Abstract
Summary: NAD is a coenzyme for redox reactions and a substrate of NAD-consuming enzymes, including ADP-ribose transferases, Sir2-related protein lysine deacetylases, and bacterial DNA ligases. Microorganisms that synthesize NAD from as few as one to as many as five of the six identified biosynthetic precursors have been identified. De novo NAD synthesis from aspartate or tryptophan is neither universal nor strictly aerobic. Salvage NAD synthesis from nicotinamide, nicotinic acid, nicotinamide riboside, and nicotinic acid riboside occurs via modules of different genes. Nicotinamide salvage genes nadV and pncA, found in distinct bacteria, appear to have spread throughout the tree of life via horizontal gene transfer. Biochemical, genetic, and genomic analyses have advanced to the point at which the precursors and pathways utilized by a microorganism can be predicted. Challenges remain in dissecting regulation of pathways.
INTRODUCTION
Functions of NAD
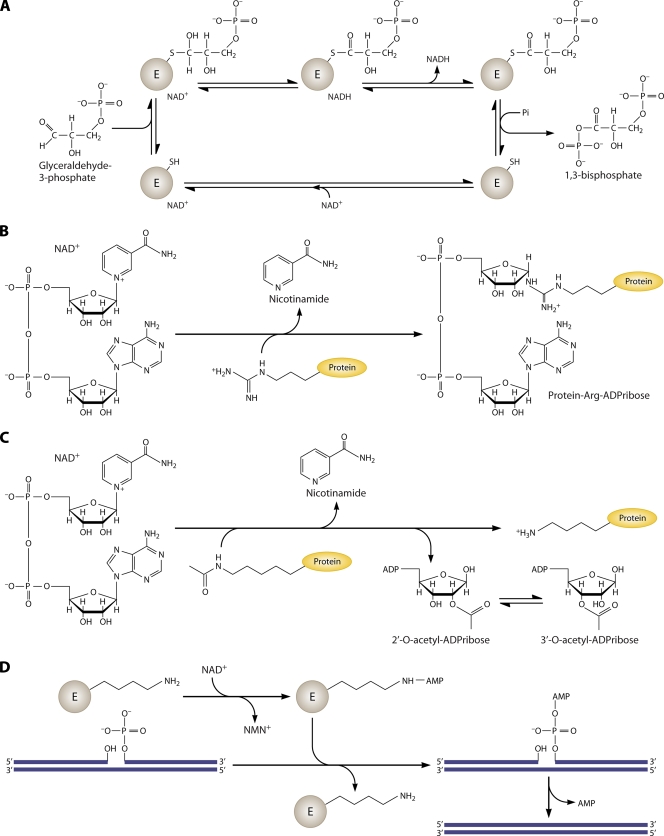
NAD and its reduced and phosphorylated derivatives, NADH, NADP, and NADPH, function as hydride acceptors and donors in a variety of cellular redox reactions (2). In addition, NAD is a consumed substrate of ADP-ribose transferases (25), Sir2- and CobB-related protein lysine deacetylases termed sirtuins (29), and bacterial DNA ligases (64). Metazoans have evolved further NAD-consuming activities, including cADP-ribose synthetases and poly(ADP-ribose) transferases (7). A sampling of major biochemical reactions requiring NAD and its derivatives is shown in Fig. 1.
FIG. 1.
Biochemical reactions of NAD. (A) NAD is utilized as a coenzyme in the conversion of glyceraldehyde 3-phosphate to 1,3-bisphosphate by glyceraldehyde phosphate dehydrogenase. The enzyme-bound NAD promotes formation of a covalent thiohemiacetal intermediate and conversion to a dehydrogenated thioester with conversion of the coenzyme to NADH. The bound thioester is then phosphorylated to the 1,3-bisphosphate product. (B) NAD is the ADP-ribose donor for ADP-ribose transfer reactions by ADP-ribose transferases. The depicted reaction involves protein arginine ADP-ribosylation with production of nicotinamide. (C) Sirtuins are NAD-dependent protein lysine deacetylases. In sirtuin reactions, NAD is the acetyl acceptor, forming 2′- and 3′-acetylated ADP-ribose plus nicotinamide and the nonmodified protein lysine, from an acetylated protein Lys. (D) Bacterial DNA ligases utilize NAD to adenylylate the ligase active-site lysine residue, which activates the 5′ phosphate of a nicked DNA substrate, forming an adenylylated nicked substrate. The enzyme then promotes the attack of the 3′ hydroxyl on the 5′ phosphate, releasing AMP and forming a DNA phosphodiester bond.
Canonical De Novo and Salvage Biosynthetic Pathways
Though NAD metabolism is presented in textbooks as a universal process, there is remarkable diversity in cellular approaches to NAD synthesis. The two basic types of NAD synthesis consist of de novo biosynthetic pathways and salvage biosynthetic pathways. As shown in Fig. 2, in de novo biosynthesis, nicotinic acid mononucleotide (NaMN) is synthesized in three enzymatic steps from Asp or in five steps from Trp, followed by two enzymatic steps to complete synthesis of NAD. In salvage biosynthesis, nicotinic acid, nicotinamide, nicotinic acid riboside, or nicotinamide riboside—NAD breakdown products that contain a pyridine ring—are either imported from outside of cells or recycled from inside cells and converted in a few steps to intact NAD (11).
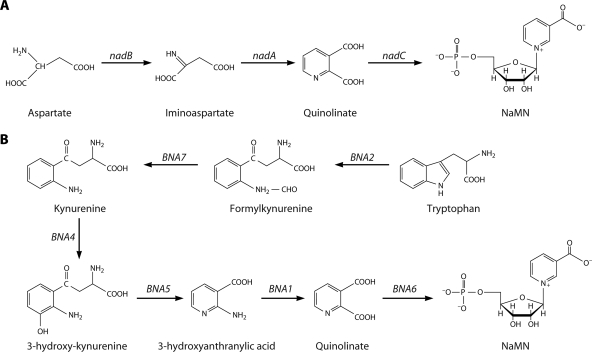
FIG. 2.
De novo biosynthesis of NAD. (A) Schematic diagram of NAD biosynthesis from aspartate to NaMN (E. coli gene names). In some archaea and thermotoga, the first step is catalyzed by aspartate dehydrogenase rather than aspartate oxidase (67). (B) Schematic diagram of NAD biosynthesis from tryptophan to NaMN (S. cerevisiae gene names).
In the absence of evidence for an abiotic source of nicotinamide or nicotinic acid, it is reasoned that ancestral cells synthesized NAD de novo (13). Canonically, the three-step pathway from Asp is found in monera, whereas the five-step pathway from Trp is found in eukaryotes, and both de novo pathways are commonly termed aerobic. However, no aspect of the canon is without exception. There are bacteria such as Haemophilus influenzae, which do not carry genes for a de novo pathway (21), and bacteria such as Cytophaga hutchinsonii, which carry the genes for a de novo pathway from Trp (32). Similarly, there are fungi such as Candida glabrata, which do not carry the genes for a de novo pathway (18), and plants such as Arabidopsis thaliana, which carry the genes for a de novo pathway from Asp (30). Finally, there are anaerobes such as the hyperthermophilic archaeon Pyrococcus horikoshii OT-3, which carry genes for a de novo pathway (53).
