Abstract
Subtyping was conducted in late 2007 on 57 Cryptosporidium specimens from sporadic cases in Colorado, Idaho, New Mexico, and Iowa. One previously rare Cryptosporidium hominis subtype was indentified in 40 cases (70%) from all four states, and the Cryptosporidium horse genotype was identified in a pet shop employee with severe clinical symptoms.
The protozoan pathogen Cryptosporidium continues to be an important cause of waterborne disease outbreaks and gastrointestinal illness in the United States (4, 6, 21-23). While few recent cryptosporidiosis outbreaks have been associated with drinking water, the incidence of recreational water-associated outbreaks is increasing. In the United States, specimens from patients with outbreak-related cases have been extensively characterized by molecular epidemiology methods (20), but few specimens from patients with sporadic disease have been similarly analyzed (5). As a result, U.S. endemic transmission of Cryptosporidium spp. is not well understood. Conversely, genotyping of a large number of specimens from sporadic cases has facilitated understanding of endemic cryptosporidiosis transmission in the United Kingdom (3, 9, 10).
In this study, we genotyped and subtyped specimens from patients with sporadic cryptosporidiosis detected in Colorado, Idaho, New Mexico, and Iowa during the late summer and fall of 2007. The former three states border Utah, where a statewide outbreak of cryptosporidiosis involving 1,902 laboratory-confirmed cases occurred during June to December 2007 (13). Nevertheless, investigations of the sporadic Colorado, Idaho, New Mexico, and Iowa cases revealed no epidemiologic links with any known outbreaks of cryptosporidiosis. These states, and the United States as a whole, reported increased numbers of sporadic cryptosporidiosis cases during 2007 (2). We also report on a symptomatic case of human infection with the Cryptosporidium horse genotype.
Specimens and laboratory and epidemiological investigations.
A total of 57 Cryptosporidium-positive stool specimens were collected from patients with sporadic cryptosporidiosis, including 5, 10, 11, and 31 specimens from New Mexico, Iowa, Colorado, and Idaho residents, respectively. During 2007, totals of 125, 610, 211, and 464 laboratory-confirmed cases were reported in New Mexico, Iowa, Colorado, and Idaho, respectively, and 11,170 cases were reported in the country (2). Only one specimen per patient was used in the study. Specimens were confirmed to be Cryptosporidium positive by submitting laboratories using direct immunofluorescence microscopy or enzymatic immunoassays.
Cryptosporidium DNA was extracted from 0.2 ml of each stool specimen, as described previously (7). Parasite DNA was genotyped by nested PCR-restriction fragment length polymorphism (RFLP) analysis of the small-subunit (SSU) rRNA gene (8). PCR products of the Cryptosporidium horse genotype were sequenced to verify the identification. All Cryptosporidium spp. were further subtyped by DNA sequencing of the 60-kDa glycoprotein (gp60) gene amplified by a nested PCR (1), using modified primary PCR primers LX0374 (5′-TTA CTC TCC GTT ATA GTC TCC-3′) and LX0375 (5′-GGA AGG AAC GAT GTA TCT GA-3′), which reduced the overlapping sequence shared between primary and secondary forward primers. To confirm the results, analysis at each locus was repeated at least three times using 2 μl of DNA per PCR. The established subtype nomenclature was used in identifying subtype families and subtypes (15).
The patient infected with the Cryptosporidium horse genotype was interviewed via telephone by a state-based epidemiologist to collect clinical and exposure data, using a structured questionnaire, 2.5 months after the initial cryptosporidiosis diagnosis. Informed consent was obtained from the adult patient before the interview.
Distribution of Cryptosporidium species/genotypes and Cryptosporidium hominis and C. parvum subtypes.
PCR-RFLP analysis of the SSU rRNA gene identified C. hominis in 51 stool specimens and C. parvum in 5. PCR amplicons from a specimen obtained in New Mexico showed an RFLP banding pattern nearly identical to that of the previously reported Cryptosporidium horse genotype (14). DNA sequencing of the gp60 gene confirmed the identification of C. hominis and C. parvum in the stool specimens (Table 1). The C. hominis specimens belonged to five subtypes in two subtype families, whereas the C. parvum specimens belonged to four subtypes in the well-recognized zoonotic subtype family IIa. The most commonly identified subtype was the C. hominis subtype IaA28R4, which was found in 40 specimens (70%) from all four states sampled. This was followed by the IbA10G2 subtype, found in eight specimens (14%) from two states. Other C. hominis and C. parvum subtypes were each found only in one or two specimens (Table 1).
