Abstract
Human rhinovirus C (HRV-C) is a newly identified genotype of HRV found in patients with respiratory tract infections (RTIs); however, its epidemiological profile and clinical characteristics are not well understood. In this study, Chinese children with RTIs were screened for HRV-C and their epidemiological and clinical characteristics were analyzed. From December 2006 to November 2007, 406 nasopharyngeal aspirates from children younger than 14 years of age with RTIs were screened for HRV and other common respiratory viruses by PCR or reverse transcription-PCR. Two-hundred twenty-four (55.2%) of the specimens were infected with at least one virus, including 53 patients with HRV (13%). HRV-A, HRV-B, and HRV-C were detected in 22, 12, and 19 specimens, respectively. HRV-C was detected mainly from December 2006 to April 2007 and from October to November 2007, with peaks in December and April (10/19). Acute upper respiratory infection and bronchopneumonia were observed in 53 and 37% of the cases, respectively. The most common symptoms were cough (82%), runny nose (53%), and fever (37%). Wheezing and bronchiolitis were less common in patients infected with HRV-C than in those infected with respiratory syncytial virus (RSV). Partial sequencing of the genes coding for VP4 and VP2 revealed that the HRV-C strains were 56 to 62% identical at the amino acid level to HRV-B and HRV-A reference strains and 80 to 99% identical to HRV-C reference strains. In conclusion, HRV-C is an important cause of RTIs in children, and highly diversified strains of HRV-C are prevalent in China. HRV-C may produce different epidemiological features, and patients infected with HRV-C may exhibit different clinical features from patients infected with RSV or HRV-A/B.
Viruses are the most frequent cause of acute respiratory infections and are a leading cause of childhood mortality (23). Human rhinoviruses (HRVs) are not only the most common causative agents of mild upper respiratory tract infections (RTIs) but are also associated with more serious diseases such as pneumonia or acute wheezing episodes associated with bronchiolitis and acute asthma in children (2, 4, 11, 15). More than 100 HRV serotypes have been identified. Traditionally, HRVs have been divided into two strains: HRV-A and HRV-B. Considerable variation may exist in the genetic characteristics of different HRV strains, and the viruses seem to circulate without any identifiable pattern (15).
Over the past few years, a new HRV variant has been detected in patients with acute RTIs in America, Australia, and Hong Kong that shares 53 to 57% homology at the amino acid level with HRV-A and HRV-B (13, 14, 17). The chief symptoms of patients infected with this new strain vary by location: patients from Australia and Hong Kong present mainly with bronchiolitis, wheezing, and asthmatic exacerbation, whereas those from America present mainly with flu-like symptoms (13, 14, 17). However, the clinical impact of this new viral strain in children is unclear. In this 1-year prospective study conducted during 2006 and 2007, 406 children younger than 14 years of age with an RTI were screened for this new viral strain and other respiratory viruses. The presenting symptoms and epidemiological characteristics associated with the strain were also analyzed.
MATERIALS AND METHODS
Study subjects, specimen collection, and processing.
From 1 December 2006 to 31 November 2007, 406 children 14 years of age or younger who presented to the Department of Pediatrics at the First Hospital of Lanzhou University with acute respiratory symptoms were recruited for this study.
Nasopharyngeal aspirates (NPAs) were collected from each patient. The demographic data, clinical findings, and severity of disease for each patient are presented in Table 1. Informed consent was obtained from the parents of the children. The study protocol was approved by the hospital ethics committee.
TABLE 1.
