Abstract
Enzyme immunoassays (EIA) to detect glutamate dehydrogenase or toxins A (TcdA) and B (TcdB), a cytotoxicity assay, and bacteriologic culture have disadvantages when applied individually to diagnosis of Clostridium difficile infections. Stool specimens (n = 1,596) were subjected to toxin detection via an enzyme-linked fluorescent immunoassay (ELFA; Vidas CDAB assay) and bacteriologic culture for toxigenic C. difficile in a three-step algorithm with additional toxigenic culture. Isolates (n = 163) from ELFA-negative stool specimens were examined via ELFA for toxin production. We amplified tcdA and tcdB from C. difficile isolates and tcdB from stool specimens that were ELFA positive or equivocal and culture negative, and we compared the results to those obtained with the three-step algorithm. More than 26% of stool specimens (419/1,596) were culture positive, yielding 248 isolates (59.2%) with both toxin genes (tcdA- and tcdB-positive isolates), 88 isolates (21.0%) with either tcdA or tcdB, and 83 (19.8%) that had no toxin genes (tcdA- and tcdB-negative isolates). Among 49 (culture-negative/ELFA-positive or -equivocal) stool specimens, 53.1% (26/49) represented tcdB-positive isolates. Therefore, the total number of PCR-positive cases was 362, and 27.1% (98/362) of these were detected through toxigenic culture. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were 63.3%, 96.7%, 90.5%, and 92.4% (ELFA alone); 92.8%, 93.3%, 80.2%, and 97.8% (culture); and 70.7%, 91.4%, 95.5%, and 100% (three-step algorithm ELFA and bacterial culture with toxigenic culture), respectively, with culture and PCR for tcdA and tcdB as the standards. Thus, sensitivity and specificity were highest using culture and ELFA, respectively, but we recommend the three-step algorithm comprising EIA to detect both toxins and toxigenic culture for C. difficile as a practical method for achieving better PPV and NPV.
Clostridium difficile is an important nosocomial pathogen, causing antimicrobial-associated diarrhea and pseudomembranous colitis. Toxins A (TcdA) and B (TcdB) mediate the pathogenesis of C. difficile infection (CDI), and toxin detection is an important part of diagnosis. A cytotoxicity neutralization assay (CNA) is the reference method for toxin detection, but it is expensive and time-consuming and requires tissue culture facilities (34, 35). Most laboratories now use a commercial enzyme immunoassay (EIA) to detect TcdA and/or TcdB, with the benefits of rapid turnaround time and ease of use (3, 21, 22, 23, 26, 27, 33, 35). The putative >90% sensitivity of toxin EIAs is not often realized in practice, but EIA is the only toxin detection method available to many routine medical laboratories. The demand for EIA kits detecting both TcdA and TcdB has increased due to increased worldwide prevalence of TcdA-negative, TcdB-positive (TcdA− TcdB+) strains (1, 12, 24, 29, 32).
A two-step algorithm, based upon EIA-based detection of species-specific antigen glutamate dehydrogenase (GDH-Ag) and toxin detection via CNA, was suggested to have improved sensitivity and specificity in the detection of toxigenic C. difficile (34). However, the GDH-Ag assay detects both nontoxigenic and toxigenic strains, and the aforementioned shortcomings of the CNA assay make it unavailable to many routine laboratories.
Bacteriologic culture can be time-consuming, but it is more straightforward and sensitive than CNA for the detection of toxigenic C. difficile. Furthermore, it provides isolates for characterization, yielding information about CDI epidemiology and antimicrobial susceptibility (11, 28, 36). We evaluated the combination of bacteriologic culture and EIA-based detection of TcdA and TcdB as a new strategy for diagnosis of CDI, especially in areas where TcdA− TcdB+ strains are prevalent.
MATERIALS AND METHODS
Specimens.
Fecal specimens (n = 1,596) were collected, between April 2007 and July 2008, from patients admitted to a tertiary teaching hospital in Seoul, South Korea, with clinical signs compatible with CDI.