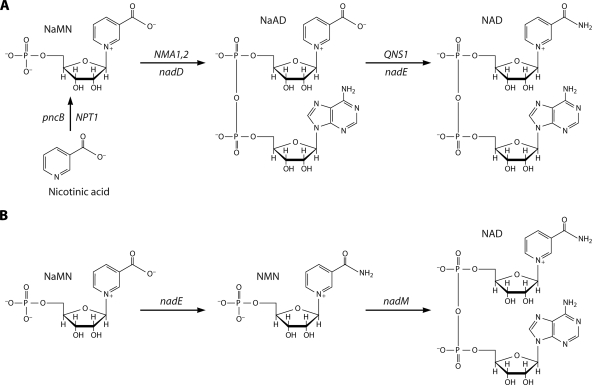
Most of the literature on NAD salvage pathways concerns the two salvageable pyridine bases, nicotinic acid and nicotinamide, which are collectively termed niacin. These compounds were discovered as vitamins by Elvehjem et al. as anti-black tongue factors for malnourished dogs (19). Nicotinic acid salvage was solved by Preiss and Handler with their description of the three-step pathway through NaMN and nicotinic acid dinucleotide (NaAD) (Fig. 3A) (46, 47). Because de novo NAD biosynthesis produces NaMN, the second and third steps of the Preiss-Handler pathway are described as common to nicotinic acid salvage and de novo biosynthesis. As will be discussed below, the conversion of NaMN to NAD, once thought to be either absent from particular microbes, such as H. influenzae, or present and conducted by Preiss-Handler enzymes, is accomplished differently in Francisella tularensis (Fig. 3B) (59a).
FIG. 3.
Synthesis of NAD through the NaMN intermediate. (A) In the Preiss-Handler pathway, nicotinic acid is salvaged to NAD via NaMN and NaAD intermediates. The pathway is depicted with S. cerevisiae gene names over the arrows and E. coli gene names under the arrows. (B) In F. tularensis, the de novo pathway depicted in Fig. 2A was found, but the nadD gene was found to be missing. Utilization of NaMN depends on NaMN amidation to NMN by a unique nadE gene, followed by NMN adenylylation by nadM.
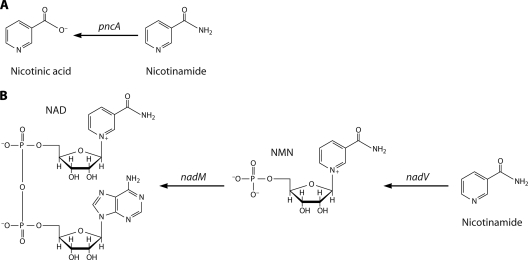
The widespread phylogenetic occurrence of NAD-consuming enzymes such as sirtuins (22), which break down NAD to nicotinamide, creates a demand for nicotinamide salvage. Thus, nicotinamide salvage would seem to be a universally conserved enzyme function but it is not. Though Escherichia coli and Saccharomyces cerevisiae both encode homologous nicotinamidases (24), which convert nicotinamide to nicotinic acid for Preiss-Handler salvage (Fig. 4A), vertebrates lack a homologous gene. Instead, nicotinamide salvage in a divergent set of bacteria and eukaryotes is accomplished by nicotinamide phosphoribosyltransferase (Fig. 4B). Opining on nature, Jacques Monod famously claimed that what was true for E. coli would be true for the elephant. In fact, the tools of molecular biology and comparative genomics allow one to delimit which organisms and viruses perform a homologous function and which organisms and viruses do not. Attempting to account for the phylogenetic distribution of the two different nicotinamide salvage systems, this review will make the case for multiple horizontal gene transfer events in the dissemination of nicotinamidases and nicotinamide phosphoribosyltransferases within bacteria and between bacteria and other domains.
FIG. 4.
Nicotinamide salvage. (A) Nicotinamidases homologous to E. coli pncA convert nicotinamide to nicotinic acid for Preiss-Handler salvage. (B) Nicotinamide phosphoribosyltransferases homologous to the H. ducreyi and F. tularensis nadV products convert nicotinamide to NMN for adenylylation to NAD. Vertebrate nadV homologs are also termed NAMPT, PBEF, and Visfatin. Bacterial NMN adenylyltransferase, encoded by nadM, converts NMN to NAD. Eukaryotic enzymes do not discriminate well between NaMN and NMN adenylylation, such that most eukaryotic NaMN adenylyltransferases, such as S. cerevisiae Nma1 and Nma2 (depicted in Fig. 3A), are also NMN adenylyltransferases.
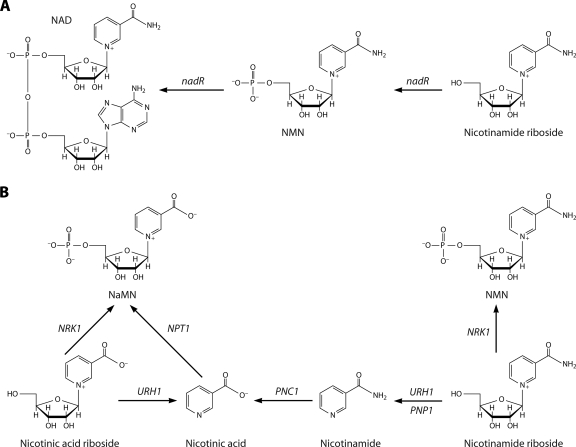
Nicotinamide riboside was identified as a salvageable NAD precursor in Haemophilus influenzae (26, 35, 57) and more recently was discovered as an NAD precursor in yeast (8). The enzymatic basis for utilization of nicotinamide riboside by Haemophilus influenzae depends on a specific nicotinamide riboside kinase fused to a specific nicotinamide mononucleotide (NMN) adenylyltransferase encoded by the nadR gene (58). The eukaryotic nicotinamide riboside kinases (8) are also specific enzymes, possessing sequences different from those of bacterial nicotinamide riboside kinases, which phosphorylate nicotinamide riboside and nicotinic acid riboside (62). However, in addition to salvage of nicotinamide riboside by production of NMN, nicotinamide riboside salvage to nicotinamide, mediated by enzymes previously characterized as uridine hydrolase (Urh1) and purine nucleoside phosphorylase (Pnp1), has been demonstrated in yeast (3, 4, 36). Finally, recent work indicates that nicotinic acid riboside can be converted to NaMN by nicotinamide riboside kinase and can also be utilized by the nucleosidase activity of Urh1, followed by nicotinic acid salvage (3, 62). Nicotinamide riboside and nicotinic acid riboside salvage pathways are depicted in Fig. 5.
FIG. 5.
Nicotinamide riboside and nicotinic acid riboside salvage. (A) Nicotinamide riboside salvage in H. influenzae is mediated by the nicotinamide riboside kinase domain and the NMN adenylyltransferase domain of nadR. (B) Nicotinamide riboside and nicotinic acid riboside salvage in S. cerevisiae is mediated by nicotinamide riboside kinase (NRK1)-dependent and nucleoside-splitting pathways.
Thus, to summarize current knowledge on canonical NAD biosynthetic pathways, organisms can be considered to carry or not to carry the genetic modules depicted in Fig. 2 to 5, which carry out two possible de novo biosynthetic pathways and salvage pathways from as many as four different salvageable precursors. In contrast to a restaurant offering a prix fixe dinner, organisms can be considered to prepare NAD using a “Chinese menu” approach in which there can be a pathway from Asp (Fig. 2A) or Trp (Fig. 2B) or none, enzymes for nicotinic acid utilization (Fig. 3A) or none, a possible alternative route from NaMN to NAD (Fig. 3B), and/or enzymes for nicotinamide (Fig. 4) and/or nicotinamide riboside/nicotinic acid riboside utilization (Fig. 5).
