of an immunological BoNT assay, therapy for botulism, and recombinant vaccines for BoNT (34, 35).
In HCC, the level of amino acid replacement between BoNT/Osaka05 and BoNT/Okra was 15 residues, that between BoNT/Osaka05 and BoNT/111 was 14 residues, and that between BoNT/Okra and BoNT/111 was 23 residues. Our previous studies revealed that the toxicity of BoNT/111 was lower than that of BoNT/Okra because of the replacement of 2 residues in HCC which contribute to binding to the receptors (ganglioside GT1b and the synaptotagmin2-GT1b complex) (15, 20, 22). The other 8 residues essential for receptor binding in HCC were also confirmed: 4 residues for binding to ganglioside GT1b and 4 residues for binding to the synaptotagmin2-GT1b complex (20, 32, 38). In BoNT/Osaka05 and BoNT/Okra, those 10 residues were identical. The antigenicity and genetic characteristics of BoNT/Osaka05 were related to those of BoNT/111 rather than to those of BoNT/Okra, but the residues contributing to receptor binding in BoNT/Osaka05 were identical to those in BoNT/Okra. Further investigation into the biological character of BoNT/Osaka05 would give new insight into the receptor binding system of BoNT/B.
In this study, we developed new methods for the subtyping of BoNT/B: phylogenetic analysis of partial boNT/B gene sequences and a multiplex PCR assay. The former was also suitable for the identification of a new BoNT/B subtype, and the latter represents the first report of a PCR-based method for the identification of BoNT/B subtypes. The correlation between the BoNT/B subtype and the source of isolation was reported by Hill et al. (13); B1 strains likely originated in the United States and are associated with food-borne disease due to improperly processed vegetables, while B2 strains exist mostly in Europe and are associated with animal cases or meat. The distribution of BoNT/B subtypes in Japan was found to be distinct from the distributions in both the United States and Europe; isolates associated with infant botulism were classified into subtype B2 and the newly identified Osaka05 subtype, and food samples were divided into subtypes B1 and B2. Therefore, further molecular genotyping of type B C. botulinum isolates by our BoNT/B subtyping methods will contribute to understanding of the epidemiology of C. botulinum and the infectious diseases that it causes.
Acknowledgments
We thank T. Asao and S. Nakamura for the gifts of the C. botulinum strains.
This work was partially supported by a grant for research on food safety from the Ministry of Health, Labor and Welfare, Japan.
Footnotes
Published ahead of print on 1 July 2009.
REFERENCES
- 1.Abe, S., M. Nishino, O. Miyake, N. Hashimoto, T. Nakagawa, E. Satake, T. Asao, Y. Kumada, T. Kawai, and M. Takahashi. 2006. An infant botulism case in which detected were Clostridium botulinum type B and its toxin, May 2006—Osaka. Infect. Agents Surveill. Rep. 27275-276. (In Japanese.) [Google Scholar]
- 2.Arndt, J. W., M. J. Jacobson, E. E. Abola, C. M. Forsyth, W. H. Tepp, J. D. Marks, E. A. Johnson, and R. C. Stevens. 2006. A structural perspective of the sequence variability within botulinum neurotoxin subtypes A1-A4. J. Mol. Biol. 362733-742. [DOI] [PubMed] [Google Scholar]
- 3.Arnon, S. S., K. Damus, and J. Chin. 1981. Infant botulism: epidemiology and relation to sudden infant death syndrome. Epidemiol. Rev. 345-66. [DOI] [PubMed] [Google Scholar]
- 4.Binz, T., H. Kurazono, M. Wille, J. Frevert, K. Wernars, and H. Niemann. 1990. The complete sequence of botulinum neurotoxin type A and comparison with other clostridial neurotoxins. J. Biol. Chem. 2659153-9158. [PubMed] [Google Scholar]
- 5.Cai, S., B. R. Singh, and S. Sharma. 2007. Botulism diagnostics: from clinical symptoms to in vitro assays. Crit. Rev. Microbiol. 33109-125. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y., H. Korkeala, J. Aarnikunnas, and M. Lindstrom. 2007. Sequencing the botulinum neurotoxin gene and related genes in Clostridium botulinum type E strains reveals orfx3 and a novel type E neurotoxin subtype. J. Bacteriol. 1898643-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, N., and R. Hinkle. 2002. Infant botulism. Am. Fam. Physician 651388-1392. [PubMed] [Google Scholar]
- 8.Efron, B., E. Halloran, and S. Holmes. 1996. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 9313429-13434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, C. K., C. A. Keet, and J. B. Strober. 2005. Recent advances in infant botulism. Pediatr. Neurol. 32149-154. [DOI] [PubMed] [Google Scholar]