Assay of stool specimens for TcdA and TcdB.
Stool specimens were examined for TcdA and TcdB via an enzyme-linked fluorescent immunoassay (ELFA; Vidas CDAB assay; bioMérieux SA, Marcy-l'Etoile, France). An aliquot (200 μl) of well-mixed liquid stool was added to 1 ml diluent and centrifuged for 5 min at 12,000 × g. Supernatant fluid (300 μl) was added to the sample well of the CDAB kit, according to the manufacturer's instructions. Results were reported as positive, equivocal, or negative, according to the intensity of the fluorescence. Specimens with a test value of ≥0.37 were considered positive, and those with a test value of <0.13 were considered negative. Specimens with a test value between 0.13 and 0.37 were considered equivocal.
Semiquantitative culture for C. difficile.
Fecal specimen (1.0 ml) was mixed with an equal volume of 70% isopropanol and incubated at room temperature for 30 min. One drop (∼100 μl) was then inoculated onto prereduced Clostridium difficile selective agar (Becton Dickinson, Sparks, MD) and incubated at 37°C under anaerobic conditions (anaerobic pouch; Becton Dickinson, Sparks, MD) for 48 to 72 h. Suspect C. difficile isolates were identified by colony morphology on C. difficile selective agar, spore staining, and biochemical characteristics (ANA identification kit; bioMérieux, Marcy-l'Etoile, France). Extent of growth was scored as grade 1 (<10 colonies), grade 2 (10 to 50 colonies), grade 3 (>50 to 100 colonies), and grade 4 (>100 colonies).
Toxigenic culture of C. difficile isolates for TcdA and TcdB.
Isolates of C. difficile from toxin-negative stool specimens were examined for toxin production via ELFA. Colonies (n = 2 to 3) from selective medium were suspended in 2.0 ml of sterile 0.45% saline to achieve a concentration approximating McFarland standard no. 4. An aliquot (200 μl) was diluted in 1 ml diluent buffer, and 300 μl was inoculated into a well. The assay was performed and interpreted as described above.
PCR assay for tcdA and tcdB.
Clostridium difficile strains (n = 419) were examined for tcdA and tcdB by a modification of a previously reported PCR method (15). Template DNA was prepared by suspending 20 colonies in 200 μl 5% (wt/vol) Chelex-100 (Bio-Rad), boiling for 12 min, and then centrifuging at 12,000 × g for 5 min. To detect tcdB in stool specimens (n = 49), template DNA was extracted from 200 mg of stool by use of an AccuPrep stool extraction kit (Bioneer, Daejeon, South Korea). The PCR volume was 100 μl, containing ∼30 ng of template DNA, 0.15 μg of each primer, the four deoxynucleoside triphosphates (200 μM each), 10 mM Tris HCl (pH 8.3), 2.5 mM MgCl2, 50 mM KCl, and 2.5 U Taq polymerase. Amplification was performed via primers NK9 (5′-CCACCAGCTGCAGCCATA-3′) and NK11 (5′-TGATGCTAATAATGAATCTAAAATGGTAAC-3′), derived from the repeating sequence of tcdA, and primers NK104 (5′-GTGTAGCAATGAAAGTCCAAGTTTACGC-3′) and NK105 (5′-CACTTAGCTCTTTGATTGCTGCACCT-3′), derived from the nonrepeating sequence of tcdB. ATCC strain 43596 was used as the tcdA- and tcdB-positive control, and ATCC strain 43598 was used as the tcdA-negative, tcdB-positive variant control. Amplification of tcdA was performed in a thermal cycler (Perkin-Elmer, CT) via 40 cycles of 95°C for 15 s, 62°C for 120 s, and 72°C for 40 s. The thermal profile for the amplification of tcdB was 40 cycles at 95°C for 20 s, 62°C for 60 s, and 74°C for 40 s. In both cases, amplification was followed by incubation at 74°C for 5 min to complete extension. Amplified products (10 μl) were electrophoresed in a 2% agarose gel, and bands were visualized by UV transillumination after staining with ethidium bromide. Strains in which tcdA was intact yielded 1,200-bp amplicons, but in those with a variant tcdA (tcdAv), the products were 700 bp or 500 bp. Strains in which tcdB was intact yielded a 204-bp product.