Comparison of the clinical characteristics between groups of children with acute respiratory infections
| Patient characteristic | Patient groupa
|
P value for:
|
|||||
|---|---|---|---|---|---|---|---|
| HRV-C
|
HRVA/B monoinfection (n = 19) | RSV monoinfection (n = 73) | Group 1 vs group 2 | Group 1 vs group 3 | Group 1 vs group 4 | ||
| Monoinfection (group 1; n = 11) | Coinfection (group 2; n = 8) | ||||||
| General | |||||||
| Male | 7 (64) | 5 (63) | 12 (63) | 44 (60) | 1 | 1 | 1 |
| Age of ≤3 yr | 6 (55) | 7 (88) | 12 (63) | 58 (80) | 0.121 | 0.712 | 0.374 |
| Hospitalization | 5 (46) | 4 (50) | 17 (90) | 46 (63) | 0.328 | 0.028 | 1 |
| Median duration of hospitalization (days)b | 5 [2-26] | 3 [3-7] | 7 [2-17] | 10 [2-36] | 0.377 | 0.946 | 0.752 |
| Clinical diagnosis | |||||||
| AURI | 5 (46) | 5 (63) | 3 (16) | 7 (10) | 0.007 | 0.104 | 1 |
| Acute laryngitis | 1 (9) | 0 | 1 (5) | 1 (1) | 0.246 | 1 | |
| Suppurative tonsillitis | 1 (9) | 0 | 0 | 0 | |||
| Bronchitis | 0 | 0 | 3 (15) | 3 | |||
| Bronchiolitis | 0 | 0 | 2 (10) | 20 | |||
| Bronchopneumonia | 4 (36) | 3 (38) | 9 (47) | 42 (58) | 0.212 | 0.708 | 1 |
| Clinical manifestations | |||||||
| Runny nose | 4 (36) | 6 (75) | 2 (11) | 37 (52) | 1 | 1 | 1 |
| Fever of >38°C | 3 (27) | 4 (50) | 5 (26) | 34 (47) | 0.332 | 1 | 0.642 |
| Cough | 9 (82) | 7 (88) | 14 (74) | 64 (88) | 0.632 | 1 | 1 |
| Wheezing | 0 | 3 (38) | 5 (26) | 24 (33) | |||
| Cyanosis | 1 (9) | 1 (13) | 3 (15) | 8 (11) | 1 | 1 | 1 |
| Dyspnea | 1 (9) | 1 (13) | 2 (10) | 15 (20) | 0.682 | 1 | 1 |
| Crackles | 5 (46) | 5 (63) | 10 (53) | 45 (62) | 0.34 | 1 | 1 |
| Heart failure | 0 | 0 | 0 | 11 | |||
Except as noted, values represent the numbers of patients with the characteristic shown, with percentages shown in parentheses.
Values in this row represent median numbers of days hospitalized (with ranges shown in brackets), in addition to the associated P values.
After collection, the NPAs were stored at −80°C until further processing. DNA and RNA were extracted from 0.2 ml of each NPA using a QIAamp viral DNA minikit and QIAamp viral RNA minikit (Qiagen, Beijing, China), respectively. cDNA synthesis was carried out with SSIII viral reverse transcriptase and random hexamer primers (Invitrogen), and PCR amplification was performed with rTaq DNA polymerase (Takara, Beijing, China). All laboratory tests were conducted at the National Institute for Viral Disease Control and Prevention, China Center for Disease Control and Prevention.
HRV detection.
The primers used to amplify a 549-bp fragment of HRV (forward, [5′-GGGACCAACTACTTTGGGTGTCCGTGT-3′]; reverse, [5′-GCATCIGGYARYTTCCACCACCANCC-3′]) were described previously (19). PCR was performed under the following conditions: 94°C for 8 min followed by 35 cycles of 94°C for 45 s, 60°C for 45 s, and 72°C for 45 s, with a final extension at 72°C for 8 min.
Detection of other respiratory viruses.
Human metapneumovirus (hMPV), respiratory syncytial virus (RSV), influenza viruses A and B (IFVA and IFVB, respectively), parainfluenza virus types 1 to 3 (PIV-1 to -3, respectively), and human coronavirus (HCoV; strains 229E, OC43, NL63, and HKU1) were detected by reverse transcription-PCR (5, 6, 21, 22), and adenovirus (AdV) and human bocavirus (HBoV) were detected by PCR (1, 12). All positive PCR products were purified using a QIAquick PCR purification kit (Qiagen) and sequenced by Invitrogen (Shanghai, China) to confirm their specificity.
Sequence analysis.
The nucleotide and deduced amino acid sequences of the VP4 and VP2 genes were compared with reference HRV strains available from GenBank. Phylogenetic analysis was conducted using MEGA version 3.1.
Statistical analysis.
The statistical significance of differences between the various groups was tested using the χ2 test and Fisher's exact test. The durations of patient hospitalization were compared between different virus-infected groups using an independent sample t test. A P value of < 0.05 was considered statistically significant. All analyses were performed using SPSS version 13.0.
Nucleotide sequence accession number.
The 53 partial sequences of VP4/VP2 have been submitted to GenBank under accession no. EU822829 to EU822880 and EU822882.