- 10.Franciosa, G., F. Floridi, A. Maugliani, and P. Aureli. 2004. Differentiation of the gene clusters encoding botulinum neurotoxin type A complexes in Clostridium botulinum type A, Ab, and A(B) strains. Appl. Environ. Microbiol. 707192-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujinaga, Y., K. Inoue, S. Watarai, Y. Sakaguchi, H. Arimitsu, J. C. Lee, Y. Jin, T. Matsumura, Y. Kabumoto, T. Watanabe, T. Ohyama, A. Nishikawa, and K. Oguma. 2004. Molecular characterization of binding subcomponents of Clostridium botulinum type C progenitor toxin for intestinal epithelial cells and erythrocytes. Microbiology 1501529-1538. [DOI] [PubMed] [Google Scholar]
- 12.Hielm, S., J. Bjorkroth, E. Hyytia, and H. Korkeala. 1998. Genomic analysis of Clostridium botulinum group II by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 64703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, K. K., T. J. Smith, C. H. Helma, L. O. Ticknor, B. T. Foley, R. T. Svensson, J. L. Brown, E. A. Johnson, L. A. Smith, R. T. Okinaka, P. J. Jackson, and J. D. Marks. 2006. Genetic diversity among botulinum neurotoxin producing clostridial strains. J. Bacteriol. 189818-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutson, R. A., M. D. Collins, A. K. East, and D. E. Thompson. 1994. Nucleotide sequence of the gene coding for non-proteolytic Clostridium botulinum type B neurotoxin: comparison with other clostridial neurotoxins. Curr. Microbiol. 28101-110. [DOI] [PubMed] [Google Scholar]
- 15.Ihara, H., T. Kohda, F. Morimoto, K. Tsukamoto, T. Karasawa, S. Nakamura, M. Mukamoto, and S. Kozaki. 2003. Sequence of the gene for Clostridium botulinum type B neurotoxin associated with infant botulism, expression of the C-terminal half of heavy chain and its binding activity. Biochim. Biophys. Acta 162519-26. [DOI] [PubMed] [Google Scholar]
- 16.Infectious Disease Surveillance Center, National Institute of Infectious Diseases. 2008. Botulism in Japan as of January 2008. Infect. Agents Surveill. Rep. 2935-36. (In Japanese.) [Google Scholar]
- 17.Johnson, E. A., and M. Bradshaw. 2001. Clostridium botulinum and its neurotoxins: a metabolic and cellular perspective. Toxicon 391703-1722. [DOI] [PubMed] [Google Scholar]
- 18.Kakinuma, H., H. Maruyama, H. Takahashi, K. Yamakawa, and S. Nakamura. 1996. The first case of type B infant botulism in Japan. Acta Paediatr. Jpn. 38541-543. [DOI] [PubMed] [Google Scholar]
- 19.Kirma, N., J. L. Ferreira, and B. R. Baumstark. 2004. Characterization of six type A strains of Clostridium botulinum that contain type B toxin gene sequences. FEMS Microbiol. Lett. 231159-164. [DOI] [PubMed] [Google Scholar]
- 20.Kohda, T., H. Ihara, Y. Seto, H. Tsutsuki, M. Mukamoto, and S. Kozaki. 2007. Differential contribution of the residues in C-terminal half of the heavy chain of botulinum neurotoxin type B to its binding to the ganglioside GT1b and the synaptotagmin 2/GT1b complex. Microb. Pathog. 4272-79. [DOI] [PubMed] [Google Scholar]
- 21.Kozaki, S., S. Nakaue, and Y. Kamata. 1995. Immunological characterization of the neurotoxin produced by Clostridium botulinum type A associated with infant botulism in Japan. Microbiol. Immunol. 39767-774. [DOI] [PubMed] [Google Scholar]
- 22.Kozaki, S., Y. Kamata, T. Nishiki, H. Kakinuma, H. Maruyama, H. Takahashi, T. Karasawa, K. Yamakawa, and S. Nakamura. 1998. Characterization of Clostridium botulinum type B neurotoxin associated with infant botulism in Japan. Infect. Immun. 664811-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J. C., K. Yokota, H. Arimitsu, H. J. Hwang, Y. Sakaguchi, J. Cui, K. Takeshi, T. Watanabe, T. Ohyama, and K. Oguma. 2005. Production of anti-neurotoxin antibody is enhanced by two subcomponents, HA1 and HA3b, of Clostridium botulinum type B 16S toxin-haemagglutinin. Microbiology 1513739-3747. [DOI] [PubMed] [Google Scholar]
- 24.Lindstrom, M., and H. Korkeala. 2006. Laboratory diagnostics of botulism. Clin. Microbiol. Rev. 19298-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall, K. M., M. Bradshaw, S. Pellett, and E. A. Johnson. 2007. Plasmid encoded neurotoxin genes in Clostridium botulinum serotype A subtypes. Biochem. Biophys. Res. Commun. 36149-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumura, T., Y. Jin, Y. Kabumoto, Y. Takegahara, K. Oguma, W. I. Lencer, and Y. Fujinaga. 2008. The HA proteins of botulinum toxin disrupt intestinal epithelial intercellular junctions to increase toxin absorption. Cell. Microbiol. 10355-364. [DOI] [PubMed] [Google Scholar]