RESULTS
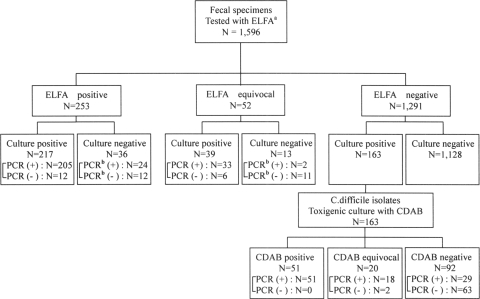
Figure 1 presents the three-step algorithm as a flow chart, including 163 toxigenic C. difficile cultures. Among 1,596 stool specimens, 253 (15.9%), 52 (3.2%), and 1,291 (80.9%) cases were ELFA positive, equivocal, and negative, respectively. The culture positivity rate was 26.3% (419/1,596). Culture positivity was 85.5% (217/253), 75.0% (39/52), and 12.6% (163/1,291) in ELFA-positive, -equivocal, and -negative groups, respectively. Isolates from Clostridia species other than C. difficile were collected from 18.7% (220/1,177) of C. difficile culture-negative cases. Toxigenic culture (n = 163) detected 69 additional cases among ELFA-positive or -equivocal specimens.
FIG. 1.
Flow diagram depicting the three-step algorithm, finally confirmed by tcdA and tcdB PCR assays.
Among 419 total isolates, including 163 toxigenic culture-positive strains (i.e., those obtained from ELFA-negative specimens), 336 strains (80.2%) were PCR positive. Ninety-eight (60.1%) of the 163 toxigenic culture cases were tcdA and/or tcdB positive. Direct PCR for tcdB on 49 stool specimens (ELFA-positive or -equivocal/culture-negative cases) revealed 26 tcdB-positive cases (53.1%). Therefore, the total number of PCR-positive cases was 362, and 27.1% (98/362) of these were detected through toxigenic culture.
We placed tcdA-positive and tcdB-positive; tcdA-negative, tcdB-positive; tcdA-positive, tcdB-negative; and tcdAv, tcdB-positive strains in a hypothetical ELFA-positive group, and tcdA- and tcdB-negative strains in a hypothetical ELFA-negative group (Table 1). We also placed culture/ELFA conegative cases (n = 1,128) in a hypothetical ELFA-negative group. We could not absolutely rule out the presence of TcdA or TcdB in culture-negative stool specimens, but it seems justified to conclude that culture is more sensitive than fecal CNA for detection of infection by toxigenic C. difficile (11, 28, 35). Among 419 C. difficile culture-positive cases, tcdA-positive and tcdB-positive strains were most common (n = 248 [59.2%]), while those which were tcdA-negative and tcdB-positive (n = 9 [2.1%]), tcdA-positive and tcdB-negative (n = 2 [0.5%]), tcdAv and tcdB-positive (n = 77 [18.4%]), and tcdA- and tcdB-negative (n = 83 [19.8%]) were detected less frequently. Among 88 (24.3% of 362 hypothetical positive cases) tcdAv/tcdB-positive, tcdA-negative/tcdB-positive, and tcdA-positive/tcdB-negative C. difficile strains, 59 (67.0%) and eight (9.1%) strains were from cases with positive and equivocal results with ELFA, respectively (Table 1).
TABLE 1.
