Hepatitis B virus (HBV) genotypes (2, 4, 5, 7-9) and subgenotypes (11, 13, 14, 16-18) are important factors that influence the course of the disease and the outcome of treatment. Because of the clinical importance of the HBV classification, an accurate assignation of newly found subgenotypes to their respective clades and an unambiguous nomenclature are needed. A new HBV subgenotype should differ from previously established by at least 4%, and the segregation into a new clade should be supported by robust bootstrapping data (3).
In the June 2009 issue of the Journal of Clinical Microbiology, a new C6 subgenotype of HBV found in the Papua province of Indonesia (C6 Papua-Indonesia) is described (15). However, a tentative C6 subgenotype found in the Philippines (C6 Philippines) has already been published contemporaneously (1). To keep up with an unambiguous system of HBV subgenotypes, it is important to analyze if C6 Papua-Indonesia and C6 Philippines are identical or map to different clades.
Using methods described by Cavinta et al. (1) and published sequences from Utsumi et al. (15), we made a phylogenetic analysis using whole genomes that clearly showed that both C6 groups mapped to different clades (Fig. 1).
FIG. 1.
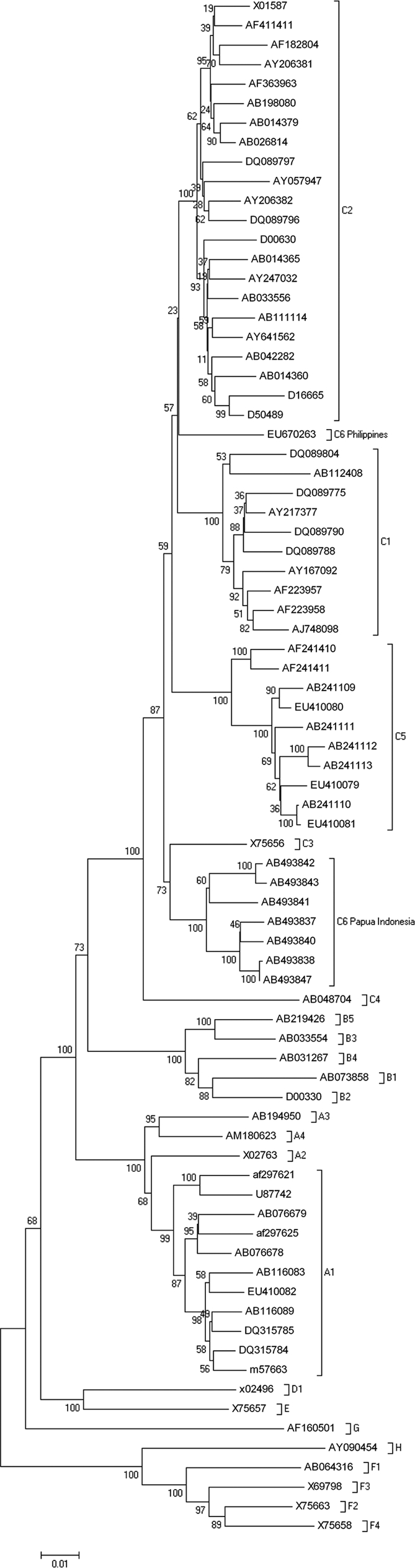
Phylogenetic tree of complete HBV genomes of genotypes A to H. An alignment of complete sequences was performed with ClustalW using the program DNASTAR. The alignment was further analyzed by bootstrapping with 500 replicates, using the neighborhood-joining method contained in MEGA version 4.0 (12).
C6 Papua-Indonesia and C6 Philippines differed by 5.1% from each other and diverged by at least 4.1% from the other HBV subgenotypes (Table 1).
TABLE 1.