TABLE 1.
Distribution of Cryptosporidium genotypes and subtypes in 57 sporadic cases in Colorado, Idaho, New Mexico, and Iowa in the summer and fall of 2007
| Genotype/subtype | No. of specimens analyzed | No. of positive specimens by state
|
|||
|---|---|---|---|---|---|
| Colorado | Idaho | Iowa | New Mexico | ||
| C. hominis | 51 | 11 | 28 | 10 | 2 |
| IaA12R4 | 1 | 1 | 0 | 0 | 0 |
| IaA14R3 | 1 | 0 | 0 | 0 | 1 |
| IaA27R4 | 1 | 0 | 1 | 0 | 0 |
| IaA28R4 | 40 | 8 | 27 | 4 | 1 |
| IbA10G2 | 8 | 2 | 0 | 6 | 0 |
| C. parvum | 5 | 0 | 3 | 0 | 2 |
| IIaA15G2R1 | 2 | 0 | 1 | 0 | 1 |
| IIaA16G1R1 | 1 | 0 | 1 | 0 | 0 |
| IIaA17G2R1 | 1 | 0 | 0 | 0 | 1 |
| IIaA17G4R1 | 1 | 0 | 1 | 0 | 0 |
| Horse genotype | 1 | 0 | 0 | 0 | 1 |
| VIbA13 | 1 | 0 | 0 | 0 | 1 |
Characteristics of the Cryptosporidium horse genotype.
The partial SSU rRNA and gp60 genes of the Cryptosporidium horse genotype from the human case were sequenced, and the results were compared with those identified in a foal in the Czech Republic and a calf in Northern Ireland (14, 16). The bovine specimen was initially identified as the IIj subtype family of C. parvum, but resequencing of the SSU rRNA gene confirmed the Cryptosporidium horse genotype identity. The ∼800-bp SSU rRNA gene sequence of the human specimen was identical to sequences obtained from specimens from the horse in the Czech Republic and the calf in Northern Ireland, with only an AG-to-GA substitution. A close examination of the electropherograms of products from all three specimens revealed the presence of mixed A and G signals at both sites.
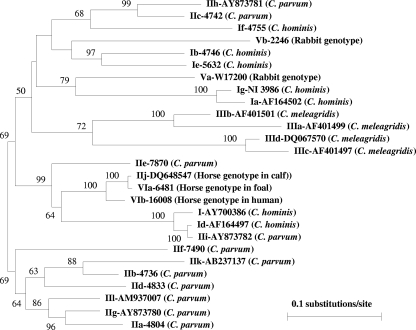
The 823-bp gp60 sequence of the human specimen of the Cryptosporidium horse genotype had high sequence similarity to that of the Czech horse (96.3% sequence identity, excluding the trinucleotide repeat region). The latter, however, had higher sequence similarity to the gp60 sequence (GenBank accession number DQ648547) of the specimen from the calf in Northern Ireland (previously identified as the C. parvum subtype family IIj) (16). The sequence difference between the two in the nonrepeat regions was smaller than 1%, with only six nucleotide substitutions. In phylogenetic analysis, the gp60 sequences of the three Cryptosporidium horse genotype specimens clustered together and were most closely related to C. hominis Id and C. parvum IIe subtype families (Fig. 1).
FIG. 1.
Phylogenetic relationship among major Cryptosporidium gp60 subtype families inferred by a neighbor-joining analysis of nucleotide sequences, using a sequence alignment generated by the ClustalX 1.81 package and the Kimura two-parameter genetic distances calculated by the Treecon W program. I, C. hominis subtype families; II, C. parvum subtype families; III, C. meleagridis subtype families; V, rabbit genotype subtype families; VI, horse genotype subtype families. The equine and bovine specimens of the Cryptosporidium horse genotype belong to subtype family VIa, while the human specimen of that genotype belongs to subtype family VIb.