Comparison of results of C. difficile culture, EIA for TcdA and TcdB, and PCR for tcdA and tcdB in the entire study group (n = 1,596)
| Culture result | PCR result fora:
|
ELFA result (no. of specimens)c
|
Total (%) | |||
|---|---|---|---|---|---|---|
| tcdA | tcdB | Positive | Equivocal | Negative | ||
| + | + | + | 146 | 25 | 77 | 248 (15.5) |
| + | + | − | 1 | 0 | 1 | 2 (0.1) |
| + | Variant | + | 51 | 7 | 19 | 77 (4.8) |
| + | − | + | 7 | 1 | 1 | 9 (0.6) |
| + | − | − | 12 | 6 | 65 | 83 (5.2) |
| − | ND | +b | 24 | 2 | 0 | 26 (1.6) |
| − | ND | −b | 12 | 11 | 0 | 23 (1.4) |
| − | ND | ND | 0 | 0 | 1,128 | 1,128 (70.8) |
| Total | 253 | 52 | 1,291 | 1,596 (100.0) | ||
ND, not done.
PCR performed with stool specimens.
n = 1,596.
Fifty-two specimens (3.2%) yielding equivocal ELFA results were not included in the calculation of concordance, sensitivity, and specificity. On the basis of these results, the sensitivity, specificity, positive predictive value, and negative predictive value of the three-step algorithm were 70.7% (256/362), 91.4% (1,128/1,234), 95.5% (256/268), and 100% (1,128/1,128), respectively. The corresponding values for the ELFA were 63.3% (229/362), 96.7% (1,193/1,234), 90.5% (229/253), and 92.4% (1,193/1,291), respectively, and for bacteriologic culture were 92.8% (336/362), 93.3% (1,151/1,234), 80.2% (336/419), and 97.8% (1,151/1,177), respectively.
Results of semiquantitative culture were correlated with ELFA positivity (Table 2). The greater the number of colonies on a primary culture plate, the more likely a specimen was to be toxin positive. Odds ratios were 2.109, 4.674, and 5.571 in grades 2, 3, and 4, respectively, compared with grade 1 (P < 0.05). The difference among grades was statistically significant (chi-square test for trend, P < 0.0001), excepting that between grades 3 and 4.
TABLE 2.
Correlation between results of semiquantitative C. difficile culture and ELFA
| Gradea | ELFA resultb (no. of specimens [%])
|
Odds ratio | 95% Confidence interval | P value | |||
|---|---|---|---|---|---|---|---|
| Positive | Equivocal | Negative | Total | ||||
| 1 | 18 (28.6) | 3 (6.3) | 42 (66.7) | 63 (100) | 1 | ||
| 2 | 78 (44.1) | 26 (14.7) | 73 (41.2) | 177 (100) | 2.109 | 1.133-3.927 | 0.0186 |
| 3 | 43 (65.2) | 8 (12.1) | 15 (22.7) | 66 (100) | 4.674 | 2.219-9.846 | <0.0001 |
| 4 | 78 (69.0) | 2 (1.8) | 33 (29.2) | 113 (100) | 5.571 | 2.832-10.96 | <0.0001 |
| Total | 217 (51.8) | 39 (9.3) | 163 (38.9) | 419 (100) | |||
Extent of growth of semiquantitative culture for C. difficile. Grade 1, <10 colonies; grade 2, 10 to 50 colonies; grade 3, >50 to 100 colonies; grade 4, >100 colonies.
n = 419.
DISCUSSION
A variety of tests is available for detection of C. difficile toxins, but a lack of guidelines for appropriate use can lead to difficulties in choice of test and to uncertain outcomes. CNA is generally considered the gold standard for diagnosis of CDI but is not performed by most laboratories; the assay is tedious, requires tissue culture facilities, may lack sensitivity under some conditions, and is difficult to standardize. An assay for GDH-Ag has a high false-positive rate, perhaps due to detection of nontoxigenic C. difficile (23, 27, 33, 34). Bacteriologic culture is strongly recommended by some authors, and its advantages include availability of isolates for determination of toxinogenesis (toxigenic culture), more effective study of epidemiology, and determination of antimicrobial susceptibility (11, 27, 36). EIA for TcdA and/or TcdB is preferred in most laboratories because a positive test provides indirect evidence that toxigenic C. difficile is in stool specimens; the EIA is readily available and less time-consuming than are CNA and C. difficile culture (4, 8, 26, 28).