RESULTS
Patient characteristics.
The patients enrolled in this study were between 1 day and 168 months of age (29.9 ± 31.5 months [mean ± standard deviation]), with 88% (357/406) under 5 years of age. The boy/girl ratio was 1.5:1; the outpatient/inpatient ratio was 1:1.7.
Virological findings in children with acute RTIs.
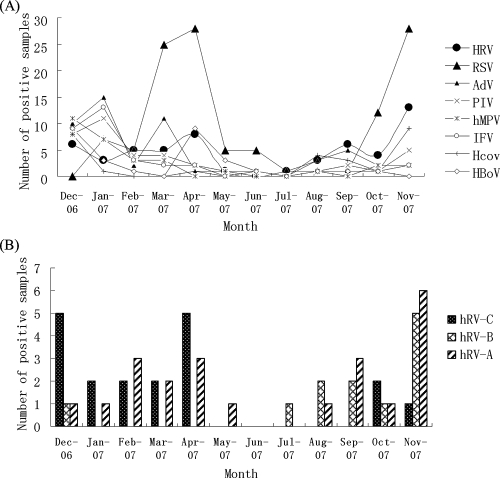
Of 406 samples tested, 224 (55.2%) were positive for at least one viral agent. RSV, which was detected in 118 children (29%), was the most common viral agent, followed by HRV (detected in 53 children [13%]), AdV (52 children [13%]), PIV-3 (38 children [9%]), hMPV (29 children [7%]), HBoV (29 children [7%]), HCoV NL63 (15 children [4%]), HCoV HKU1 (12 children [3%]), IFVA (6 children [2%]), PIV-1 (2 children [0.5%]), and HCoV 229E (1 child [0.2%]). PIV-2 and HCoV OC43 were not detected in any patients. Most virus-associated RTIs occurred during low-temperature months (winter and early spring). For example, 69% (81/118) of all RSV infections were detected during the months of March, April, and November in 2007 and 69% (36/52) of all AdV infections were detected in December of 2006 and January and March of 2007. Similarly, PIV, IFV, and hMPV were detected mainly during the months of December and January, and HBoV was detected in December and April; HCoV NL63 and HKU1 were detected mainly in autumn and winter (Fig. 1A). Of the 224 virus-positive samples, 142 were also positive for additional viruses, resulting in a coinfection rate of 63%. No significant differences in age distribution or sex ratio were found among the different viral groups.
FIG. 1.
Seasonal distribution of HRV and other common viruses (A) and HRV-A, -B, and -C during the study period (B).
Detection of HRV-C.
Phylogenetic analysis of the coding regions of the viral protein VP4/VP2 revealed that among the 53 patients infected with HRV, 22 (42%) were infected with genotype A, 12 (23%) were infected with genotype B, and 19 (36%) were infected with the new strains from HRV-C. The rates of coinfection for genotypes A, B, and HRV-C with another virus were 36% (8/22), 75% (9/12), and 42% (8/19), respectively. RSV was the most common additional respiratory virus detected in the three groups, accounting for 26% (14/53) of all HRV infections. HCoVs, including NL-63 and HKU1, were other common pathogens causing coinfections. In the HRV-C infection group, four patients were coinfected with RSV and one was coinfected with hMPV, HBoV, NL63, and HKU1.
Epidemiology of HRV.
An HRV isolate was detected each month of the year, except June, with the highest detection rates between December 2006 and April 2007 (51% [27/53]) and September to November of 2007 (40% [21/53]). HRV-C was found mainly from December 2006 to April 2007 and October to November of 2007, with peaks in December and April (10/19). HRV-A was detected throughout the study year, except in June and July. HRV-B was only detected during the autumn and winter months, with a peak in November (Fig. 1B). The age range of the 53 HRV-infected children was 1 to 108 months (30.4 ± 32.7 months [mean ± standard deviation]). The male/female ratio was 33:20. About 89% (17/19) of the children infected with HRV-C were <5 years of age, and 74% (14/19) were <36 months of age. Although HRV-A was detected in children of all age groups, HRV-B was largely detected in children of <1 year, 25 to 36 months, and >5 years of age (Fig. 2).
FIG. 2.
