Abstract
Summary: Heme is the prosthetic group for cytochromes, which are directly involved in oxidation/reduction reactions inside and outside the cell. Many cytochromes contain heme with covalent additions at one or both vinyl groups. These include farnesylation at one vinyl in hemes o and a and thioether linkages to each vinyl in cytochrome c (at CXXCH of the protein). Here we review the mechanisms for these covalent attachments, with emphasis on the three unique cytochrome c assembly pathways called systems I, II, and III. All proteins in system I (called Ccm proteins) and system II (Ccs proteins) are integral membrane proteins. Recent biochemical analyses suggest mechanisms for heme channeling to the outside, heme-iron redox control, and attachment to the CXXCH. For system II, the CcsB and CcsA proteins form a cytochrome c synthetase complex which specifically channels heme to an external heme binding domain; in this conserved tryptophan-rich “WWD domain” (in CcsA), the heme is maintained in the reduced state by two external histidines and then ligated to the CXXCH motif. In system I, a two-step process is described. Step 1 is the CcmABCD-mediated synthesis and release of oxidized holoCcmE (heme in the Fe+3 state). We describe how external histidines in CcmC are involved in heme attachment to CcmE, and the chemical mechanism to form oxidized holoCcmE is discussed. Step 2 includes the CcmFH-mediated reduction (to Fe+2) of holoCcmE and ligation of the heme to CXXCH. The evolutionary and ecological advantages for each system are discussed with respect to iron limitation and oxidizing environments.
INTRODUCTION
Heme is protoporphyrin IX with iron (Fig. 1). It is also sometimes called protoheme IX or heme b when it is associated as the prosthetic group of b-type cytochromes. A comprehensive review on the enzymes involved in heme biosynthesis and their regulation has been recently published (109). The present review covers covalent additions to the porphyrin of the heme molecule, with emphasis on heme c (cytochrome c assembly) and emerging concepts. In all cases discussed here, the natural modifications to heme occur after the ferrous iron (Fe2+) is inserted into the porphyrin by ferrochelatase, the last enzyme in heme biosynthesis. The site of action of ferrochelatase is inside the prokaryotic cell (probably at the cytoplasmic membrane) and similarly in the mitochondrial matrix or chloroplast stroma of eukaryotes. Hence, the mechanisms defining modifications must take into account how the heme travels from this location to where it is modified, what the oxidation state of the heme iron is along the way, and what accessory factors might be involved.
FIG. 1.
Types of heme (b, o, a, and c), modifications and genes/enzymes required for modification, as discussed in the text. The Fischer numbering system was used.
Heme has been used as a cofactor for over 3 billion years, and the modifications to make various types, such as c heme in a cytochrome c, have probably existed for almost as long. Consequently, these ancient molecules have stood the test of time. A cytochrome c has at least one thioether bond formed between a cysteine of the protein and a vinyl group of the heme. Almost always, two cysteines in a CXXCH “heme binding motif” of the protein are linked by two thioether bonds to the two vinyl groups of heme, where the histidine acts as one axial ligand to the heme iron (Fig. 1). In general, it is thought that such covalent linkages provide stability to the cytochrome, making them much less likely to lose their heme or denature (9). Stability might be particularly important to cytochromes which function outside of the inner membrane, where all cytochromes c are found. While the mitochondrial cytochrome c, containing a single heme group, has received the lion's share of attention in the last century, it has become clear that in the prokaryotic world there are hundreds of cytochromes c with structures and functions unlike those of mitochondrial cytochrome c (128). Some of these may have more than 10 heme groups (each attached to a distinct CXXCH motif) in a single polypeptide. Moreover, they function in diverse and sometimes extraordinary electron transport chains that have evolved in an untold number of prokaryotic species. Many species have the classic aerobic respiratory chain from which the mitochondrion is derived, but countless cytochromes c are involved in photosynthesis or reduction of terminal electron acceptors such as nitrates, dimethyl sulfoxide, metals, and more (128). These oxidation/reduction reactions have a profound impact on the planet's environment, including all of the earth's nutrient cycles.
The typical purpose of these diverse electron transport chains is the generation of a proton gradient across the inner membrane and thus energy conversion. This gradient is in turn used to make ATP, for motility, for transport, and for other processes common to all life. It is the respiratory diversity that has, in part, made the prokaryotic world so successful in inhabiting nearly every microenvironment on earth. For example, in Shewanella species, “nanowires” assembled with different multiheme cytochromes c shuttle electrons from organic substrates inside the bacterium to metal precipitates such as iron oxides or uranium outside (67, 143). In so doing, they often solubilize the metal. This molecular nanowire involves four different c-type cytochromes, three each with 10 c-type hemes (MtrA, MtrC, and OmcA) and one with 4 hemes (CymA) (143). Understanding the mechanisms by which organisms attach heme to each CXXCH motif in all cytochromes c is the major topic of this review. Of the three pathways to assemble a cytochrome c (i.e., to covalently attach the heme), prokaryotes use the pathways called systems I and II. System I is comprised of eight proteins (CcmABCDEFGH) and system II of minimally two proteins (CcsBA) that are dedicated strictly for cytochrome c biogenesis (Fig. 2). All are integral inner membrane proteins.