Nucleotide distance between proposed subgenotypes C6 Philippines and C6 Papua-Indonesia of HBV and other reference genotype strains from the database
| Genotype/subgenotype strain | Nucleotide distance between strainsa
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | A4 | B1 | B2 | B4 | B5 | C2 | C3 | C4 | C5 | C6 Papua-Indonesia | C6 Philippines | D1 | E | F1 | F2 | F3 | F4 | G | H | C1 | B3 | |
| A1 | ||||||||||||||||||||||||
| A2 | 0.049 | |||||||||||||||||||||||
| A3 | 0.051 | 0.054 | ||||||||||||||||||||||
| A4 | 0.046 | 0.046 | 0.040 | |||||||||||||||||||||
| B1 | 0.100 | 0.096 | 0.102 | 0.098 | ||||||||||||||||||||
| B2 | 0.098 | 0.096 | 0.099 | 0.095 | 0.046 | |||||||||||||||||||
| B4 | 0.100 | 0.096 | 0.100 | 0.097 | 0.055 | 0.039 | ||||||||||||||||||
| B5 | 0.102 | 0.098 | 0.103 | 0.097 | 0.065 | 0.048 | 0.046 | |||||||||||||||||
| C2 | 0.092 | 0.089 | 0.092 | 0.084 | 0.105 | 0.095 | 0.093 | 0.091 | ||||||||||||||||
| C3 | 0.095 | 0.091 | 0.093 | 0.086 | 0.104 | 0.094 | 0.091 | 0.093 | 0.044 | |||||||||||||||
| C4 | 0.106 | 0.102 | 0.103 | 0.095 | 0.118 | 0.107 | 0.105 | 0.107 | 0.069 | 0.065 | ||||||||||||||
| C5 | 0.092 | 0.092 | 0.096 | 0.089 | 0.112 | 0.100 | 0.097 | 0.098 | 0.055 | 0.060 | 0.084 | |||||||||||||
| C6 Papua-Indonesia | 0.098 | 0.097 | 0.098 | 0.091 | 0.111 | 0.101 | 0.100 | 0.099 | 0.048 | 0.045 | 0.071 | 0.066 | ||||||||||||
| C6 Philippines | 0.092 | 0.091 | 0.093 | 0.084 | 0.106 | 0.096 | 0.094 | 0.090 | 0.041 | 0.047 | 0.072 | 0.059 | 0.051 | |||||||||||
| D1 | 0.106 | 0.103 | 0.106 | 0.102 | 0.119 | 0.112 | 0.111 | 0.112 | 0.107 | 0.106 | 0.113 | 0.108 | 0.118 | 0.103 | ||||||||||
| E | 0.101 | 0.100 | 0.102 | 0.095 | 0.118 | 0.112 | 0.111 | 0.113 | 0.106 | 0.097 | 0.113 | 0.107 | 0.111 | 0.104 | 0.078 | |||||||||
| F1 | 0.148 | 0.149 | 0.154 | 0.144 | 0.156 | 0.151 | 0.151 | 0.155 | 0.146 | 0.147 | 0.151 | 0.149 | 0.147 | 0.149 | 0.153 | 0.140 | ||||||||
| F2 | 0.148 | 0.145 | 0.153 | 0.150 | 0.155 | 0.150 | 0.147 | 0.150 | 0.145 | 0.142 | 0.153 | 0.143 | 0.144 | 0.142 | 0.149 | 0.142 | 0.060 | |||||||
| F3 | 0.157 | 0.159 | 0.158 | 0.154 | 0.159 | 0.153 | 0.152 | 0.155 | 0.146 | 0.142 | 0.151 | 0.148 | 0.145 | 0.143 | 0.150 | 0.141 | 0.059 | 0.049 | ||||||
| F4 | 0.152 | 0.151 | 0.155 | 0.154 | 0.158 | 0.150 | 0.150 | 0.150 | 0.153 | 0.145 | 0.157 | 0.149 | 0.152 | 0.151 | 0.145 | 0.143 | 0.067 | 0.041 | 0.051 | |||||
| G | 0.118 | 0.118 | 0.121 | 0.115 | 0.138 | 0.133 | 0.128 | 0.129 | 0.129 | 0.131 | 0.146 | 0.129 | 0.134 | 0.132 | 0.120 | 0.115 | 0.158 | 0.155 | 0.154 | 0.157 | ||||
| H | 0.156 | 0.154 | 0.161 | 0.156 | 0.162 | 0.155 | 0.155 | 0.158 | 0.152 | 0.154 | 0.157 | 0.160 | 0.153 | 0.150 | 0.149 | 0.153 | 0.089 | 0.090 | 0.091 | 0.098 | 0.158 | |||
| C1 | 0.095 | 0.095 | 0.095 | 0.088 | 0.110 | 0.098 | 0.095 | 0.094 | 0.047 | 0.055 | 0.079 | 0.063 | 0.061 | 0.051 | 0.110 | 0.110 | 0.150 | 0.145 | 0.149 | 0.153 | 0.130 | 0.154 | ||
| B3 | 0.103 | 0.096 | 0.104 | 0.100 | 0.066 | 0.047 | 0.043 | 0.031 | 0.092 | 0.091 | 0.109 | 0.100 | 0.100 | 0.092 | 0.108 | 0.114 | 0.157 | 0.152 | 0.155 | 0.148 | 0.132 | 0.159 | 0.093 | |
The comparisons were performed over the complete genome with the Kimura two-parameter model using MEGA version 4.0 (12). Between groups, the average was calculated.
Thus, both contemporaneously described subgenotypes (1, 15) fulfill all criteria for having new and separate subgenotypes. Because the designation C6 has been claimed for partial sequences found in the Papua province of Indonesia already (6), we suggest that the subgenotype C6 found in the Papua province of Indonesia shall keep the designation C6, whereas the tentative subgenotype from the Philippines shall be called C7. To avoid similar confusion in the future, we have suggested a procedure that would involve the International Committee for Taxonomy of Viruses to allot new genotype/subgenotype designations (10).
Ed. Note: The authors of the published article declined to submit a response.