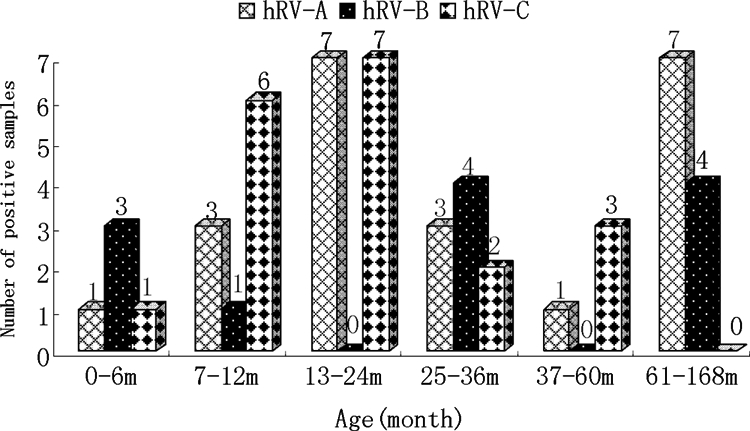
Distribution of various genotypes of HRV-positive specimens by patient age.
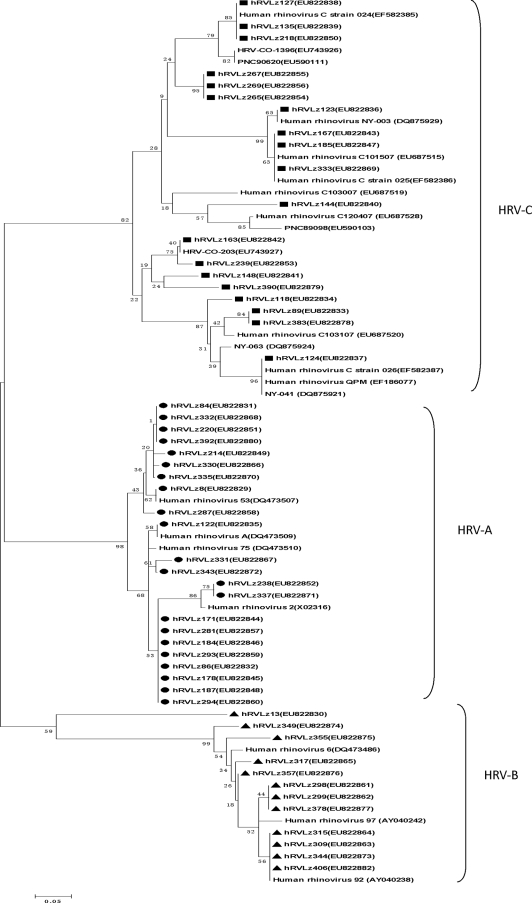
Phylogenetic analysis of HRV.
Partial sequences (549 bp) of VP4/VP2 from the HRVs obtained from children with RTIs in China were aligned with sequences from reported rhinovirus prototype strains, including the following (with accession numbers in parentheses): HRV 2 (X02316), HRV 6 (DQ473486), HRV 53 (DQ473507), HRV 75 (DQ473510), HRV 92 (AY040238), HRV 97 (AY040242), HRV-A (DQ473509), HRV NY-003 (DQ875929), HRV QPM (EF186077), HRV-C 024 (EF582385), HRV-C 025 (EF582386), HRV-C 026 (EF582387), HRV-C 101507 (687515), HRV-C 103007 (687519), HRV-C 103107 (687520), HRV-C O-203 (EU743927), HRV-C O-1396 (EU743926), HRV PNC89098 (EU590103), HRV PNC90620 (EU590111), HRV NY-041 (DQ875921), and HRV NY-063 (DQ875924). Alignment of the sequences in the present study with the HRV prototype strains resulted in three clusters: HRV-A, HRV-B, and a novel genetic cluster (Fig. 3). The sequences from the strains of the novel cluster were 56 to 62% identical at the amino acid level to those from HRV-A and HRV-B, while retaining 80 to 99% identity to HRV-C reference strains. Sequences Lz127, Lz135, and Lz218 and sequences Lz167, Lz185, and Lz333 showed marked similarity to HRV-C 024 (EF582385) and HRV-C 025 (EF582386), respectively, from Hong Kong. Sequences Lz163 and Lz239 and Lz89 and Lz383 showed similarity to HRV-C O-203 (EU743927) from Colorado and HRV-C 103107 (687520) from Beijing, respectively; the sequences from Lz123 and Lz124 shared strong similarity with HRV NY-003 (DQ875929) and HRV QPM (EF186077), respectively. The percent identity among the strains ranged from 87.9% to 100%within group A and from 90.8%to 100% within group B.