FIG. 2.
Working models for cytochrome c biogenesis by systems I, II, and III. Models include trafficking and oxidation states of heme, as well as the subpathways for apocytochrome reduction (in red for system I and system II). Representative genera possessing each system are listed under the models.
Here we bring in old and new ideas on mechanisms of cytochrome c assembly via analogies to other heme modification mechanisms. We propose future challenges in fully understanding the molecular mechanisms and evolutionary underpinnings behind the biosynthetic pathways. One of the goals of this review is to move from the traditional “protein blob” models to a membrane topological and structure/function level in describing the biosynthetic pathways. We believe that experimental topological data and recent biochemical studies of the systems facilitate this level of understanding.
HEME TYPES AND MODIFICATIONS
Experimentalists over the last century have characterized a handful of major types of heme molecules with various natural modifications (Fig. 1). These hemes were initially discovered based on their spectral characteristics and later characterized based on their unique heme chemistries that yield the distinct spectral absorptions. Over the last 2 decades, the biosynthetic mechanisms behind these heme modifications have been at the forefront (reviewed in, e.g., references 84, 115, and 154). Because there are some similarities among the chemical mechanisms and positions in the porphyrins for certain modifications, it is instructive to review the prominent types of hemes (Fig. 1).
Except for heme d, which appears to have bireductive hydroxylations at carbons 5 and 6 of the porphyrin rings, other noteworthy modifications to heme involve additions to one or both vinyl groups at the porphyrin carbons designated 2 and/or 4 (Fig. 1). The modifications can range from a single change catalyzed by a single enzyme to a complicated pathway involving many protein accessory factors. In the former, hemes o and a both have a farnesyl addition to vinyl group 2, catalyzed by the protoheme IX farnesyltransferase enzyme, also called heme o synthase. Heme o synthase is encoded by the Escherichia coli cyoE, the Bacillus ctaB, and the yeast COX10 genes, all of which are related (133, 156). Additionally, in heme a, a methyl group at carbon 8 is oxidized to an aldehyde. Heme o is the substrate for this modification, which is catalyzed by a heme-containing monooxygenase (or peroxidase) enzyme called heme a synthase, encoded by the Bacillus ctaA and the related yeast COX11 genes (31, 32, 104, 149-151). In vivo, the heme o and a synthases may form a complex for substrate channeling (30). Note that E. coli has only cyoE, and thus one of its two terminal oxidases is a cytochrome bo copper oxidase, which is related to the more publicized cytochrome aa3 copper oxidase (35, 36, 119). The other E. coli oxidase is the unrelated noncopper cytochrome bd (69).
The enzymes for modifications in heme o and heme a (Fig. 1) show some parallels to certain features of cytochrome c synthesis that are worth mentioning. For example, in vitro addition of the farnesyl pyrophosphate substrate to heme by the cyoE-encoded heme o synthase (in vesicles) absolutely required heme-iron reduction by sodium dithionite (135). In that report, Saiki et al. stated, “The resulting polyprenyl cation undergoes a nucleophilic attack by the vinyl group of ferrous protoheme IX, thus succeeding to a transfer of the farnesyl group to position 2 (beta) of the 2-vinyl group of ferrous protoheme IX with concomitant addition of a hydroxyl group to position 1. Since the electron density at the 2-position of the vinyl group is assumed to be higher in ferrous protoheme IX than in its ferric state, the former must be the real substrate for farnesylation.” Thus, as in cytochrome c synthesis, the heme iron must be in the reduced state so that the vinyl groups are chemically active for ligations (see below). Although the model described by Saiki et al. suggests that the resulting heme o product maintains iron in the reduced state after farnesylation, this was not tested because of the absolute requirement for dithionite in the in vitro reaction. CyoE was predicted and experimentally demonstrated to have seven transmembrane domains (TMDs) (34-36). Most of the highly conserved residues are on the cytoplasmic side, where one would predict that reduced heme b enters for farnesylation. CyoE also contains a conserved binding site located in a cytoplasmic loop for polyprenyl diphosphate. Saiki et al. showed the functional importance of key residues in CyoE at the cytoplasmic surface of the inner membrane, including residues that may be axial ligands to the heme iron, such as histidine and tyrosine (134).