REFERENCES
- 1.Cavinta, L., J. Sun, A. May, J. Yin, M. von Meltzer, M. Radtke, N. G. Barzaga, G. Cao, and S. Schaefer. 2009. A new isolate of hepatitis B virus from the Philippines possibly representing a new subgenotype C6. J. Med. Virol. 81983-987. [DOI] [PubMed] [Google Scholar]
- 2.Chu, C. J., and A. S. Lok. 2002. Clinical significance of hepatitis B virus genotypes. Hepatology 351274-1276. [DOI] [PubMed] [Google Scholar]
- 3.Kramvis, A., K. Arakawa, M. C. Yu, R. Nogueira, D. O. Stram, and M. C. Kew. 2008. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J. Med. Virol. 8027-46. [DOI] [PubMed] [Google Scholar]
- 4.Kramvis, A., M. Kew, and G. Francois. 2005. Hepatitis B virus genotypes. Vaccine 232409-2423. [DOI] [PubMed] [Google Scholar]
- 5.Kramvis, A., and M. C. Kew. 2005. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J. Viral Hepat. 12456-464. [DOI] [PubMed] [Google Scholar]
- 6.Lusida, M. I., V. E. Nugrahaputra, Soetjipto, R. Handajani, M. Nagano-Fujii, M. Sasayama, T. Utsumi, and H. Hotta. 2008. Novel subgenotypes of hepatitis B virus genotypes C and D in Papua, Indonesia. J. Clin. Microbiol. 462160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyakawa, Y., and M. Mizokami. 2003. Classifying hepatitis B virus genotypes. Intervirology 46329-338. [DOI] [PubMed] [Google Scholar]
- 8.Norder, H., A.-M. Couroucé, P. Coursaget, J. M. Echevarria, S.-D. Leef, I. K. Mushahwar, B. H. Robertson, S. Locarnini, and L. O. Magnius. 2004. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology 47289-309. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer, S. 2005. Hepatitis B virus: significance of genotypes. J. Viral Hepat. 12111-124. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer, S. Under construction. Classification of hepatitis B virus genotypes and subgenotypes. Intervirology, in press. [DOI] [PubMed]
- 11.Sung, J. J. Y., S. K. W. Tsui, C.-H. Tse, E. Y. T. Ng, K.-S. Leung, K.-H. Lee, T. S. K. Mok, A. Bartholomeusz, T. C. C. Au, K. K. F. Tsoi, S. Locarnini, and H. L. Y. Chan. 2008. Genotype-specific genomic markers associated with primary hepatomas, based on complete genomic sequencing of hepatitis B virus. J. Virol. 823604-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka, Y., M. Mukaide, E. Orito, M. F. Yuen, K. Ito, F. Kurbanov, F. Sugauchi, Y. Asahina, N. Izumi, M. Kato, C. L. Lai, R. Ueda, and M. Mizokami. 2006. Specific mutations in enhancer II/core promoter of hepatitis B virus subgenotypes C1/C2 increase the risk of hepatocellular carcinoma. J. Hepatol. 45646-653. [DOI] [PubMed] [Google Scholar]
- 14.Truong, B. X., Y. Yano, Y. Seo, T. M. Phuong, Y. Tanaka, H. Kato, A. Miki, T. Utsumi, T. Azuma, N. K. Trach, M. Mizokami, Y. Hayashi, and M. Kasuga. 2007. Variations in the core promoter/pre-core region in HBV genotype C in Japanese and Northern Vietnamese patients. J. Med. Virol. 791293-1304. [DOI] [PubMed] [Google Scholar]
- 15.Utsumi, T., M. I. Lusida, Y. Yano, V. E. Nugrahaputra, M. Amin, Juniastuti, Soetjipto, Y. Hayashi, and H. Hotta. 2009. Complete genome sequence and phylogenetic relatedness of hepatitis B virus isolates in Papua, Indonesia. J. Clin. Microbiol. 471842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, Z., J. Hou, G. Zeng, S. Wen, Y. Tanaka, J. Cheng, F. Kurbanov, L. Wang, J. Jiang, N. V. Naoumov, M. Mizokami, and Y. Qi. 2007. Distribution and characteristics of hepatitis B virus genotype C subgenotypes in China. J. Viral Hepat. 14426-434. [DOI] [PubMed] [Google Scholar]
- 17.Wang, Z., Y. Tanaka, Y. Huang, F. Kurbanov, J. Chen, G. Zeng, B. Zhou, M. Mizokami, and J. Hou. 2007. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J. Clin. Microbiol. 451491-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan, J., B. Zhou, Y. Tanaka, F. Kurbanov, E. Orito, Z. Gong, L. Xu, J. Lu, X. Jiang, W. Lai, and M. Mizokami. 2007. Hepatitis B virus (HBV) genotypes/subgenotypes in China: mutations in core promoter and precore/core and their clinical implications. J. Clin. Virol. 3987-93. [DOI] [PubMed] [Google Scholar]