FIG. 3.
Phylogenetic analysis of the deduced amino acid sequences of the VP4/VP2 gene (420 bp) of the 53 HRVs with reference strains. Phylogenetic trees were constructed by the neighbor-joining method by using MEGA 3.1, and bootstrap values were determined with 1,000 replicates. Virus sequence names shown without symbols were reference strains; representative HRV sequences and other rhinoviruses from GenBank are indicated by isolate names. GenBank accession numbers of each strain are given in parentheses.
Clinical characteristics of HRV-C infection in children.
The age range of the patients who tested positive for HRV-C was 4 days to 7 years. Among the 19 HRV-C-positive children, 9 were inpatients with a mean hospital stay of 6.6 days. Ten children (53%) had an upper RTI, 7 children (37%) had bronchopneumonia, 1 child (5%) had acute laryngitis, and 1 child had suppurative tonsillitis. The most common symptom was a cough (n = 16; 82%), followed by a runny nose (n = 10; 53%), crackle (n = 10; 53%), and a fever (n = 7; 37%). Other clinical presentations included wheezing (n = 3; 16%), cyanosis (n = 2; 11%), dyspnea (n = 2; 11%), vomiting (n = 1; 6%), and hoarseness (n = 1; 5%). Of 11 children who underwent chest radiography, 6 had abnormal findings and 5 were confirmed to have pneumonia. Complete blood counts were performed for 16 of the 19 children, and the mean white blood cell count was 92,500 cells/μl (range 45,000 to 178,000 cells/μl). Bacterial cultures of the NPAs were obtained for six patients: two of the cultures were Haemophilus influenzae (Lz89) and Streptococcus pneumoniae (Lz333), and four were normal flora (Lz8, Lz127, Lz167, and Lz269).
A few distinct clinical characteristics were noted when different groups of patients were compared (Table 1). HRV-C monoinfection was more common than RSV monoinfection in patients with acute upper RTI (AURI) (P = 0.007), whereas patients diagnosed with bronchiolitis were more common in the RSV monoinfection group than in the HRV-C monoinfection group. Although wheezing occurred frequently in those patients with HRV-C coinfection (33%), RSV infection (33%), and HRV-A/B monoinfection (26%), patients with HRV-C monoinfection did not present with wheezing. Among the patients with HRV-C monoinfection, HRV-C coinfection, and RSV monoinfection, no significant differences were found in terms of sex; proportion of patients <3 years of age; or presence of a fever (≥38°C), cough, cyanosis, dyspnea, and crackle. The number of patients requiring hospitalization was lower in the HRV-C monoinfection group than in the HRV-A or HRV-B monoinfection group (P = 0.028); however, the durations of hospitalization were not significantly different between the HRV-C monoinfection group, HRV-A or HRV-B monoinfection group, and RSV group.
DISCUSSION
In the present study, more than half of the children (55%) were infected with at least one viral agent, with HRV being the second most common virus. The detection rate of viruses was lower than that of other studies (9, 18, 20). One reason for that is the low PCR sensitivity with the pair of primers used in this study. Another is the low clinical visit rate of patients with minimal RTI symptoms in the general hospital; these patients usually get medical care easily from community hospitals. HRV coinfections were more frequent in the present study than in other studies (8, 10, 16). RSV and coronavirus (NL63 and HKU1) were the major coinfection viruses. The reason is partly due to the high detection rate of RSV and the similar seasonal circulation patterns of these viruses.
The detection rates for three HRV species, HRV-A, HRV-B, and the novel species HRV-C, were 5.4, 3.0, and 4.7%, respectively. AURI and bronchopneumonia were the main diagnoses for the HRV-C-positive patients. A previous report indicated that 5% of RTIs among inpatients are caused by HRV-C (7). In our study, 9 of 19 patients were hospitalized and 11 were found to have HRV-C monoinfection. These results indicate that HRV-C is an important etiological factor in children with an RTI.
In this study, The HRV-C sequences are genetically distinct, sharing only 56 to 62% of their amino acids with HRV-A and HRV-B, while retaining 77.3 to 100% identity with each other. In addition, these sequences showed 69 to 96% amino acid identity with known strains of HRV QPM. In accordance with previous data, HRV-C showed a high degree of sequence variation between individual strains. Further analysis is needed to confirm this finding.