The aforementioned heme a synthase (CtaA) is predicted to contain eight TMDs, interestingly, with five conserved histidines that are located on the periplasmic side of the membrane; these, in theory, are for coordinating two hemes that were shown to be present (76). One is a b heme that transfers electrons in the oxidative reaction, and the other is the heme o substrate (32, 149-151). Since the conserved histidines are at the outer surface of the membrane, it is suggested that the heme b substrate for CyoE (CtaB) and the heme o substrate for CtaA may be located on opposite sides of the inner membrane. As discussed below, certain analogies to CcmF in the system I cytochrome c assembly can be made for the CtaA heme a synthase.
Heme c synthesis requires involvement of the apocytochrome, that is, attachment of the CXXCH motif to the heme, so the terms “synthesis of (holo)cytochrome c” and “synthesis of heme c” are necessarily the same. Cytochrome c synthesis is by far the most complex heme modification, with three different systems having evolved to accomplish the same ligations (84, 115) (see below). The cysteine residues of the CXXCH motif must be reduced to form the thioether bonds to the alpha carbons of the heme vinyl groups. Some recent reviews on cytochrome c synthesis have included thorough discussions of the thiol reduction components (58, 157), and so the present review will only briefly describe these components. One aspect that makes cytochrome c synthesis even more phenomenal is that the polypeptide folds around the heme only after ligation, unlike the case for hemoglobins and other cytochromes, where protein folding occurs prior to heme binding. It is well documented that most apocytochromes c are degraded in vivo if the heme is not attached, yet apoglobins and apocytochromes b fold properly without heme and can be purified and reconstituted with heme in vitro (see reference 18 for a discussion).
Reactions of protein side chains to the heme vinyls have been studied in other contexts with various approaches, and the particular reactivities of the vinyl groups have been elegantly addressed, as have the ramifications of modifications (78). For instance, Metcalfe et al. studied ascorbate peroxidase, which was mutated to form a spontaneous covalent bond to the 2-vinyl group (100). Interestingly, if the amino acid residue adjacent to the heme vinyl was changed to a cysteine, the covalent adduct required iron(II) (Fe2+). If the residue was changed to a methionine, a covalent adduct required iron(III) (Fe3+) for formation, and an oxidative H2O2-catalyzed reaction was proposed. Similarly, Barker and colleagues (19, 20) and Allen et al. (4) have studied cytochrome b562 and b5, changing the amino acid residues adjacent to the vinyls (predicted from the three-dimensional structures) to cysteine to study spontaneous thioether bond formation. Again, the reactions require a reduced heme, using dithionite. Daltrop et al. have analyzed a unique cytochrome c from a thermophile, which, unlike most cytochromes c, folds properly even when the heme is not attached (39-41). They have made some important observations concerning the spontaneous nature of the thioether ligations, which again occur only when the iron is reduced (to Fe2+). A very recent study using an exceptional hemoglobin that normally forms a covalent histidine linkage to the 2-vinyl, has shown that the fifth and sixth axial ligands to the iron can also play a key role in the type of ligation, even changing preference to 4-vinyl over the 2-vinyl (108). Clearly, nothing can be left to chance in the synthesis of hemes c, o, and a, and orchestrated modifications with precise mechanisms are the rule.
CYTOCHROME c BIOGENESIS: THREE SYSTEMS AND SOME EMERGING, SPECIALIZED ONES
The three systems for cytochrome c synthesis are summarized in Fig. 2, as are representative genera that possess each system. For systems I and II, there is a good understanding of how the CXXCH cysteines are reduced, using a general transmembrane thioredox protein such as DsbD (101) (or the smaller, related CcdA [43, 139]) and dedicated thioredoxins such as CcsX (23) or CcmG (24). Figure 2 diagrams the thiol proteins (in red) which transfer reducing equivalents from inside the cell to outside. Since some aspects of assembly have been reviewed in the last 5 years (5, 9, 12, 58, 70, 157), here we focus on selected emerging concepts. Because all proteins in systems I and II are integral membrane proteins, descriptions will be presented in the context of the two-dimensional topologies of the integral membrane proteins in the pathways. Special emphasis will be placed on the completely conserved residues in this framework. For example, early studies of system I and II noted that three proteins possessed a highly conserved region called the “WWD domain,” due to the completely conserved tryptophans and an aspartate (WXWD) (25). It was proposed that this domain, in key proteins in the pathways (CcmC, CcmF, and CcsA), bound the heme that was presented to the apocytochrome c for ligation. In each protein, the WWD domain is flanked by conserved histidine residues originally predicted to ligand the heme iron and later shown to face the periplasm. The authors of a recent review on the evolution of the three members of this superfamily termed this the “heme-handling protein” family (94). The present review will spotlight the heme molecule as it moves through the systems, including the oxidation-reduction state, and it will include the very recent biochemical and spectroscopic evidence that these proteins do indeed handle heme, using the external histidines as axial ligands (59, 125). Our premise is that the WWD domain in each of the three families positions the heme molecule with the two vinyl groups surface exposed for orchestrated reactivity with the interacting partners. We next describe each of the three systems. For didactic purposes, we begin with the simplest, system III, and end with the most complicated pathway, system I. Some lessons learned about system II are also instructive for understanding system I.