One or two of these methods are usually selected as diagnostic tools for CDI, depending on the situation in each laboratory. Results of a European survey revealed that 93% of laboratories undertook direct detection of toxins in stool specimens and ∼80% used a commercial EIA; 41.6% of the laboratories used both culture and toxin detection (7). A two-step algorithm (GDH-Ag EIA and CNA) was suggested to enhance detection rates (34). It has several advantages, but also limitations, in that routine medical laboratories may not be equipped for cell culture and the GDH-Ag test does not detect TcdA or TcdB.
Therefore, we established and evaluated a three-step algorithm based upon bacteriologic culture and detection of TcdA and TcdB (in stool specimens and by isolates) by EIA, which makes it more accessible to most laboratories. This allows direct detection of toxin in stool specimens and reliable ruling out of CDI.
In fact, variant strains of C. difficile have been described with increasing prevalence worldwide, ranging from 0.2 to 56% in studies from the United States, Europe, and Asia (6, 13, 15, 16, 21, 24, 29, 30), and many recent studies have revealed that TcdA− TcdB+ strains are involved in a wide spectrum of CDI, ranging from colonization to uncomplicated diarrhea to pseudomembranous colitis (1, 3, 14, 17, 24, 31). TcdA+ TcdB− strains have been reported only rarely worldwide. However, one case was reported (10), and in this work, we encountered two cases (0.5%) involving this strain type. Therefore, a diagnostic method capable of detecting both TcdA and TcdB is likely the best choice, owing to the high prevalence of TcdA− TcdB+ strains in many countries and the possible emergence of TcdB− strains.
In our study, 59 (67.0%) and eight (9.1%) strains among 88 cases yielding tcdAv/tcdB-positive, tcdA-negative/tcdB-positive, and tcdA-positive/tcdB-negative C. difficile strains yielded positive and equivocal ELFA results, respectively. These may have been missed had we used a method detecting TcdA or TcdB alone, although 23.9% (21/88) of these variant strains were not detected with ELFA.
The sensitivities of toxin EIAs are reportedly variable, ranging from 55% to >90% (2, 22, 26, 35, 36). The limited sensitivity of both toxin EIA and CNA might be due to inhibitors in stools, by variable amounts of toxins in stools, and by instability of products and procedures (11, 22, 27, 35, 36). In this work, sensitivity of the dual-toxin EIA was not so high (63.3%). However, the rate of case detection (positive predictive value) via our three-step algorithm (using culture and ELFA) was higher than with another such algorithm, which was based upon detection of GDH-Ag and CNA (95.5% versus 82.1%). There was no significant difference in specificity between the GDH-Ag/CNA algorithm and our ELFA/culture algorithm. Moreover, the processing time with our algorithm (2 to 3 days) was comparable to that of the earlier one.
Culture also allowed identification of additional ELFA-positive cases that would otherwise have been missed, through application of ELFA to toxigenic culture (5, 18, 20). In our study, 98 (60.1%) of 163 toxigenic culture cases were tcdA and/or tcdB positive. Among them, 69 strains were ELFA positive (n = 51) or equivocal (n = 18), respectively. These may have been missed had we not used toxigenic culture, although 31.6% (31/98) of them were not detected with ELFA.
Interpretation of equivocal ELFA results is not part of the manufacturer's instructions, in contrast to the toxin A detection kit (CDA2, Vidas; bioMérieux, France), but isolates in 86.4% (51/59) of culture-positive/ELFA-equivocal cases (including ELFA-equivocal cases of toxigenic culture) were PCR positive for tcdA and tcdB. Thus, if C. difficile culture is positive, ELFA-equivocal results can likely be interpreted as toxin positive. Therefore, if we regarded equivocal ELFA results as positive, the sensitivity of the three-step algorithm would increase to 84.8% (307/362).