Previous studies indicated that HRV was detected throughout the year, with peaks in spring and autumn in temperate regions (15). Different rhinoviral species occurred in different seasons. The three HRV species in our study were detected predominately in winter and spring. The most frequently detected species was HRV-A, which occurred in all months except June and July. HRV-B circulated during summer, autumn, and winter. It was reported that all HRV-B species in Queensland, Australia, were from specimens collected during the winter months, whereas the new HRV-C was found mostly during spring (3). In our study, HRV-C infections peaked in early winter (December) and late spring (April). It was previously reported that HRV QPM infections peaked in winter but were also detected in spring and summer (17). In Hong Kong, all HRV-C infections were found between October and February, with peaks in autumn and winter (14). Thus, HRV subgroups cocirculate throughout the year, and the predominant species vary with location and year.
HRV is the second most frequently recognized agent associated with pneumonia and bronchiolitis in infants and young children, and infection with HRV frequently exacerbates preexisting airway diseases such as asthma (15). In this study, HRVs were the second most frequently detected respiratory virus, and the HRV-infected children suffered from a wide range of upper and lower respiratory tract symptoms. The most common diagnoses among the children infected with HRV-C were upper RTI and bronchopneumonia, indicating that HRV-C contributes substantially to RTIs, which frequently require hospitalization. The number of cases requiring hospitalization in the HRV-C monoinfection group was smaller than that in the HRV-A or HRV-B monoinfection group, and AURI occurred more often in the HRV-C monoinfection group than in the RSV infection group. Although the clinical presentations of HRV-C infection were similar to those of RSV, bronchiolitis, wheezing, cyanosis, dyspnea, and heart failure, which are common in cases of HRV-C infection, were not detected as frequently as in RSV infection. These observations support the view that HRV-C is associated with relatively mild upper respiratory infections, rather than being typical for lower respiratory tract disease.
The overall severity of disease, median duration of hospitalization, and frequency of major symptoms between the HRV-C monoinfection and HRV-C coinfection groups were similar. Moreover, no difference in disease severity was observed between the HRV-A/B and HRV-C groups. A study from Hong Kong showed that febrile wheezing and asthma were the most common presentations of HRV-C infection (76%) (14), and children infected with HRV QPM most commonly present with bronchiolitis and wheezing (17). In the present study, the prevalence of wheezing and asthma was not as high as that reported in Hong Kong or Queensland. A possible explanation for this result is that the specimens used in the present study were obtained from both inpatients and outpatients, and the number of patients with underlying chronic diseases was lower than in the samples from Hong Kong or Queensland.
In conclusion, our data indicate that the novel genotype HRV-C exhibits significant genetic variation and a distinct epidemiological profile from HRV-A and HRV-B. It can also cause upper and lower respiratory diseases in children and is frequently associated with AURI and bronchopneumonia. Wheezing and bronchiolitis in the HRV-C group were less common than in patients infected with RSV. Additional studies are needed to elucidate the relationship between HRV-C and wheezing and to define the epidemiological profile and genetic characteristics of the newly recognized species of HRV.
Acknowledgments
This work was partly supported from the 973 National Key Basic Research Program of China (grant no. 2007CB310500).
None of the authors has a conflict of interest.