System III: CCHL
In 1987, Dumont et al. reported the first identification of the gene encoding the enzyme that attaches heme to the two isoforms of yeast cytochrome c (49). This gene was able to complement the cytochrome c deficiency of the Saccharomyces cerevisiae CYC3 mutant, and it encoded a polypeptide with 293 amino acid residues. Designated the cytochrome c heme lyase (CCHL), it has since been shown to be related to the CCHLs of humans and some other eukaryotes (Fig. 2). CCHL has been recently reviewed (70), so here we provide only an overview.
A CCHL expressed with its cognate cytochrome c in the E. coli cytoplasm is all that is necessary to synthesize recombinant holocytochrome c, proving the enzyme's cytochrome c synthetase activity (see, e.g., reference 118). [A note on enzyme nomenclature: currently it is common to use the term (holo)cytochrome c synthetase (or synthase) for the enzyme that directly catalyzes the heme attachment to apocytochrome c. As discussed above, the term CCHL was originally used in the yeast mitochondrial studies (see, e.g., reference 49). In this review we retain the use of CCHL for system III for historic and descriptive purposes. Clearly the CCHL is a cytochrome c synthetase.] In Fig. 2 we color code orange the proteins in each system which have been designated with the cytochrome c synthetase activity. Each cytochrome c synthetase must bind heme and the apocytochrome c substrates. In some organisms, such as yeast, there are two CCHLs, one for cytochrome c (CCHL) and a related one for cytochrome c1 (CC1HL) (162). In other organisms, such as humans, a single CCHL recognizes and synthesizes both cytochromes c and c1 (26). Thus, there appears to be more apocytochrome c substrate specificity than just the CXXCH motif for system III; systems I and II typically recognize only the CXXCH (see below). There must be features other than CXXCH within the cognate apocytochromes that the CCHL enzymes recognize. Defining the apocytochrome c substrate features and the CCHL recognition determinants remain important aims. Surprisingly, even though the CCHLs are soluble enzymes in the intermembrane space and have been functionally expressed in the E. coli cytoplasm, a structure has yet to emerge.
Although the CCHL gene in yeast was discovered 2 years earlier (48-50, 105-107) than the first system I ccm genes (85), there have been no reports of any other proteins necessary for system III, with the exception of Cyc2p (27). When the gene encoding Cyc2p is mutated, some cytochrome c, albeit lower levels, is still produced (27). Cyc2p is a flavoprotein, and it was proposed that it might reduce either the heme-iron or cysteines for ligation by the CCHL. It is also possible that dedicated disulfide-forming and thiol-reducing pathways are present in mitochondria (17, 79, 82, 96, 144), but the exact mechanism(s) to maintain reduction of the apocytochrome c cysteines for system III is unknown.
A heme binding site in CCHL has likewise been elusive. A particular sequence, CPX, was implicated as a heme binding motif and is required for CCHL function (145), but it has been noted that some CCHLs do not have this sequence (70). Also unknown is how the mitochondria employing system III traffic heme to the CCHL (Fig. 2) (99). Three possibilities come to mind: (i) a specific, yet-unidentified heme transporter such as described below for system II; (ii) simple diffusion through the inner membrane, with heme equilibrating between the membrane and intermembrane space, where CCHL binds it; and (iii) a broad substrate “flippase” like those used to “flip” phospholipids from the inside leaflet to the outside leaflet (33, 46, 77, 83), followed by binding to the CCHL. Recall that like phospholipids, heme is an amphipathic molecule (Fig. 1). Indirect support for the second or third mechanism comes from the heterologous synthesis of hemoglobins and cytochromes b in the E. coli periplasm (64). As long as the heme is maintained in the reduced state, the second or third mechanism could suffice for CCHL function and cytochrome c synthesis.