- 27.Midura, T. F., and S. S. Arnon. 1976. Infant botulism. Identification of Clostridium botulinum and its toxins in faeces. Lancet ii934-936. [DOI] [PubMed] [Google Scholar]
- 28.Nishida, H., M. Shiota, K. Nakagawa, T. Minamigata, M. Takuwa, T. Morishima, H. Kumakura, T. Yoshioka, A. Uematsu, N. Haneda, D. Hata, K. Umeda, J. Ogasawara, and M. Takahashi. 2007. An infant botulism case due to Clostridium botulinum type B toxin, October 2005—Osaka City. Infect. Agents Surveill. Rep. 28168-169. (In Japanese.) [Google Scholar]
- 29.Noda, H., K. Sugita, A. Koike, T. Nasu, M. Takahashi, T. Shimizu, K. Ooi, and G. Sakaguchi. 1988. Infant botulism in Asia. Am. J. Dis. Child. 142125-126. [DOI] [PubMed] [Google Scholar]
- 30.Oguma, K., Y. Fujinaga, and K. Inoue. 1995. Structure and function of Clostridium botulinum toxins. Microbiol. Immunol. 39161-168. [DOI] [PubMed] [Google Scholar]
- 31.Pickett, J., B. Berg, E. Chaplin, and M. A. Brunstetter-Shafer. 1976. Syndrome of botulism in infancy: clinical and electrophysiologic study. N. Engl. J. Med. 295770-772. [DOI] [PubMed] [Google Scholar]
- 32.Rummel, A., S. Mahrhold, H. Bigalke, and T. Binz. 2004. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Mol. Microbiol. 51631-643. [DOI] [PubMed] [Google Scholar]
- 33.Santos-Buelga, J. A., M. D. Collins, and A. K. East. 1998. Characterization of the genes encoding the botulinum neurotoxin complex in a strain of Clostridium botulinum producing type B and F neurotoxins. Curr. Microbiol. 37312-318. [DOI] [PubMed] [Google Scholar]
- 34.Shone, C., J. Ferreira, A. Boyer, N. Cirino, C. Egan, E. Evans, J. Kools, and S. Sharma. 2006. The 5th International Conference on Basic and Therapeutic Aspects of Botulinum and Tetanus Neurotoxins. Workshop review: assays and detection. Neurotox. Res. 9205-216. [DOI] [PubMed] [Google Scholar]
- 35.Smith, L. A. 1998. Development of recombinant vaccines for botulinum neurotoxin. Toxicon 361539-1548. [DOI] [PubMed] [Google Scholar]
- 36.Smith, T. J., K. K. Hill, B. T. Foley, J. C. Detter, A. C. Munk, D. C. Bruce, N. A. Doggett, L. A. Smith, J. D. Marks, G. Xie, and T. S. Brettin. 2007. Analysis of the neurotoxin complex genes in Clostridium botulinum A1—A4 and B1 strains: BoNT/A3, /Ba4 and /B1 clusters are located within plasmids. PLoS One 2e1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, T. J., J. Lou, I. N. Geren, C. M. Forsyth, R. Tsai, S. L. Laporte, W. H. Tepp, M. Bradshaw, E. A. Johnson, L. A. Smith, and J. D. Marks. 2005. Sequence variation within botulinum neurotoxin serotypes impacts antibody binding and neutralization. Infect. Immun. 735450-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swaminathan, S., and S. Eswaramoorthy. 2000. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat. Struct. Biol. 7693-699. [DOI] [PubMed] [Google Scholar]
- 39.Szabo, E. A., J. M. Pemberton, and P. M. Desmarchelier. 1993. Detection of the genes encoding botulinum neurotoxin types A to E by the polymerase chain reaction. Appl. Environ. Microbiol. 593011-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeshi, K., Y. Fujinaga, K. Inoue, H. Nakajima, K. Oguma, T. Ueno, H. Sunagawa, and T. Ohyama. 1996. Simple method for detection of Clostridium botulinum type A to F neurotoxin genes by polymerase chain reaction. Microbiol. Immunol. 405-11. [DOI] [PubMed] [Google Scholar]
- 41.Whelan, S. M., M. J. Elmore, N. J. Bodsworth, J. K. Brehm, T. Atkinson, and N. P. Minton. 1992. Molecular cloning of the Clostridium botulinum structural gene encoding the type B neurotoxin and determination of its entire nucleotide sequence. Appl. Environ. Microbiol. 582345-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamakawa, K., T. Karasawa, H. Kakinuma, H. Maruyama, H. Takahashi, and S. Nakamura. 1997. Emergence of Clostridium botulinum type B-like nontoxigenic organisms in a patient with type B infant botulism. J. Clin. Microbiol. 352163-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, Y., H. Sugiyama, H. Nakano, and E. A. Johnson. 1995. The genes for the Clostridium botulinum type G toxin complex are on a plasmid. Infect. Immun. 632087-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]