Whereas the rules for following a Chinese menu might dictate a choice between one dish or another, the diversity in NAD biosynthetic processes is highly combinatorial and embraces examples in which organisms encode multiple salvage pathways from the same precursor. As shown in Fig. 6, the betaproteobacteria Ralstonia solanacearum and Chromobacterium violaceum are remarkable for encoding nicotinamidase and nicotinamide phosphoribosyltransferase activities. As discussed above, two redundant types of nicotinamide riboside/nicotinic acid riboside salvage have been demonstrated functionally in S. cerevisiae (3, 4).
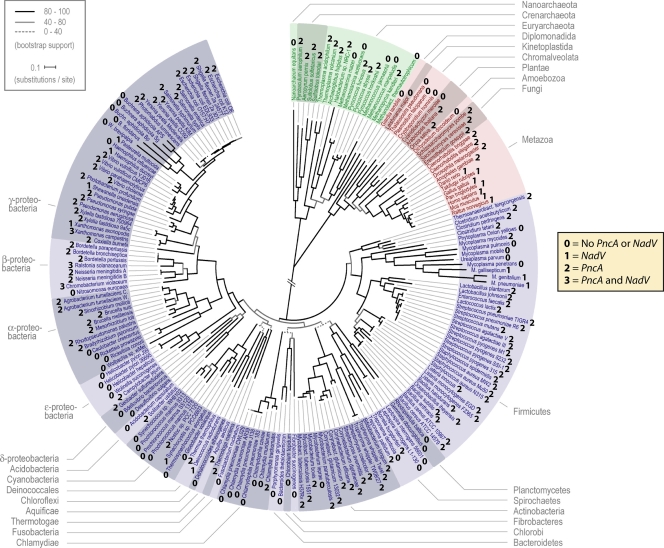
FIG. 6.
Nicotinamide metabolism throughout the tree of life. The tree of life calculated with universally conserved proteins (17) was annotated on the basis of possession of apparent pncA and nadV orthologs. For each fully sequenced genome, we scored the organism as carrying nadV (1), pncA (2), both genes (3), or neither gene (0). (Adapted from reference 17 with permission from AAAS.)
MICROBES LACKING DE NOVO NAD BIOSYNTHESIS
Whereas salvage pathways are nonessential in model organisms such as E. coli and S. cerevisiae, there are organisms that have evolved to depend entirely on salvage of NAD precursors from other cells.
Haemophilus influenzae, a Nicotinamide Riboside Auxotroph
Formerly called Pfeiffer's bacillus, Haemophilus influenzae is an opportunistic gram-negative pathogen causing otitis media and respiratory tract infection in humans, and it was the first free-living organism to have its genome sequenced and assembled (21). As is apparent from the genus name, culture of H. influenzae initially required blood, such that fractionation programs were used to identify the molecules for which the bacterium is auxotrophic. Two essential factors, termed X factor and V factor, were identified as essential for growth of most strains of H. influenzae on synthetic media. The X factor is heme, and the V factor, which cannot be replaced by amino acids, nicotinic acid, or nicotinamide, was discovered to be NAD, NMN, or nicotinamide riboside (26, 35, 57). The nadR gene, which is required for growth of H. influenzae on nicotinamide riboside, is a bifunctional nicotinamide riboside kinase/NMN adenylyltransferase (58). The nicotinamide riboside kinase domain of nadR is found in additional bacteria and is diagnostic for bacterial nicotinamide riboside utilization (33).
Candida glabrata, a Vitamin Auxotroph
Candida glabrata is an opportunistic yeast fungal pathogen and the second-leading cause of candidiasis in humans. In a remarkable study, C. glabrata was reported to be a nicotinic acid auxotroph because it lacks de novo biosynthetic genes and because nicotinic acid limitation derepresses expression of adhesin genes, thereby linking Sir2-dependent gene silencing to repression of a pathway important for urinary tract infection (18). Genes encoding pathways from nicotinamide riboside and nicotinamide were observed in the C. glabrata genome (2) and demonstrated to function in fungal growth in culture as well as in murine infection (36). Thus, like H. influenzae, the eukaryotic pathogen C. glabrata has a lifestyle requiring supply of specific NAD precursor vitamins from its environment.
HORIZONTAL GENE TRANSFERS OF BOTH TYPES OF NICOTINAMIDE SALVAGE
NAD consumption creates a requirement for de novo and/or salvage biosynthesis. Additionally, because NAD-consuming enzymes such as sirtuins are inhibited by nicotinamide (10), there may be a selection for evolution or acquisition of enzymes performing nicotinamide salvage. Nicotinamide salvage differs between vertebrates, fungi, and protostomes in that vertebrates carry a nadV-like nicotinamide phosphoribosyltransferase gene and no nicotinamidase gene, whereas fungi and protostomes typically carry a pncA-like nicotinamidase gene but no nicotinamide phosphoribosyltransferase gene. Bacteria usually carry one gene or the other and occasionally carry both.
To determine how these genes came to be present in extant species, we collected apparent nadV and pncA genes from public databases and estimated phylogenetic trees based upon the predicted protein sequences. The phylogenetic trees for these two nicotinamide-metabolizing enzymes are strikingly discordant with simple descent and are distinct from each other. Phylogenetic analyses suggest that nadV genes originated either in an isolated eubacterium or near the base of the vertebrate tree and that viruses and plasmids were vectors for multiple, relatively recent horizontal transfers of nicotinamide phosphoribosyltransferase genes among eubacteria. A Francisella tularensis-like nadV gene is branched with metazoan nicotinamide phosphoribosyltransferase genes, which are absent from the sequenced genomes of protists, protostomes, fungi, and plants. In contrast, the data suggest that nicotinamidase genes emerged relatively early in bacterial history and that that distinct bacterial pncA genes were transferred to archaea, plants, fungi, and protostomes.
To provide a global view of the distribution of nicotinamide salvage enzymes, we annotated an automatically constructed tree of life that was calculated with universally conserved proteins (17). Figure 6 depicts many examples in which nearest neighbors in cellular evolution utilize different genes for nicotinamide salvage.
Evidence for Virus- and Plasmid-Mediated Gene Transfers of nadV
From searches of GenBank (5) and the Sargasso Sea metagenomic translations (63), we identified apparently full-length nadV-homologous nicotinamide phosphoribosyltransferase sequences from 12 animals, 23 bacteria, and 5 viruses. We excluded sequences with more than 94% identity to another isolate and those with more similarity to nicotinic acid phosphoribosyltransferase sequences. Six nadV sequences from the Sargasso Sea database, which are represented in GenBank with known phylogenetic origins, were discarded. Two putative bacterial nadV sequences from the Sargasso Sea that were only 47% and 87% identical to a previously reported sequence were included in further phylogenetic analysis.