System II: CcsB and CcsA Form a Heme Channel and Cytochrome c Synthetase
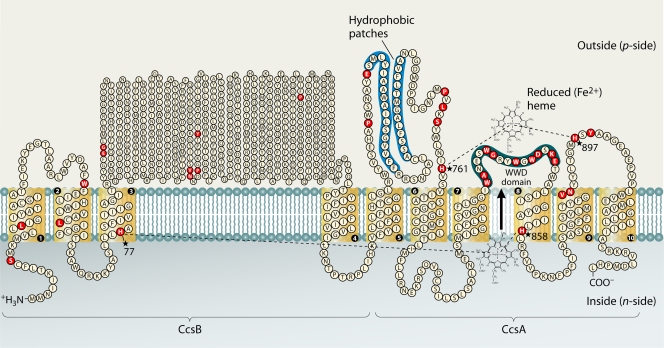
The system II pathway emerged in the mid-1990s through genetic analyses of Chlamydomonas (47, 80, 159, 160) and selected bacteria such as Bacillus (92, 139, 140) and Bordetella (23, 55, 86) (Fig. 2). There were some inklings in 1992 that a system different from systems I or III existed when a comparison to the CcmC and CcmF proteins yielded a chloroplast open reading frame (ORF) which comprised a different WWD subfamily (25). This ORF was later called ccsA (cytochrome c synthesis) when it was deleted from the chloroplast genome of Chlamydomonas, resulting in a strain that could not make cytochrome b6f (160). Cytochrome f is analogous to cytochrome c1 of the bc1 complex and contains the CXXCH motif. System II uses reducing equivalents from inside the cell to reduce DsbD (or a similar, smaller version called CcdA) (23, 139). A dedicated thioredoxin called CcsX is involved in reducing the cysteines of the CXXCH motif directly (23). Note that exogenous thiol-reducing agents such as dithiothreitol can substitute for DsbD and CcsX activity; thus, they are not essential. The CcsA protein has the WWD domain and three conserved histidines, two predicted to be on the outside (H761 and H897 in Fig. 3) and one in TMD8 (H858). Concerning the topology of this protein, although hydropathy programs typically predict eight TMDs (94), two experimental analyses suggest six (62, 71). In the experimentally derived model, there are two hydrophobic patches that are placed in the bacterial periplasm rather than as TMDs, as shown in Fig. 3. The clear placement of these two conserved hydrophobic patches will await three-dimensional structural determination. Nevertheless, note that whether six or eight TMDs are present, no conserved residues change in location (topology), and so it is worthwhile to consider the roles of conserved residues in topological context.
FIG. 3.
Topology of the system II CcsBA fusion protein from Helicobacter hepaticus and completely conserved residues (red). This topology is based on the experimentally determined topologies for the CcsBA fusion (59), the CcsB protein of B. pertussis, and the CcsA proteins of Mycobacterium leprae and Chlamydomonas reinhardtii as described in the text. The delineation between the CcsB and CcsA portions of the fusion protein is noted. Absolutely conserved (identical) residues in bacterial system II are colored red. Four absolutely conserved histidines that are potential axial ligands to the heme iron are starred. The highly conserved WWD domain in CcsA is shaded, as are the hydrophobic patches. Reduced heme is proposed to move through CcsBA (using H77 and H858 as initial axial ligands) to the WWD domain where H761 and H897 ligand the heme iron. Conservation was determined by alignments of the CcsB and CcsA proteins from the following organisms, which were chosen as representatives across the kingdoms and genera shown in Fig. 2: Arthrobacter aurescens, Arthrobacter butzleri, Bdellovibrio bacteriovorus HD100, Bordetella pertussis Tohama I, Campylobacter jejuni NCTC 11168, Chlamydomonas reinhardtii, Chlorobium tepidum TLS, Desulfitobacterium hafniense Y51, Geobacter sulfurreducens PCA, Helicobacter acinonychis Sheeba, Helicobacter hepaticus ATCC 51449, Helicobacter pylori 26695, Leifsonia xyli CTCB07, Mycobacterium leprae TN, Neisseria gonorrhoeae FA1090, Ralstonia metallidurans CH34, Synechocystis sp. strain PCC6803, Thiobacillus denitrificans ATCC 25259, Thiomicrospira denitrificans ATCC 33889, and Wolinella succinogenes DSMZ 1740.
The other absolutely essential protein for system II is called CcsB (80). In most organisms the CcsB and CcsA proteins are present as two separate polypeptides, usually encoded in an operon in bacteria. Experimental topological studies indicate that CcsB has four TMDs, with the third and fourth separated by a large periplasmic domain (200 to 500 residues) (Fig. 3) (23). Conserved residues in CcsB are very sparse, although the predicted secondary structures are maintained. The CcsB and CcsA proteins form a tight complex in Bordetella pertussis, and CcsB is completely degraded in the absence of CcsA (55). Likewise, in ccsB (ccs1) mutants of Chlamydomonas, the CcsA protein is not detectable (159). This may mean that critical interactions between TMDs of CcsB and CcsA exist. Early proposals suggested that the WWD domain bound heme, liganded by flanking histidines that would maintain the heme in the reduced state for presentation to the CXXCH motif (Fig. 3). The large periplasmic domain in CcsB would feature the apocytochrome c CXXCH recognition function, presenting the apocytochrome to the heme in CcsA.