Positivity rates in the EIA were highly associated with the outcome of semiquantitative culture—the higher the grade, the higher the positivity rates in the ELFA (chi-square test for trend, P < 0.0001). This suggests that detection rates in the EIA depend on the quantity of TcdA or TcdB produced by C. difficile, and a false-negative EIA result is associated, at least in part, with lower numbers of C. difficile isolates (and, thus, its toxins) in stool. The manufacturer suggests that the limit of detection in the ELFA is 3 ng/ml of TcdA and 1 ng/ml of TcdB, and samples in which these limits were not reached would be reported as negative. However, culture positivity in EIA-negative stools, with subsequent demonstration of tcdA and/or tcdB by a PCR assay, may increase sensitivity. Culture was the most sensitive method for detection of toxigenic C. difficile, but it also missed 7.2% (26/362) of hypothetical positive cases. The prevalence of such cases was very low (3.0%), but it suggested that ELFA-positive or -equivocal/culture-negative cases may be associated with CDI.
In conclusion, we recommended a three-step algorithm comprising EIA (detecting TcdA and TcdB) and toxigenic bacteriologic culture. This approach, which is supported by good positive and negative predictive values, allows direct and rapid assay for toxins and toxigenic C. difficile in stool specimens to confirm or rule out CDI in a reasonable time period. The method is readily accessible for routine microbiology laboratories, and the availability of C. difficile isolates will facilitate epidemiologic study and determination of antimicrobial susceptibility.
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.al-Barrak, A., J. M. Embil, B. Dyck, K. Olekson, D. Nicoll, M. Alfa, and A. Kabani. 1999. An outbreak of toxin A negative, toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can. Commun. Dis. Rep. 2565-69. [PubMed] [Google Scholar]
- 2.Aldeen, W. E., M. Bingham, A. Aiderzada, J. Kucera, S. Jense, and K. C. Carroll. 2000. Comparison of the Tox A/B test to a cell culture cytotoxicity assay for the detection of Clostridium difficile in stools. Diagn. Microbiol. Infect. Dis. 36211-213. [DOI] [PubMed] [Google Scholar]
- 3.Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. al-Barrack, G. K. H. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 382706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, M. A., and J. G. Songer. 2008. Evaluation of two enzyme immunoassay for detection of Clostridium difficile toxin A and B in swine. Vet. Microbiol. 128204-206. [DOI] [PubMed] [Google Scholar]
- 5.Barbut, F., C. Kajzer, N. Planas, and J. C. Petit. 1993. Comparison of three enzyme immunoassays, a cytotoxicity assay, and toxigenic culture for diagnosis of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 31963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbut, F., V. Lalande, B. Burghoffer, H. V. Thien, E. Grimprel, and J. C. Petit. 2002. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J. Clin. Microbiol. 402079-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbut, F., M. Delmee, and J. S. Brazier. 2003. A European survey of diagnostic methods and testing protocols for Clostridium difficile. Clin. Microbiol. Infect. 9989-996. [DOI] [PubMed] [Google Scholar]
- 8.Barbut, F., P. Mastrantonio, M. Delmee, J. S. Brazier, E. Kuijper, I. Poxton, and European Study Group on Clostridium Difficile 2007. Prospective study of Clostridium difficile infection in Europe with phenotypic and genotypic characterization of the isolates. Clin. Microbiol. Infect. 131048-1057. [DOI] [PubMed] [Google Scholar]
- 9.Reference deleted.