Beyond the two unknown Sargasso Sea samples, bacteria carrying an apparent nadV gene include 15 proteobacteria, 4 firmicutes, 1 deinococcus, 2 cyanobacteria, and 1 bacteriodete. Metazoa with a full-length nadV homolog include human, chimp, dog, rat, mouse, chicken, two frogs, three fish, and a sponge. Though protostome genomes, including those of Drosophila melanogaster and Caenorhabditis elegans, are complete, we found no apparent nadV ortholog in any invertebrate other than one attributed to the sponge, Suberites domuncula. The only fungal nadV-related sequences were genes encoding nicotinic acid phosphoribosyltransferase (15). Similarly, despite extensive genomic data from plants and protists, we found no evidence for nadV-orthologous sequences in the eukaryotic domain outside of metazoa. In the case of nicotinic acid phosphoribosyltransferase from Saccharomyces cerevisiae, physiological experiments have proven that this enzyme cannot utilize nicotinamide to support the growth of a glutamine-dependent NAD synthetase mutant, thereby establishing the in vivo substrate specificity of the enzyme (8). Thus, nadV homologs have an unusual phylogeny consisting of eubacteria, bacteriophage, sponge, and vertebrates.
Inspection of Fig. 6 indicates that NadV is infrequently encoded by the first 150 sequenced bacterial genomes. The gene is infrequently carried in beta- or gammaproteobacteria and was not observed in any of the first alpha-, delta-, or epsilonproteobacteria with sequenced genomes. nadV sequences were identified in one cyanobacterium, one deinococcus, and three mycobacteria. At the time of our analysis, nadV was absent from all archaeal genomes and absent from all sequenced eukaryotic genomes except those of vertebrates, in which it has been always found. These data provide an impression of late appearance and/or frequent elimination of nadV sequences, as the gene is rarely present in multiple members of any particular clade other than vertebrates, the xanthomonads Xanthomonas axonopodis and X. campestris, and the mycoplasmas Mycoplasma gallisepticum, M. genitalium, and M. pneumoniae.
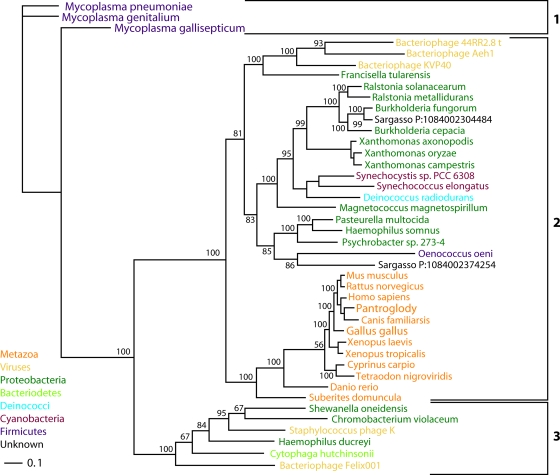
A nadV phylogenetic tree (Fig. 7) calculated by Bayesian estimation of protein phylogeny (28) has three primary protein clades. Three nadV sequences from Mycoplasma populate the first clade. Animal sequences are in a second clade with nadV sequences from cyanobacteria, deinococci, two Sargasso Sea isolates, and the majority of nadV-positive proteobacteria, including Francisella tularensis, Haemophilus somnus, and Ralstonia solanacearum, which carries nadV on a megaplasmid (54). The virally carried nadV genes from bacteriophage KVP40, which infects Vibrio (38), and bacteriophages Aeh1 and 44RR2.8t, which infect Aeromonas (16), are each other's nearest neighbors in the second clade. In the third protein clade are sequences from the bacteriodete Cytophaga hutchinsonii and three proteobacteria with sequences similar to that of the plasmid-carried nadV gene of Haemophilus ducreyi (37) and the virally carried nadV genes from bacteriophage Felix 01 and staphylococcus phage K (42). The plasmid, pNAD1, which carries H. ducreyi nadV and is integrated into some strains of H. ducreyi, has been shown to carry three additional open reading frames similar to those in cholera toxin phage and enterobacterial phage I2-2 (41).
FIG. 7.
nadV phylogeny. The nadV phylogenetic tree was calculated with Bayeseian analysis (28, 52) using the sequence from Mycoplasma pneumoniae as the outgroup. Protein clades 1 to 3 are indicated. Bayesian posterior probabilities are indicated for nodes. Branch lengths are proportional to evolutionary distance.
Protein clades 1 and 3 appear to have a common ancestor. However, our data are inconsistent with the speculation that the H. ducreyi nadV gene was horizontally transferred to Mycoplasma genitalium (37). Rather, it appears that a nadV gene with an ancestor common to protein clade 3 was transferred to Mycoplasma prior to the speciation of M. pneumoniae, M. genitalium, and M. gallisepticum. The plasmid-carried nadV gene of H. ducreyi (37) and the virally carried nadV genes from bacteriophage Felix 01 and staphylococcus phage K (42) are reasonable candidates for vectors transmitting nadV sequences within protein clade 3.
The H. ducreyi nadV sequence is not the only type of nadV gene found in Pasteurella. Instead, Pasteurella multocida and Haemophilus somnus have a protein clade 2 gene more similar to nadV sequences found in F. tularensis, Ralstonia solanacearum, and metazoans. Bacteriophages similar to KVP40, Aeh1, and 44RR2.8t may have had a role in transmitting nadV to F. tularensis. The available data are consistent with an F. tularensis-type nadV sequence having entered the metazoan lineage horizontally after the separation from protists, plants, and fungi. If such a sequence entered the ancient animal lineage only once, prior to the emergence of the sponge Suberites domuncula, then the gene was lost in the lineage leading to protostomes. Alternatively, two F. tularensis-type nadV sequences may have been transferred to animal lineages, with one event giving rise to the nadV-homologous gene of Suberites domuncula and one event giving rise to the nadV-homologous gene of vertebrates. A third possibility consistent with available data is that a metazoan, potentially near the base of the vertebrate tree, produced the first nicotinamide-specific phosphoribosyltransferase, which was transferred to eubacteria such as F. tularensis.
Horizontal Transfers of pncA in Three Kingdoms
Apparent moneran PncA peptide sequences were identified from genome annotations at http://www.theseed.org, where gene assignments make use of conservation of gene order and metabolic inferences (44). Apparent eukaryotic homologs of S. cerevisiae Pnc1 were obtained from public databases. Whereas it is relatively simple to determine by relative similarity and alignment that nadV genes are not pncB, i.e., encoding nicotinic acid phosphoribosyltransferase, it is relatively difficult to rely on sequence alone to discern authentic pncA genes from the related isochorismatase-encoding genes. Thus, caution must be exercised with sequences not yet validated by biochemical and/or genetic data. Inspection of Fig. 6 reveals that pncA genes are more common than nadV genes and are distributed more deeply in the tree of life. For example, pncA was found in the first 13 sequenced enterobacteria, embracing Photorhabdus, Yersinia, Salmonella, Escherichia, and Shigella, and was found in 13 of the first 14 sequenced actinobacteria. pncA was found in 8 of the first 12 sequenced alphaproteobacteria and in 7 of the first 8 sequenced betaproteobacteria, two of which were distinguished by carrying both pncA and nadV. Within the archaea, pncA was found in the first 12 of 18 sequenced genomes. There are plants, fungi, worms, and insects that carry pncA homologs, and all known vertebrate genomes are negative. (It should be noted, however, that despite the absence of vertebrate-encoded nicotinamidases, bacteria resident in the vertebrate gut are likely to provide nicotinamidase function for provision of nicotinic acid to nicotinic acid-utilizing tissues.)