In a few bacteria such as Helicobacter, Bacteroides, and Wolinella, the CcsB and CcsA proteins are naturally fused into one large ORF (called ccsBA). This ORF (from Helicobacter), when overexpressed in E. coli deleted of its ccm (system I) genes, was all that was necessary to synthesize a periplasmic cytochrome c (57). In this recombinant reconstitution, the CXXCH motif is reduced by the housekeeping reductants (e.g., DsbD and DsbC [E. R. Frawley and R. G. Kranz, unpublished data]). Thus, it is clear that CcsBA represents the system II cytochrome c synthetase. Topological studies of CcsBA give results similar to those for the separated CcsB and CcsA, as shown in Fig. 3, with the same caveat discussed above concerning the two hydrophobic patches in CcsA (59). Recently, Frawley and Kranz purified milligram quantities of the CcsBA protein with heme bound (59). Spectral analyses indicate that this heme is b type and is purified in the reduced state (Fe2+). Importantly, when the two flanking periplasmic histidines (H761 and H897) were individually mutated to alanines, the purified proteins still had heme, but it was in the oxidized state (Fe3+) and the spectral absorptions were significantly perturbed (59). We conclude that the function of each periplasmic histidine is to ligand the iron and maintain the heme in the reduced state for covalent thioether bond formation. This represents the “external heme binding domain,” which probably defines the active site of the CcsBA cytochrome c synthetase (Fig. 3).
The third conserved histidine (H858) in TMD8 near the bacterial cytoplasm was also mutated to an alanine. Notably, purified CcsBA (H858A) does not have heme in its external heme binding domain, yet the protein is stably synthesized (59). Another conserved histidine (H77) is located in the CcsB portion of CcsBA (and in all CcsB proteins). The CcsBA (H77A) protein contains fourfold less heme in its external heme binding domain. All four histidines in the recombinant CcsBA are required for synthesis of a cytochrome c (59), and they are also required in the Chlamydomonas CcsA (71) and CcsB (Ccs1) (47). Remarkably, the H77A and H858A TMD mutants are complemented for function by adding exogenous imidazole, but the histidine mutants that comprise the external heme binding domain are not (59). Complementation by imidazole has been demonstrated for recombinant myoglobin when the proximal histidine is mutated, wherein it was corrected for heme binding (21, 22). For this myoglobin “cavity” mutant, crystallographic results indicated that the cavity contains the exogenous imidazole, allowing the myoglobin to bind heme (22). Our working model is that two heme binding sites are present in CcsBA, one at the channel entry (via H77 and H858) that facilitates delivery of reduced heme through a CcsBA channel, and a second site, the external heme binding domain, that is composed of the WWD domain and the two external histidine ligands that protect heme from oxidation. The results with CcsBA H77A and H858A also suggest that CcsB and CcsA are integrally associated, with a single heme binding site formed from the respective TMDs 3 and 8 (see Fig. 3).
Once the CXXCH motif is brought near the heme tethered within the WWD domain of CcsA, we predict that the spontaneous ligation to form the thioether bonds may induce the ligand switching to occur (i.e., the histidine of CXXCH for either the H761 or H897 in CcsA). Subsequently, we envision that this ligand switching and folding of the holocytochrome c polypeptide provides the energy to release the heme from the CcsA WWD domain. Note that one idea for system II is that it binds heme with a lower affinity than system I, so that the energy needed to release holocytochrome c may be less (see Evidence for Why Three Systems Exist: Evolution and Ecology below).
System I: CcmABCDEFGH
Genetic studies in the late 1980s and early 1990s started to unravel a complicated pathway for cytochrome c synthesis in a few model alphaproteobacteria such as Rhodobacter (24, 25, 85, 91), Paracoccus (111-114), and Rhizobium (120, 131). Although the players (now called the ccmABCDEFGH genes) were well defined genetically and were certainly dedicated specifically to cytochrome c synthesis, biochemical studies have only recently begun to elucidate the mechanisms involved. In short, cytochrome c synthesis by system I includes some fascinating chemistry and elegant molecular mechanisms for heme trafficking. It is instructive to describe system I as a two-step process, with oxidation and reduction of heme as it moves through the pathway (Fig. 2). One of the proteins in system I is a periplasmic heme chaperone called CcmE, discovered by Schulz et al. (142), and Reid et al. (122). Step 1 is the formation and release of oxidized (Fe3+) holoCcmE (by CcmABCD), while step 2 ends with a cytochrome c synthetase complex (CcmF/H) that is proposed to use quinols to rereduce the iron of heme from holoCcmE, bring the CXXCH to this heme, and thus catalyze the ligation reaction.
Step 1: heme delivery (to form oxidized holoCcmE) and the holoCcmE release subpathway, CcmABCD.