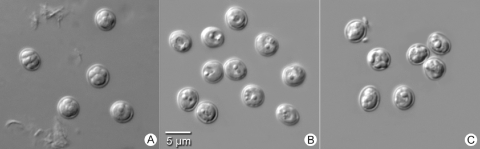
The Cryptosporidium horse genotype oocysts from the human patient were morphologically similar to those of C. parvum and C. hominis under light microscopy (Fig. 2). They were 4.63 (±0.25) by 4.21 (±0.19) μm in size (n = 63), with a length/width ratio of 1.10.
FIG. 2.
Oocysts of C. parvum (A), C. hominis (B), and the Cryptosporidium horse genotype (C) under differential interference contrast (magnification of ×1,000) on a Zeiss Axiophot microscope (Carl Zeiss MicroImaging Inc., Thornwood, NY).
Case history of human infection with the Cryptosporidium horse genotype.
The patient was an 18-year-old Caucasian female living in a suburban area. She had the onset of diarrhea on 1 December 2007, which lasted for 14 days. At the peak of her illness, she had nine loose or watery stools per day. Other clinical symptoms included fever (38.3°C for 1 day), loss of appetite and body aches (for 2 to 5 days), nausea, abdominal cramps, and fatigue (for 6 to 14 days). The patient visited an emergency room because of the illness and received intravenous fluids. She was prescribed antibiotics for presumed giardiasis and had taken over-the-counter medicines for diarrhea. A stool specimen was collected 8 days after the onset of diarrhea, and a diagnosis of cryptosporidiosis was confirmed. The patient was prescribed nitazoxanide and recovered from cryptosporidiosis within a few days. Over the course of the illness, the patient missed work for 5 days, lost 2.3 kg of weight, and reported being unable to perform her usual daily activities for 10 days. The patient reported developing irritable bowel disease after the resolution of cryptosporidiosis.
The patient reported no travel during the 2 weeks prior to illness onset. She also did not report any apparent food-borne, waterborne, or person-to-person risk factors for potential exposure to Cryptosporidium. The patient worked in a pet store and was involved in the care of pet animals, including kittens, puppies, ferrets, rabbits, rodents, chinchillas, hedgehogs, birds, reptiles, and amphibians. Some of the puppies, rabbits, and hedgehogs had diarrhea, and one hedgehog died of dehydration as a result of the diarrhea. The patient did not visit or work on farms or visit petting zoos but owned fish and a puppy as household pets. The puppy had diarrhea, starting approximately on 15 November 2007 and was diagnosed as having giardiasis.
Public health significance of the Cryptosporidium horse genotype.
The Cryptosporidium horse genotype has been identified in only one person (a recently reported case), a 30-year-old immunocompetent woman with diarrhea in a rural area of southwest England (3, 12). Findings of unusual Cryptosporidium species/genotypes in humans have been reported occasionally, such as C. andersoni, C. muris, C. suis, and Cryptosporidium of the cervine, skunk, and chipmunk I genotypes (10, 12, 20). Recent reviews have identified at least 85 cases of C. felis, 24 cases of C. canis, and 21 cases of the Cryptosporidium cervine genotype infections in immunocompetent and immunocompromised persons around the world, in addition to the three common Cryptosporidium spp., as follows: C. hominis, C. parvum, and C. meleagridis (17, 19). Reports of human infections caused by other Cryptosporidium spp. were identified in only one to a few cases. The Cryptosporidium horse genotype represents another among the increasing number of unusual Cryptosporidium species and genotypes identified in human stool specimens.
The pathogenicity of the unusual Cryptosporidium spp. infecting humans is generally unclear, as most reports lack detailed data on the clinical history of the affected patients. Detailed clinical manifestations of C. muris and C. suis infections have been reported for only two persons, both human immunodeficiency virus-positive adults in Lima, Peru (11, 18). The patient infected with the Cryptosporidium horse genotype had no recognized immunocompromising conditions. Nevertheless, she experienced severe clinical symptoms consistent with cryptosporidiosis and required emergency room care, including intravenous fluids, repeated antimicrobial prescriptions, and sick leave from work. No detailed clinical data are available on the human case of infection with the Cryptosporidium horse genotype in England, although the patient was assumed to have diarrhea (12).