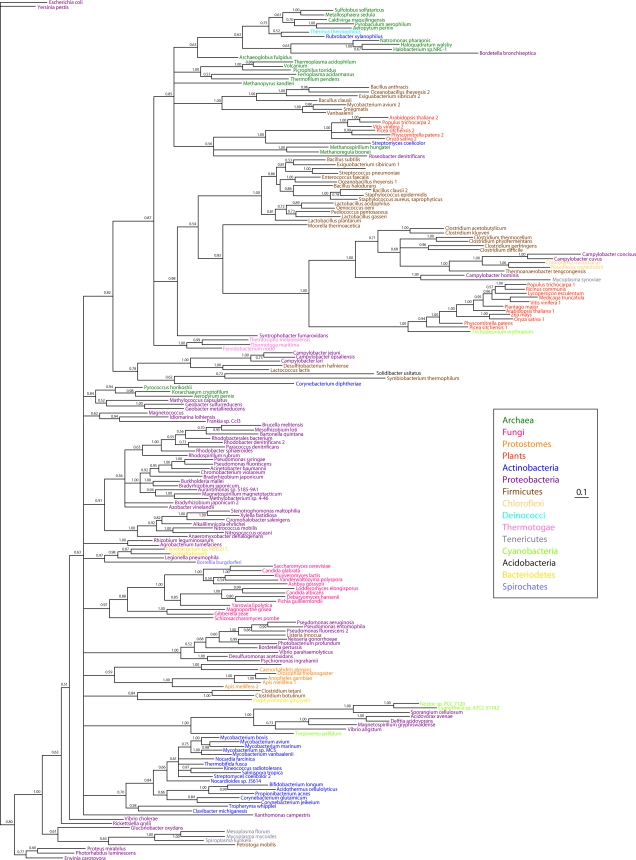
A pncA phylogenetic tree (Fig. 8) calculated by Bayesian estimation of protein phylogeny (28) contains some protein clades that are deeply rooted in proteobacterial, actinobacterial, fungal, and protostome phylogenies and other protein clades that strongly suggest horizontal gene transfer events. For example, the methanogenic archaeon Methanosprillia has a PncA sequence similar to that of Streptomyces, an actinobacterium, and to a set of plant nicotinamidases. However, the thermophilic archaeon Pyrococcus has a PncA sequence closely related to that of the proteobacterium Geobacter. Inspection of Fig. 8 suggests that PncA sequences may have been transferred to eukaryotic lineages multiple times.
FIG. 8.
pncA phylogeny. The pncA phylogenetic tree was calculated with Bayesian analysis (28, 52) using the sequence from Escherichia coli as the outgroup. Nearly identical phylogenetic tree topologies were obtained with other sequences as the outgroup. Bayesian posterior probabilities are indicated for nodes. Branch lengths are proportional to evolutionary distance.
COMPARATIVE GENOMICS UNDERSCORES THE RULES AND EXCEPTIONS IN NAD BIOSYNTHESIS
The Limitations of EC Numbers
Initially, NAD biosynthetic enzymes were discovered as enzyme activities and named accordingly. For example, in the Preiss-Handler pathway of nicotinic acid utilization, the nicotinic acid phosphoribosyltransferase, the NaMN adenylyltransferase, and the glutamine-dependent NAD synthetase were purified and characterized from yeast and from human blood (47). These enzymes were given EC numbers, EC 2.4.2.11, EC 2.7.7.18, and EC 6.3.5.1. Working independently, Arthur Kornberg purified and characterized NMN adenylyltransferase (31), which was given EC number EC 2.7.7.1. In yeast, the two “different” enzyme activities, i.e., NaMN adenylyltransferase and NMN adenylyltransferase, are both catalyzed by the same gene products, Nma1 and Nma2 (1, 20). In vertebrate systems, there are differences in the degrees of NaMN versus NMN specificity of three different gene products. More importantly, the genes are differentially regulated and differentially expressed, and their gene products are not expressed in the same cellular compartments (6, 59). Thus, there are many cases in which a single EC number corresponds to two or more genes and cases in which a single gene product corresponds to multiple EC numbers.
In the case of nicotinamide riboside metabolism, there is also a poor correspondence between EC number and gene product. EC 2.7.1.22 refers to nicotinamide riboside kinase, an enzyme that phosphorylates nicotinamide riboside to NMN. The sequence and structure responsible for this activity were identified as the metabolite kinase domain of NadR in H. influenzae, Salmonella enterica, and E. coli (33, 58). However, since eukaryotes have no NadR ortholog, a specific nicotinamide riboside kinase activity was not anticipated to exist in yeast or humans. Indeed, the yeast and human NRK enzymes (8) are structurally distinct from bacterial nicotinamide riboside kinases, though they are all members of the metabolite kinase superfamily (62).
We argue that more functional, structural, and mechanistic information can be gleaned from referring to “homologs of the metabolite kinase domain of Haemophilus influenzae NadR” or to “homologs of S. cerevisiae Nrk1” than to EC 2.7.1.22, which is a non-sequence-specific designation. Indeed, remarkable progress in predictive biochemical reconstruction of microbial NAD metabolism has been made, principally using the tools of sequence similarity and conservation of gene organization (23, 44, 50, 51). Recently, four new discoveries in microbial NAD metabolism have emerged from the examination of genome sequences and functional characterization.
De Novo Synthesis in Anaerobic Archaea
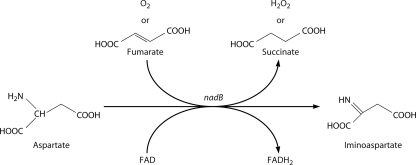
The de novo pathways of NAD synthesis are frequently termed aerobic because oxygen is an electron acceptor for l-aspartate oxidase, which catalyzes the first step in the canonical bacterial de novo pathway, and oxygen is consumed by indoleamine 2,3-dioxygenase in the first step of the canonical eukaryotic de novo pathway. However, despite being an anaerobic archaeon, Pyrococcus horikoshii encodes a homolog of E. coli l-aspartate oxidase (53). As shown in Fig. 9, it has been known for more than a decade that fumarate is an alternative electron acceptor for E. coli l-aspartate oxidase (61), which potentially explains how extant and ancient anaerobes make NAD de novo (53). An interesting implication of this work is that whereas an ancient cell presumably made NAD in five enzymatic steps from Asp without oxygen, l-aspartate oxidase requires flavin adenine dinucleotide (FAD) as a cofactor. This suggests that riboflavin synthesis, a process regulated by RNA (39, 66), may have predated NAD synthesis.
FIG. 9.
l-Aspartate oxidase can function without oxygen. The FAD-dependent reaction catalyzed by l-aspartate oxidase can utilize fumarate as the electron acceptor, thereby producing iminoaspartate, succinate, and reduced FAD under anaerobic conditions. The molecular oxygen-utilizing reaction produces iminoaspartate, hydrogen peroxide, and reduced FAD.
Salvage Synthesis in Mycobacterium tuberculosis
Tuberculosis remains the most lethal infectious pathogen and may latently infect one-third of all living people. NAD metabolic genes have been identified in M. tuberculosis, and the de novo genes were proposed to be essential for viability based on transposon studies (55). Though the pncA gene, encoding nicotinamidase, is required for sensitivity to pyrazinamide, a frontline treatment for tuberculosis (56), no one had ever demonstrated nicotinamide or nicotinic acid salvage in M. tuberculosis. The pncB gene, encoding nicotinic acid phosphoribosyltransferase, is duplicated, and deletion of the two homologous genes abolishes conversion of nicotinic acid to NAD (12). pncB2 expression is induced under hypoxic conditions and upon chronic infection of a mouse. Moreover, because a strain with a mutation in de novo biosynthesis is capable of replicating in mouse and killing its host, it is now clear that tuberculosis infection makes use of salvage synthesis (12).