Briefly, step 1 (Fig. 2) involves the flux of reduced heme (from ferrochelatase) to the periplasmic WWD domain of CcmC, concomitant with binding of CcmE to this heme/CcmC/CcmE ternary complex (Fig. 4 shows the proteins in step 1). The spontaneous covalent attachment of the 2-vinyl group of heme to the CcmE histidine (H130) then occurs (Fig. 4), which we refer to here as “adduct” formation (142). Adduct formation results in oxidized heme. A “release complex” comprised of the CcmABCD proteins uses ATP hydrolysis to release the oxidized (Fe3+) holoCcmE, freeing it to react with the CcmF/H complex (37, 56). CcmA is the ATP binding cassette (ABC) protein, while CcmB is a six-TMD protein (62) always encoded adjacent to ccmA (85, 120). Note that as diagrammed in Fig. 2, this step (formation of released holoCcmE) is likely very rapid in vivo. However, we have recently been able to trap, purify, and characterize intermediates in this step using various genetic strategies (125). These intermediates are reviewed here, and we discuss the chemistries underlying adduct formation and heme iron redox states.
FIG. 4.
Topologies of the CcmABCDE integral membrane proteins required for system I cytochrome c biogenesis step 1, i.e., heme delivery to apoCcmE and release of holoCcmE by CcmABCD. Completely conserved residues are in red. The indicated sequences are from Escherichia coli. The topologies of CcmB, CcmC, CcmD, and CcmE are based on experimentally determined topologies as described in the text (see, e.g., reference 125). Completely conserved (i.e., identical) amino acid residues (red) were identified by individual protein alignments with bacterial sequences from genera shown in Fig. 2. Histidine residues that potentially act as axial heme iron ligands are starred, and some are shown with dashed lines to specific heme molecules. CcmE has a completely conserved His or Cys (deinococci) residue (green) for covalent heme attachment and a Tyr (Y134) residue as an axial heme iron ligand (dashed line). The oxidation states of heme are shown, and other important residues, domains, or motifs are labeled for descriptions used in the text. Genes from the following organisms were used for determining conserved residues: the alphaproteobacteria, Agrobacterium tumefaciens C58, R. capsulatus, Caulobacter crescentus CB15, and Bradyrhizobium japonicum (blr 0467); the betaproteobacteria, Nitrospira multiformis ATCC 25196 and Nitrosomonas europaea ATCC 19718; the gammaproteobacteria, E. coli K-12 MG1655, Pseudomonas fluorescens Pf01, Shewanella oneidensis MR-1, and Vibrio parahaemolyticus RIMD 2210633; the deltaproteobacteria, Myxococcus xanthus and Desulfovibrio desulfuricans; and the deinococci, Deinococcus geothermalis and Thermus thermophilus.
(i) Formation of the holoCcmCDE “preadduct” intermediate complex.
It has been shown genetically that the six-TMD protein CcmC (Fig. 4) (62) is sufficient to bring heme to CcmE (56, 141). By removing the ccmA and ccmB genes, we have been able to trap heme in stable CcmCDE complexes (56, 125). Our recent results indicate that CcmC alone will not bind heme and that apoCcmE is necessary; heme is also necessary for CcmC to interact with apoCcmE, and so we propose that determinants on the heme molecule are recognized by both CcmC and apoCcmE, facilitating the formation of this stable ternary complex (125; C. Richard-Fogal and R. G. Kranz, unpublished data). If wild-type CcmCDE is overexpressed (without ccmA or ccmB), all of the heme is covalently linked to CcmE via the H130 residue (see “(ii) Formation of the oxidized holoCcmCDE ‘postadduct’ intermediate complex” below). However, in a CcmE(H130A) mutant the heme is b type in the holoCcmCDE preadduct complex (125). Thus, heme binding determinants in both CcmC and CcmE (other than H130) are important for initial binding of heme. This is consistent with results of site-directed mutagenesis of CcmE, where it was suggested that hydrophobic residues may be involved in binding heme (54, 75). The intermediate holoCcmCDE complexes use two histidines in CcmC flanking the WWD domain as axial ligands to the heme (Fig. 4) (see below). Importantly, at least half of the heme in the preadduct holoCcmCDE complex is purified in the reduced state (125).
One of the important questions in the field has been how heme accesses each of its external sites (i.e., WWD domain), either through a protein channel (e.g., as shown above for CcsBA) or through the inner or outer leaflet of the membrane (see, e.g., references 62 and 155). For CcmC, there are no conserved histidines within TMDs, but other residues such as tyrosine are conserved (Fig. 4). Now that heme can be assayed in CcmC (as a complex) (125), it will be important to determine if a specific binding site in TMDs (and thus a channel from inside) is suggested. Because the covalent bond formation to CcmE H130 does not necessarily require Fe2+ (see below), a nonspecific channel that does not protect the heme may be sufficient. Alternatively, without a need for protection from oxidation, heme could enter the CcmC WWD domain from the membrane directly.
(ii) Formation of the oxidized holoCcmCDE “postadduct” intermediate complex.