The source of the Cryptosporidium horse genotype infection is not clear, although results from the traditional epidemiologic investigation indicate that it was probably of animal origin. Because the patient had no contact with horses and was in close contact with various animals at work and home, it was impossible to identify the animal species involved in this probable case of zoonotic transmission of Cryptosporidium. Few studies have investigated the infection source associated with unusual Cryptosporidium species and genotypes in humans. The Peruvian infected with C. suis had no exposure to pigs or other farm animals. In that case, anthroponotic rather than zoonotic transmission was a strong possibility, as the patient was a homosexual man with multiple sex partners and participated in anal intercourse (18). The person infected with the Cryptosporidium horse genotype in England reported swimming and foreign travel but no contact with animals (12).
Rare C. hominis subtypes and cryptosporidiosis outbreaks.
Another unique finding of this study is the predominance of the IaA28R4 subtype among the tested specimens. Prior to 2007, IaA28R4 was a rare subtype, having been implicated only in two swimming pool-associated outbreaks of cryptosporidiosis in Ohio (2005) and South Carolina (2006) amid the 28 waterborne and food-borne cryptosporidiosis outbreaks in the United States, whose investigation included genotyping (20). This subtype was found in 40 of the 57 specimens (70%) from sporadic cases detected in late 2007 from all four states sampled and was also responsible for two of the seven cryptosporidiosis outbreaks investigated in the summer and fall of 2007, as follows: a swimming pool-associated outbreak in Pennsylvania (n = 730) and an interactive splash park-associated outbreak in Idaho (n = 51). In contrast, the C. hominis subtype IbA10G2, commonly implicated in cryptosporidiosis outbreaks, especially major ones in North America and Europe (20), was seen only in 8/57 cases. It might have been a more frequently identified subtype if the sample size were bigger and other states participated in this study. Nevertheless, results of this small-scale study with limited sampling indicate that some “sporadic” cases might have been part of larger multistate outbreaks caused by IaA28R4. This is a unique but expected finding in the context of increased cryptosporidiosis reporting in the United States in 2007 and coincides with a large statewide outbreak in Utah (13), a state that borders three of the four states sampled in this study. Unfortunately, no specimens from Utah were available for Cryptosporidium typing, despite numerous attempts to acquire positive stool specimens. The number of sporadic cryptosporidiosis cases increased dramatically in the United States during 2007, and without molecular characterization, it is impossible to know whether this increase represented large, unrecognized cryptosporidiosis outbreaks. To better understand the transmission of Cryptosporidium in the United States, a national system is needed to systematically characterize Cryptosporidium in sporadic and outbreak-related cases over an extended period of time.
Nucleotide sequence accession numbers.
The partial SSU rRNA and gp60 gene sequences of the Cryptosporidium horse genotype were deposited in the GenBank database under accession numbers FJ435960 to FJ435964.
Acknowledgments
We thank local and state health workers for technical assistance.
The findings and conclusions in this study are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Alves, M., L. Xiao, I. Sulaiman, A. A. Lal, O. Matos, and F. Antunes. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 412744-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2009. Notifiable diseases/deaths in selected cities weekly information. MMWR Morb. Mortal. Wkly. Rep. 571420-1431. [Google Scholar]
- 3.Chalmers, R. M., K. Elwin, A. L. Thomas, E. C. Guy, and B. Mason. 2009. Long-term Cryptosporidium typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Euro Surveill. 1419086. [DOI] [PubMed] [Google Scholar]
- 4.Dziuban, E. J., J. L. Liang, G. F. Craun, V. Hill, P. A. Yu, J. Painter, M. R. Moore, R. L. Calderon, S. L. Roy, and M. J. Beach. 2006. Surveillance for waterborne disease and outbreaks associated with recreational water—United States, 2003-2004. MMWR Surveill. Summ. 551-30. [PubMed] [Google Scholar]
- 5.Feltus, D. C., C. W. Giddings, B. L. Schneck, T. Monson, D. Warshauer, and J. M. McEvoy. 2006. Evidence supporting zoonotic transmission of Cryptosporidium in Wisconsin. J. Clin. Microbiol. 444303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hlavsa, M. C., J. C. Watson, and M. J. Beach. 2005. Cryptosporidiosis surveillance—United States 1999-2002. MMWR Surveill. Summ. 541-8. [PubMed] [Google Scholar]
- 7.Jiang, J., K. A. Alderisio, A. Singh, and L. Xiao. 2005. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ. Microbiol. 711135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang, J., K. A. Alderisio, and L. Xiao. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 714446-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 383984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols, G. L., R. M. Chalmers, W. Sopwith, M. Regan, C. A. Hunter, P. Grenfell, F. Harrison, and C. Lane. 2006. Cryptosporidiosis: a report on the surveillance and epidemiology of Cryptosporidium infection in England and Wales. Drinking Water Directorate Contract Number DWI 70/2/201. Drinking Water Inspectorate, London, United Kingdom.