Glutamine-Dependent and Glutamine-Independent NAD Synthetases
Jack Preiss first described eukaryotic NAD synthetase, the enzyme which converts NaAD to NAD through the adenylylated intermediate NaAD-AMP, as a glutamine-dependent enzyme. His work indicated that the enzyme activates the nicotinic acid moiety of NaAD by adenylylation and hydrolyzes Gln to Glu as a source of ammonia to generate NAD plus AMP (47). Years later, he found that E. coli NAD synthetase, now termed NadE, is an ammonia-dependent and Gln-independent enzyme (60). However, to assume that prokaryotic enzymes would be exclusively ammonia dependent would be incorrect. The NAD synthetase from M. tuberculosis is a Gln-dependent enzyme with an N-terminal extension (14) with respect to the sequence of E. coli nadE (65). We recognized that the N-terminal sequence is a nitrilase-related domain found in all eukaryotic NAD synthetases and in those prokaryotic NAD synthetases reported to be Gln dependent (45). True to prediction, the N-terminal domain is a thiol Gln amidotransferase domain that mediates transfer of ammonia from Gln to form the NAD product (9). According to Interpro database entry IPR014445, the nitrilase-related Gln amidotransferase domain of NAD synthetases has been found in over 500 eubacteria and in 3 archaea in addition to eukaryotes (40).
NMN Synthesis in Francisella tularensis
The causative agent of tularemia, or rabbit fever, F. tularensis is a gram-negative gammaproteobacterium whose genome sequence, reported in 2005 (34), seemed to have a puzzling omission of nadD. Eukaryotic NaMN/NMN adenyltransferases are bifunctional enzymes, which adenylylate NaMN to NaAD (Fig. 3A) and also adenylylate NMN to NAD (Fig. 4B). In contrast, eubacterial NaMN adenylyltransferases are nadD homologs with specific recognition of NaMN (43, 68), whereas specific NMN adenylyltransferases are nadR (49) or nadM homologs (27, 48) with specific recognition of NMN. Examination of the F. tularensis genome revealed the de novo module from Asp to NaMN depicted in Fig. 2A and the nadV and nadM genes, which would convert nicotinamide to NAD as depicted in Fig. 4A. Because F. tularensis can grow on synthetic media without nicotinamide, Andrei Osterman and coworkers hypothesized that the Asp to NaMN module is functional and that there must be a nadD-independent route from NaMN to NAD in this organism (59a). Peculiarly, however, F. tularensis has a homolog of nadE, which, in other organisms, amidates NaAD to NAD. Why would F. tularensis have nadE, encoding an apparent NaAD-requiring enzyme, if it does not have nadD, encoding the NaAD-producing enzyme? Reasoning that nadE would have been lost along with nadD if it did not function in NAD metabolism, the investigators tested two hypotheses. First, they examined whether F. tularensis nadM encodes a bifunctional NaMN/NMN adenylyltransferase more similar in activity to eukaryotic enzymes than to bacterial nadM homologs. Second, they tested the radical hypothesis that the F. tularensis nadE product amidates NaMN to NMN for subsequent adenylylation by the nadM product. Remarkably, the nadM product is simply NMN adenylyltransferase, whereas the F. tularensis nadE product has the novel activity of NaMN amidation (59a). Thus, F. tularensis apparently completes de novo NAD synthesis without the second and third steps of the Preiss-Handler pathway and instead follows the scheme shown in Fig. 3B.
CONCLUSIONS
In any rapidly evolving research area, it is dangerous to draw too many conclusions. However, three appear to be warranted. First, NAD metabolism is best described as a series of modules, none of which are universally conserved. Second, using homology and synteny-based annotation, strong predictions of the available pathways can be made based on genome sequences. A group led by Ross Overbeek has established “The SEED” (http://www.theseed.org), a web-accessible database of genome annotations which are expertly curated on a subsystem-by-subsystem basis (44). SEED predictions can be considered to be state-of-the-art guides to the NAD gene sets of organisms with sequenced genomes. Third, though recent progress has been made (23, 44, 50, 51), we consider that regulatory processes, which control the expression of particular gene modules and link NAD metabolism to dynamic cellular and ecological processes, are poorly understood. We predict that genomic information gathered in the last 15 years combined with molecular genetic methods developed in the last 30 years will be used to determine how NAD metabolic gene modules and enzymes are regulated to respond to and to drive microbiological functions.
Acknowledgments
This work was supported by grant MCB-0822581 from the National Science Foundation to C.B. F.G., R.S., and S.Z.C. were supported by Presidential Scholarships from Dartmouth College.
We are grateful for extensive discussions with Andrei Osterman and for Norman Oppenheimer's thoughtful review of the figures.
Biography
 Francesca Gazzaniga earned her bachelor's degree at Dartmouth College, where she worked on experimental and computational aspects of NAD metabolism with Charles Brenner. She is currently a second-year graduate student with Elizabeth Blackburn at the University of California, San Francisco, working on telomerase.
Francesca Gazzaniga earned her bachelor's degree at Dartmouth College, where she worked on experimental and computational aspects of NAD metabolism with Charles Brenner. She is currently a second-year graduate student with Elizabeth Blackburn at the University of California, San Francisco, working on telomerase.
Rebecca Stebbins and Sheila Z. Chang are Dartmouth undergraduates bound for graduate school and medical school, respectively.
 Mark A. McPeek earned his bachelor's and master's degrees at the University of Kentucky and his Ph.D. at Michigan State University. He was a postdoctoral fellow at the Archbold Biological Station, Lake Placid, FL. His first academic position was at Bowling Green State University before he moved to Dartmouth, where he is currently Professor of Biological Sciences. His research interests center on understanding the diversification and adaptation of species and their resulting interactions that shape biological communities.
Mark A. McPeek earned his bachelor's and master's degrees at the University of Kentucky and his Ph.D. at Michigan State University. He was a postdoctoral fellow at the Archbold Biological Station, Lake Placid, FL. His first academic position was at Bowling Green State University before he moved to Dartmouth, where he is currently Professor of Biological Sciences. His research interests center on understanding the diversification and adaptation of species and their resulting interactions that shape biological communities.
 Charles Brenner earned his bachelor's degree at Wesleyan University and his Ph.D. at Stanford University. He was a postdoctoral fellow at Brandeis University and held academic positions at Thomas Jefferson University and Dartmouth before moving to the University of Iowa to take the position of Head of the Department of Biochemistry. His research centers on understanding the function of tumor suppressor genes and on the dissection of NAD metabolism as it relates to cellular function and aging.
Charles Brenner earned his bachelor's degree at Wesleyan University and his Ph.D. at Stanford University. He was a postdoctoral fellow at Brandeis University and held academic positions at Thomas Jefferson University and Dartmouth before moving to the University of Iowa to take the position of Head of the Department of Biochemistry. His research centers on understanding the function of tumor suppressor genes and on the dissection of NAD metabolism as it relates to cellular function and aging.