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 10212891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrewes, C. H. 1964. The complex epidemiology of respiratory virus infections. Science 1461274-1277. [DOI] [PubMed] [Google Scholar]
- 3.Arden, K. E., P. McErlean, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2006. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 781232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arruda, E., A. Pitkäranta, T. J. Witek, Jr., C. A. Doyle, and F. G. Hayden. 1997. Frequency and natural history of rhinovirus infections in adults during autumn. J. Clin. Microbiol. 352864-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastien, N., K. Anderson, L. Hart, P. Van Caeseele, K. Brandt, D. Milley, T. Hatchette, E. C. Weiss, and Y. Li. 2005. Human coronavirus NL63 infection in Canada. J. Infect. Dis. 191503-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellau-Pujol, S., A. Vabret, L. Legrand, J. Dina, S. Gouarin, J. Petitjean-Lecherbonnier, B. Pozzetto, C. Ginevra, and F. Freymuth. 2005. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods 12653-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briese, T., N. Renwick, M. Venter, R. G. Jarman, D. Ghosh, S. Kondgen, S. K. Shrestha, A. M. Hoegh, I. Casas, E. V. Adjogoua, C. Akoua-Koffi, K. S. Myint, D. T. Williams, G. Chidlow, R. van den Berg, C. Calvo, O. Koch, G. Palacios, V. Kapoor, J. Villari, S. R. Dominguez, K. V. Holmes, G. Harnett, D. Smith, J. S. Mackenzie, H. Ellerbrok, B. Schweiger, K. Schonning, M. S. Chadha, F. H. Leendertz, A. C. Mishra, R. V. Gibbons, E. C. Holmes, and W. I. Lipkin. 2008. Global distribution of novel rhinovirus genotype. Emerg. Infect. Dis. 14944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheuk, D. K., I. W. Tang, K. H. Chan, P. C. Woo, M. J. Peiris, and S. S. Chiu. 2007. Rhinovirus infection in hospitalized children in Hong Kong: a prospective study. Pediatr. Infect. Dis. J. 26995-1000. [DOI] [PubMed] [Google Scholar]
- 9.Choi, E. H., H. J. Lee, S. J. Kim, B. W. Eun, N. H. Kim, J. A. Lee, J. H. Lee, E. K. Song, S. H. Kim, J. Y. Park, and J. Y. Sung. 2006. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000-2005. Clin. Infect. Dis. 43585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez, S. R., T. Briese, G. Palacios, J. Hui, J. Villari, V. Kapoor, R. Tokarz, M. P. Glode, M. S. Anderson, C. C. Robinson, K. V. Holmes, and W. I. Lipkin. 2008. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J. Clin. Virol. 43219-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden, F. G. 2004. Rhinovirus and the lower respiratory tract. Rev. Med. Virol. 1417-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hierholzer, J. C., P. E. Halonen, P. O. Dahlen, P. G. Bingham, and M. M. McDonough. 1993. Detection of adenovirus in clinical specimens by polymerase chain reaction and liquid-phase hybridization quantitated by time-resolved fluorometry. J. Clin. Microbiol. 311886-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamson, D., N. Renwick, V. Kapoor, Z. Liu, G. Palacios, J. Ju, A. Dean, K. St. George, T. Briese, and W. I. Lipkin. 2006. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J. Infect. Dis. 1941398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau, S. K. P., C. C. Y. Yip, H.-W. Tsoi, R. A. Lee, L.-Y. So, Y.-L. Lau, K.-H. Chan, P. C. Y. Woo, and K.-Y. Yuen. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 453655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackay, I. M. 2008. Human rhinoviruses: the cold wars resume. J. Clin. Virol. 42297-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McErlean, P., L. A. Shackelton, E. Andrews, D. R. Webster, S. B. Lambert, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2008. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C). PLoS ONE 3e1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McErlean, P., L. A. Shackelton, S. B. Lambert, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2007. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J. Clin. Virol. 3967-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nokso-Koivisto, J., A. Pitkaranta, S. Blomqvist, J. Jokinen, M. Kleemola, A. Takala, T. Kilpi, and T. Hovi. 2002. Viral etiology of frequently recurring respiratory tract infections in children. Clin. Infect. Dis. 35540-546. [DOI] [PubMed] [Google Scholar]
- 19.Savolainen, C., M. N. Mulders, and T. Hovi. 2002. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 8541-46. [DOI] [PubMed] [Google Scholar]
- 20.Tsolia, M. N., S. Psarras, A. Bossios, H. Audi, M. Paldanius, D. Gourgiotis, K. Kallergi, D. A. Kafetzis, A. Constantopoulos, and N. G. Papadopoulos. 2004. Etiology of community-acquired pneumonia in hospitalized school-age children: evidence for high prevalence of viral infections. Clin. Infect. Dis. 39681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vabret, A., J. Dina, S. Gouarin, J. Petitjean, S. Corbet, and F. Freymuth. 2006. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin. Infect. Dis. 42634-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vabret, A., F. Mouthon, T. Mourez, S. Gouarin, J. Petitjean, and F. Freymuth. 2001. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J. Virol. Methods 9759-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams, B. G., E. Gouws, C. Boschi-Pinto, J. Bryce, and C. Dye. 2002. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect. Dis. 225-32. [DOI] [PubMed] [Google Scholar]