- 10.Cohen, S. H., J. Y. Tang, B. Hansen, and J. Silva. 1998. Isolation of a toxin B deficient mutant strain of Clostridium difficile in a case of recurrent C. difficile-associated diarrhea. Clin. Infect. Dis. 26410-412. [DOI] [PubMed] [Google Scholar]
- 11.Delmee, M., J. Van Broeck, A. Simon, M. Janssens, and V. Avesani. 2005. Laboratory diagnosis of Clostridium difficile associated diarrhea: a plea for culture. 54187-191. [DOI] [PubMed] [Google Scholar]
- 12.Drudy, D., N. Harnedy, S. Fanning, R. O'Mahony, and L. Kyne. 2007. Isolation and characterization of toxin A negative, toxin B positive Clostridium difficile in Dublin, Ireland. Clin. Microbiol. Infect. 13298-304. [DOI] [PubMed] [Google Scholar]
- 13.Geric, B., M. Rupnik, D. Gerding, M. Grabnar, and S. Johnson. 2004. Distribution of Clostridium difficile variant toxinotypes and strains with binary toxin genes among clinical isolates in an American hospital. J. Med. Microbiol. 53887-894. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, S., S. A. Kent, K. J. O'Leary, M. M. Merrigan, S. P. Sambol, L. R. Peterson, and D. N. Gerding. 2001. Fatal pseudomembranous colitis associated with a variant Clostridium difficile strain not detected by toxin A immunoassay. Ann. Intern. Med. 18434-438. [DOI] [PubMed] [Google Scholar]
- 15.Kato, H., N. Kato, K. Watanabe, N. Iwai, H. Nakamura, T. Yamamoto, S. Suzuki, S. M. Kim, Y. Chong, and E. B. Wasito. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 362178-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komatsu, M., H. Kato, M. Aihara, K. Shimakawa, M. Iwasaki, Y. Nagasaka, S. Fukuda, S. Matsuo, Y. Arakawa, M. Watanabe, and Y. Iwatani. 2003. High prevalence of antibiotic-associated diarrhea due to toxin A negative, toxin B positive Clostridium difficile in a hospital in Japan and risk factors for infection. Eur. J. Clin. Microbiol. Infect. Dis. 22525-529. [DOI] [PubMed] [Google Scholar]
- 17.Kuijper, E. J., J. Weerdt, H. Kato, N. Kato, A. P. Dam, E. R. Vorm, J. Weel, C. Rheenen, and J. Dankert. 2001. Nosocomial outbreak of Clostrodium difficile associated diarrhea due to a clindamycin resistant enterotoxin A negative strain. Eur. J. Clin. Microbiol. Infect. Dis. 20528-534. [DOI] [PubMed] [Google Scholar]
- 18.Lemee, L., A. Dhalluin, S. Testelin, M. A. Mattrat, K. Maillard, J. F. Lemeland, and J. L. Pons. 2004. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J. Clin. Microbiol. 425710-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Lozniewski, A., C. Rabaud, E. Dotto, M. Weber, and F. Mory. 2001. Laboratory diagnosis of Clostridium difficile-associated diarrhea and colitis: usefulness of premier cytoclone A+B enzyme immunoassay for combined detection of stool toxins and toxigenic C. difficile strains. J. Clin. Microbiol. 391996-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyerly, D. M., L. M. Neville, D. T. Evans, J. Fill, and S. Allen. 1998. Multicenter evaluation of the Clostidium difficile TOX A/B test. J. Clin. Microbiol. 36184-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musher, D. M., A. Manhas, P. Jain, F. Nuila, A. Waqar, N. Logan, B. Marino, and E. A. Graviss. 2007. Detection of Clostridium difficile toxin: comparison of enzyme immunoassay results with results obtained by cytotoxicity assay. J. Clin. Microbiol. 452737-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson, L. R., M. M. Olson, C. J. Shanholtzer, and D. N. Gerding. 1988. Results of a prospective, 18-month clinical evaluation of culture, cytotoxin testing, and culturette brand (CDT) latex testing in the diagnosis of Clostridium difficile-associated diarrhea. Diagn. Microbiol. Infect. Dis. 1085-91. [DOI] [PubMed] [Google Scholar]
- 24.Pituch, H., N. van den Braak, W. van Leeuwen, A. van Belkem, G. Martirosian, P. Obuch-Woszczatynski, M. Luczak, and F. Meisel-Mikolajczyk. 2001. Clonal dissemination of a toxin A negative/toxin B positive Clostridium difficile strain from patients with antibiotic associated diarrhea in Poland. Clin. Microbiol. Infect. 7442-446. [DOI] [PubMed] [Google Scholar]
- 25.Reference deleted.