REFERENCES
- 1.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, H. Cohen, S. S. Lin, J. K. Manchester, J. I. Gordon, and D. A. Sinclair. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 27718881-18890. [DOI] [PubMed] [Google Scholar]
- 2.Belenky, P., K. L. Bogan, and C. Brenner. 2007. NAD(+) metabolism in health and disease. Trends Biochem. Sci. 3212-19. [DOI] [PubMed] [Google Scholar]
- 3.Belenky, P., K. C. Christensen, F. S. Gazzaniga, A. Pletnev, and C. Brenner. 2009. Nicotinamide riboside and nicotinic acid riboside salvage in fungi and mammals: Quantitative basis for Urh1 and purine nucleoside phosphorylase function in NAD+ metabolism. J. Biol. Chem. 284158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belenky, P., F. G. Racette, K. L. Bogan, J. M. McClure, J. S. Smith, and C. Brenner. 2007. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD(+). Cell 129473-484. [DOI] [PubMed] [Google Scholar]
- 5.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2005. GenBank. Nucleic Acids Res. 33D34-D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, F., C. Lau, M. Dahlmann, and M. Ziegler. 2005. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 28036334-36341. [DOI] [PubMed] [Google Scholar]
- 7.Berger, F., M. H. Ramirez-Hernandez, and M. Ziegler. 2004. The new life of a centenarian: signalling functions of NAD(P). Trends Biochem. Sci. 29111-118. [DOI] [PubMed] [Google Scholar]
- 8.Bieganowski, P., and C. Brenner. 2004. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117495-502. [DOI] [PubMed] [Google Scholar]
- 9.Bieganowski, P., H. C. Pace, and C. Brenner. 2003. Eukaryotic NAD+ synthetase Qns1 contains an essential, obligate intramolecular thiol glutamine amidotransferase domain related to nitrilase. J. Biol. Chem. 27833049-33055. [DOI] [PubMed] [Google Scholar]
- 10.Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J. Biol. Chem. 27745099-45107. [DOI] [PubMed] [Google Scholar]
- 11.Bogan, K. L., and C. Brenner. 2008. Nicotinic acid, nicotinamide and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28115-130. [DOI] [PubMed] [Google Scholar]
- 12.Boshoff, H. I., X. Xu, K. Tahlan, C. S. Dowd, K. Pethe, L. R. Camacho, T. H. Park, C. S. Yun, D. Schnappinger, S. Ehrt, K. J. Williams, and C. E. Barry, 3rd. 2008. Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J. Biol. Chem. 28319329-19341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner, C. 2005. Evolution of NAD Biosynthetic Enzymes. Structure (Cambridge) 131239-1240. [DOI] [PubMed] [Google Scholar]
- 14.Cantoni, R., M. Branzoni, M. Labo, M. Rizzi, and G. Riccardi. 1998. The MTCY428.08 gene of Mycobacterium tuberculosis codes for NAD+ synthetase. J. Bacteriol. 1803218-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chappie, J. S., J. M. Canaves, G. W. Han, C. L. Rife, Q. Xu, and R. C. Stevens. 2005. The structure of a eukaryotic nicotinic acid phosphoribosyltransferase reveals structural heterogeneity among type II PRTases. Structure 131385-1396. [DOI] [PubMed] [Google Scholar]
- 16.Chow, M. S., and M. A. Rouf. 1983. Isolation and partial characterization of two Aeromonas hydrophila bacteriophages. Appl. Environ. Microbiol. 451670-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 3111283-1287. [DOI] [PubMed] [Google Scholar]
- 18.Domergue, R., I. Castano, A. De Las Penas, M. Zupancic, V. Lockatell, R. J. Hebel, D. Johnson, and B. P. Cormack. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308866-870. [DOI] [PubMed] [Google Scholar]
- 19.Elvehjem, C. A., R. J. Madden, F. M. Strong, and D. W. Woolley. 1938. The isolation and identification of the anti-black tongue factor. J. Biol. Chem. 123137-149. [PubMed] [Google Scholar]
- 20.Emanuelli, M., F. Carnevali, M. Lorenzi, N. Raffaelli, A. Amici, S. Ruggieri, and G. Magni. 1999. Identification and characterization of YLR328W, the Saccharomyces cerevisiae structural gene encoding NMN adenylyltransferase. Expression and characterization of the recombinant enzyme. FEBS Lett. 45513-17. [DOI] [PubMed] [Google Scholar]
- 21.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269496-512. [DOI] [PubMed] [Google Scholar]
- 22.Frye, R. A. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273793-798. [DOI] [PubMed] [Google Scholar]
- 23.Gerdes, S. Y., O. V. Kurnasov, K. Shatalin, B. Polanuyer, R. Sloutsky, V. Vonstein, R. Overbeek, and A. L. Osterman. 2006. Comparative genomics of NAD biosynthesis in cyanobacteria. J. Bacteriol. 1883012-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghislain, M., E. Talla, and J. M. Francois. 2002. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 19215-224. [DOI] [PubMed] [Google Scholar]
- 25.Gill, D. M., A. M. Pappenheimer, Jr., R. Brown, and J. T. Kurnick. 1969. Studies on the mode of action of diphtheria toxin. VII. Toxin-stimulated hydrolysis of nicotinamide adenine dinucleotide in mammalian cell extracts. J. Exp. Med. 1291-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingrich, W., and F. Schlenk. 1944. Codehydrogenase I and other pyridinium compounds as V factor for Haemophilus influenzae and Haemophilus parainfluenzae. J. Bacteriol. 47535-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, N., L. Sorci, X. Zhang, C. A. Brautigam, X. Li, N. Raffaelli, G. Magni, N. V. Grishin, A. L. Osterman, and H. Zhang. 2008. Bifunctional NMN adenylyltransferase/ADP-ribose pyrophosphatase: structure and function in bacterial NAD metabolism. Structure 16196-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huelsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bollback. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2942310-2314. [DOI] [PubMed] [Google Scholar]
- 29.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403795-800. [DOI] [PubMed] [Google Scholar]
- 30.Katoh, A., K. Uenohara, M. Akita, and T. Hashimoto. 2006. Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol. 141851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornberg, A. 1950. Reversible enzymatic synthesis of diphosphopyridine nucleotide and inorganic pyrophosphate. J. Biol. Chem. 182779-793. [PubMed] [Google Scholar]
- 32.Kurnasov, O., V. Goral, K. Colabroy, S. Gerdes, S. Anantha, A. Osterman, and T. P. Begley. 2003. NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 101195-1204. [DOI] [PubMed] [Google Scholar]
- 33.Kurnasov, O. V., B. M. Polanuyer, S. Ananta, R. Sloutsky, A. Tam, S. Y. Gerdes, and A. L. Osterman. 2002. Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J. Bacteriol. 1846906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37153-159. [DOI] [PubMed] [Google Scholar]
- 35.Leder, I. G., and P. Handler. 1951. Synthesis of nicotinamide mononucleotide by human erythrocytes in vitro. J. Biological Chemistry. 189889-899. [PubMed] [Google Scholar]
- 36.Ma, B., S. J. Pan, M. L. Zupancic, and B. P. Cormack. 2007. Assimilation of NAD(+) precursors in Candida glabrata. Mol. Microbiol. 6614-25. [DOI] [PubMed] [Google Scholar]