- 11.Palmer, C. J., L. Xiao, A. Terashima, H. Guerra, E. Gotuzzo, G. Saldias, J. A. Bonilla, L. Zhou, A. Lindquist, and S. J. Upton. 2003. Cryptosporidium muris, a rodent pathogen, recovered from a human in Peru. Emerg. Infect. Dis. 91174-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson, G., K. Elwin, and R. M. Chalmers. 2008. Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg. Infect. Dis. 141800-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rolfs, R. T., M. J. Beach, M. C. Hlavsa, and R. M. Calanan. 2008. Communitywide cryptosporidiosis outbreak—Utah, 2007. MMWR Morb. Mortal. Wkly. Rep. 57989-993. [PubMed] [Google Scholar]
- 14.Ryan, U., L. Xiao, C. Read, L. Zhou, A. A. Lal, and I. Pavlasek. 2003. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl. Environ. Microbiol. 694302-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulaiman, I. M., P. R. Hira, L. Zhou, F. M. Al-Ali, F. A. Al-Shelahi, H. M. Shweiki, J. Iqbal, N. Khalid, and L. Xiao. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 432805-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, H. P., J. S. Dooley, J. Kenny, M. McCoy, C. J. Lowery, J. E. Moore, and L. Xiao. 2007. Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitol. Res. 100619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, R., J. Wang, M. Sun, H. Dang, Y. Feng, C. Ning, F. Jian, L. Zhang, and L. Xiao. 2008. Molecular characterization of the Cryptosporidium cervine genotype from a sika deer (Cervus nippon Temminck) in Zhengzhou, China and literature review. Parasitol. Res. 103865-869. [DOI] [PubMed] [Google Scholar]
- 18.Xiao, L., C. Bern, M. Arrowood, I. Sulaiman, L. Zhou, V. Kawai, A. Vivar, A. A. Lal, and R. H. Gilman. 2002. Identification of the Cryptosporidium pig genotype in a human patient. J. Infect. Dis. 1851846-1848. [DOI] [PubMed] [Google Scholar]
- 19.Xiao, L., and Y. Feng. 2008. Zoonotic cryptosporidiosis. FEMS Immunol. Med. Microbiol. 52309-323. [DOI] [PubMed] [Google Scholar]
- 20.Xiao, L., and U. M. Ryan. 2008. Molecular epidemiology, p. 119-171. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press and IWA Publishing, Boca Raton, FL.
- 21.Yoder, J. S., and M. J. Beach. 2007. Cryptosporidiosis surveillance—United States, 2003-2005. MMWR Surveill. Summ. 561-10. [PubMed] [Google Scholar]
- 22.Yoder, J. S., B. G. Blackburn, G. F. Craun, V. Hill, D. A. Levy, N. Chen, S. H. Lee, R. L. Calderon, and M. J. Beach. 2004. Surveillance for waterborne-disease outbreaks associated with recreational water-United States, 2001-2002. MMWR Surveill. Summ. 531-22. [PubMed] [Google Scholar]
- 23.Yoder, J. S., M. C. Hlavsa, G. F. Craun, V. Hill, V. Roberts, P. A. Yu, L. A. Hicks, N. T. Alexander, R. L. Calderon, S. L. Roy, and M. J. Beach. 2008. Surveillance for waterborne disease and outbreaks associated with recreational water use and other aquatic facility-associated health events-United States, 2005-2006. MMWR Surveill. Summ. 571-29. [PubMed] [Google Scholar]