- 26.Post, K. W., B. H. Jost, and J. G. Songer. 2002. Evaluation of a test for Clostridium difficile toxin A and B for the diagnosis of neonatal swine enteritis. J. Vet. Diagn. Investig. 14258-259. [DOI] [PubMed] [Google Scholar]
- 27.Reller, M. E., C. A. Lema, T. M. Perl, M. Cai, T. L. Ross, K. A. Speck, and K. C. Carroll. 2007. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J. Clin. Microbiol. 453601-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes, R. C., M. A. John, D. L. Ayotte, A. Covacich, S. Milburn, and Z. Hussain. 2007. Performance of TechLab C.DIFF QUIK CHEK and TechLab C.DIFFICILE TOX A/B II for the detection of Clostridium difficile in stool samples. Diagn. Microbiol. Infect. Dis. 5933-37. [DOI] [PubMed] [Google Scholar]
- 29.Rupnik, M., N. Kato, M. Grabnar, and H. Kato. 2003. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J. Clin. Microbiol. 411118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samra, Z., S. Talmor, and J. Bahar. 2002. High prevalence of toxin A negative toxin B positive Clostridium difficile in hospitalized patients with gastrointestinal disease. Diagn. Microbiol. Infect. Dis. 43189-192. [DOI] [PubMed] [Google Scholar]
- 31.Shin, B. M., E. Y. Kuak, S. J. Yoo, W. C. Shin, and H. M. Yoo. 2008. Emerging toxin A−B+ variant strains of Clostridium difficile responsible for pseudomembranous colitis at a tertiary care hospital. Diagn. Microbiol. Infect. Dis. 60333-337. [DOI] [PubMed] [Google Scholar]
- 32.Shin, B. M., E. Y. Kuak, H. M. Yoo, E. C. Kim, K. Lee, K. O. Kang, D. H. Whang, and J. H. Shin. 2008. Multicentre study of the prevalence of toxigenic Clostridium difficile in Korea; results of a retrospective study 2000-2005. J. Med. Microbiol. 57697-701. [DOI] [PubMed] [Google Scholar]
- 33.Staneck, J. L., L. S. Weckbach, S. D. Allen, J. A. Siders, P. H. Gilligan, G. Coppitt, J. A. Kraft, and D. H. Willis. 1996. Multicenter evaluation of four methods for Clostridium difficile detection: ImmunoCard C. difficile, cytotoxin assay, culture, and latex agglutination. J. Clin. Microbiol. 342718-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ticehurst, J. R., D. Z. Aird, L. M. Dam, A. P. Borek, J. T. Hargrove, and K. C. Carroll. 2006. Effective detection of toxigenic Clostridium difficile by a two-step algorithm including tests for antigen and cytotoxin. J. Clin. Microbiol. 441145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turgeon, D. K., T. J. Novicki, J. Quick, L. Carlson, P. Miller, B. Ulness, A. Cent, R. Ashley, A. Larson, M. Coyle, A. P. Limaye, B. T. Cookson, and T. R. Fritsche. 2003. Six rapid tests for direct detection of Clostridium difficile and its toxins in fecal samples compared with the fibroblast cytotoxicity assay. J. Clin. Microbiol. 41667-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Berg, R. J., L. S. B. van Coppernraet, H. J. Gerritsen, H. P. Endtz, E. R. van der Vorm, and E. J. Kuijper. 2005. Prospective multicenter evaluation of a new immunoassay and real-time PCR for rapid diagnosis of Clostridium difficile-associated diarrhea in hospitalized patients. J. Clin. Microbiol. 435338-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]