- 37.Martin, P. R., R. J. Shea, and M. H. Mulks. 2001. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 1831168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 1855220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mironov, A. S., I. Gusarov, R. Rafikov, L. E. Lopez, K. Shatalin, R. A. Kreneva, D. A. Perumov, and E. Nudler. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111747-756. [DOI] [PubMed] [Google Scholar]
- 40.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bork, V. Buillard, L. Cerutti, R. Copley, E. Courcelle, U. Das, L. Daugherty, M. Dibley, R. Finn, W. Fleischmann, J. Gough, D. Haft, N. Hulo, S. Hunter, D. Kahn, A. Kanapin, A. Kejariwal, A. Labarga, P. S. Langendijk-Genevaux, D. Lonsdale, R. Lopez, I. Letunic, M. Madera, J. Maslen, C. McAnulla, J. McDowall, J. Mistry, A. Mitchell, A. N. Nikolskaya, S. Orchard, C. Orengo, R. Petryszak, J. D. Selengut, C. J. Sigrist, P. D. Thomas, F. Valentin, D. Wilson, C. H. Wu, and C. Yeats. 2007. New developments in the InterPro database. Nucleic Acids Res. 35D224-D228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munson, R. S., Jr., H. Zhong, R. Mungur, W. C. Ray, R. J. Shea, G. G. Mahairas, and M. H. Mulks. 2004. Haemophilus ducreyi strain ATCC 27722 contains a genetic element with homology to the Vibrio RS1 element that can replicate as a plasmid and confer NAD independence on Haemophilus influenzae. Infect. Immun. 721143-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Flaherty, S., A. Coffey, R. Edwards, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 1862862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olland, A. M., K. W. Underwood, R. M. Czerwinski, M. C. Lo, A. Aulabaugh, J. Bard, M. L. Stahl, W. S. Somers, F. X. Sullivan, and R. Chopra. 2002. Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase. J. Biol. Chem. 2773698-3707. [DOI] [PubMed] [Google Scholar]
- 44.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 335691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pace, H. C., and C. Brenner. 2001. The nitrilase superfamily: classification, structure and function. Genome Biol. 2reviews0001.1-0001.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preiss, J., and P. Handler. 1958. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J. Biol. Chem. 233488-492. [PubMed] [Google Scholar]
- 47.Preiss, J., and P. Handler. 1958. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J. Biol. Chem. 233493-500. [PubMed] [Google Scholar]
- 48.Raffaelli, N., T. Lorenzi, A. Amici, M. Emanuelli, S. Ruggieri, and G. Magni. 1999. Synechocystis sp. slr0787 protein is a novel bifunctional enzyme endowed with both nicotinamide mononucleotide adenylyltransferase and ‘Nudix’ hydrolase activities. FEBS Lett. 444222-226. [DOI] [PubMed] [Google Scholar]
- 49.Raffaelli, N., T. Lorenzi, P. L. Mariani, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1999. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 1815509-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodionov, D. A., J. De Ingeniis, C. Mancini, F. Cimadamore, H. Zhang, A. L. Osterman, and N. Raffaelli. 2008. Transcriptional regulation of NAD metabolism in bacteria: NrtR family of Nudix-related regulators. Nucleic Acids Res. 362047-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodionov, D. A., X. Li, I. A. Rodionova, C. Yang, L. Sorci, E. Dervyn, D. Martynowski, H. Zhang, M. S. Gelfand, and A. L. Osterman. 2008. Transcriptional regulation of NAD metabolism in bacteria: genomic reconstruction of NiaR (YrxA) regulon. Nucleic Acids Res. 362032-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 191572-1574. [DOI] [PubMed] [Google Scholar]
- 53.Sakuraba, H., T. Satomura, R. Kawakami, S. Yamamoto, Y. Kawarabayasi, H. Kikuchi, and T. Ohshima. 2002. l-Aspartate oxidase is present in the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii OT-3: characteristics and role in the de novo biosynthesis of nicotinamide adenine dinucleotide proposed by genome sequencing. Extremophiles 6275-281. [DOI] [PubMed] [Google Scholar]
- 54.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415497-502. [DOI] [PubMed] [Google Scholar]
- 55.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 4877-84. [DOI] [PubMed] [Google Scholar]
- 56.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2662-667. [DOI] [PubMed] [Google Scholar]
- 57.Shifrine, M., and E. L. Biberstein. 1960. A growth factor for Haemophilus species secreted by a Pseudomonad. Nature 187623. [Google Scholar]
- 58.Singh, S. K., O. V. Kurnasov, B. Chen, H. Robinson, N. V. Grishin, A. L. Osterman, and H. Zhang. 2002. Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities. J. Biol. Chem. 27733291-33299. [DOI] [PubMed] [Google Scholar]
- 59.Sorci, L., F. Cimadamore, S. Scotti, R. Petrelli, L. Cappellacci, P. Franchetti, G. Orsomando, and G. Magni. 2007. Initial-rate kinetics of human NMN-adenylyltransferases: substrate and metal ion specificity, inhibition by products and multisubstrate analogues, and isozyme contributions to NAD(+) biosynthesis. Biochemistry 464912-4922. [DOI] [PubMed] [Google Scholar]
- 59a.Sorci, L., D. Martynowski, D. A. Rodionov, Y. Eyobo, X. Zogaj, K. E. Klose, E. V. Nikolaev, G. Magni, H. Zhang, and A. L. Osterman. 2009. Nicotinamide mononucleotide synthetase is the key enzyme for an alternative route of NAD biosynthesis in Francisella tularensis. Proc. Natl. Acad. Sci. USA 1063083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spencer, R. L., and J. Preiss. 1967. Biosynthesis of diphosphopyridine nucleotide. The purification and the properties of diphospyridine nucleotide synthetase from Escherichia coli b. J. Biol. Chem. 242385-392. [PubMed] [Google Scholar]
- 61.Tedeschi, G., A. Negri, M. Mortarino, F. Ceciliani, T. Simonic, L. Faotto, and S. Ronchi. 1996. l-Aspartate oxidase from Escherichia coli. II. Interaction with C4 dicarboxylic acids and identification of a novel l-aspartate:fumarate oxidoreductase activity. Eur. J. Biochem. 239427-433. [DOI] [PubMed] [Google Scholar]
- 62.Tempel, W., W. M. Rabeh, K. L. Bogan, P. Belenky, M. Wojcik, H. F. Seidle, L. Nedyalkova, T. Yang, A. A. Sauve, H. W. Park, and C. Brenner. 2007. Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol. 5e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 30466-74. [DOI] [PubMed] [Google Scholar]
- 64.Wilkinson, A., J. Day, and R. Bowater. 2001. Bacterial DNA ligases. Mol. Microbiol. 401241-1248. [DOI] [PubMed] [Google Scholar]
- 65.Willison, J. C., and G. Tissot. 1994. The Escherichia coli efg gene and the Rhodobacter capsulatus adgA gene code for NH3-dependent NAD synthetase. J. Bacteriol. 1763400-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkler, W. C., S. Cohen-Chalamish, and R. R. Breaker. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 9915908-15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, Z., A. Savchenko, A. Yakunin, R. Zhang, A. Edwards, C. Arrowsmith, and L. Tong. 2003. Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643. J. Biol. Chem. 2788804-8808. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, H., T. Zhou, O. Kurnasov, S. Cheek, N. V. Grishin, and A. Osterman. 2002. Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD. Structure 1069-79. [DOI] [PubMed] [Google Scholar]