Overexpression of the wild-type CcmCDE (no ccmAB) results in the postadduct intermediate complex, which has been purified and characterized (125). This holoCcmCDE complex contains all oxidized heme, with a striking UV/visible absorption spectrum when reduced with dithionite (125). A novel split alpha absorption is observed (at 553 and 559 nm), which was shown to be due to the unique chemical environment of the heme. Purification and spectral characterization of various CcmCDE mutants showed that the CcmE H130 covalent adduct and the CcmC H60 and H184 each contribute to the unique spectra. Spectra of complexes that contain alanine substitutions for either of the two external histidines in CcmC (H60 and H184) unequivocally show that these are the fifth and sixth axial ligands to the heme (125). Complexes with CcmC(H60A) and CcmC(H184A) have 50% less heme than the wild-type complex, and they exhibit major spectral perturbations. Interestingly, even with the CcmC(H60A) and CcmC(H184A) mutations, all of the heme is covalently linked to CcmE, consistent with the result that mutants with mutations in each were still partially functional (124). This also distinguishes the CcmE adduct formation from the CXXCH attachment noted earlier for analogous histidine substitutions in CcsBA. In CcsBA, the flanking histidine mutations completely abolish function, and these mutant CcsBA proteins were unable to protect the heme from oxidation (59). In this case, the wild type and CcmC histidine mutants result in an oxidized holoCcmCDE complex.
The covalent bond formed between the CcmE H130 and heme has received much attention. The unique chemistry and structure of this bond to the 2-vinyl β carbon (93) have been discussed (61, 93, 147, 148, 158). We discovered that all CcmCDE postadduct complexes and released holoCcmE (see below) contain Fe3+ heme, suggesting that in addition to covalent bond formation in CcmE, there must also be an oxidation step leading to an oxidized holoCcmE. Ferguson and colleagues have discussed the idea that the CcmE H130 imidazole would require a radical intermediate to form the covalent bond at the β carbon of the heme vinyl group (3, 93). However, the final oxidation state of the heme iron has drawn little attention. Figure 5A illustrates possible mechanisms for the adduct formation to CcmE H130. The first uses the H130 imidazole cation radical, but formation of this radical would itself require an oxidant (marked “Q” in the Fig. 5A, but its identity is unknown). The second mechanism is a nucleophilic reaction that is favored by an oxidized heme iron. The fact that heme in the preadduct CcmCDE complex is approximately 50% reduced (125) could argue for either mechanism. The in vitro synthesis of the holoCcmE* adduct appears to require the reductant dithionite (15, 42, 148), leading to the suggestion of a radical mechanism, although it is unclear whether a cation or an anion radical is proposed. The mechanism by which this radical might be formed in vitro or in vivo is still a mystery, but note that with the cation radical, the end product has oxidized (Fe3+) heme. It seems unlikely the imidazole could form a cation radical in the presence of the dithionite reductant (since the oxidation potential of heme is around −0.2 V and that of imidazole is around +0.5 to 0.8 V at neutral pH.) Imidazole reduction by dithionite to form an anion radical also seems unlikely. The alternative is to form the CcmE H130 adduct through a nucleophilic Michael addition, which would prefer oxidized (Fe3+) heme (Fig. 5A). Likewise, the oxidant to form Fe3+ heme, whether a quinone or semiquinone or other, is unknown. It is feasible that both mechanisms operate in vivo and that the partial function of the CcmC histidine mutants noted above reflects this plasticity. Clearly, the more likely chemical mechanisms for adduct formation to the β carbon predict a holoCcmE with oxidized heme, and the data on intermediate holoCcmCDE complexes and released holoCcmE (125) are in agreement with these mechanisms.
FIG. 5.
Proposed radical and nucleophilic reaction mechanisms for holoCcmE (His130) adduct (A) and cytochrome c (B) linkage to heme vinyl groups. Noted are the oxidation states of iron (Fe3+ or Fe2+); red half arrows are one-electron transfers, and full red arrows are two electron transfers. Transfer of the proton from the imidazolium to the alpha carbon is probably solvent or protein mediated (i.e., the proton may be abstracted at an early step, with a solvent- or protein-mediated protonation at the alpha carbon at a later step). The final Fe3+ state could be either reduced back to the Fe2+ state by Q− or kept in the Fe3+ state to stabilize the imidazole adduct until it is ejected at the next step (reverse blue arrow). Only a single vinyl of heme and, for the radical step, only one set of one-electron arrows are shown for simplicity.
Mutation of tyrosine 134 in CcmE(Y134A) was shown previously to have a dramatic effect on the spectra of purified holoCcmE* (no TMD), and thus tyrosine 134 is an axial ligand in released CcmE (158). In the intermediate CcmCDE complex with heme, a CcmE(Y134A) mutant showed a spectrum no different from that of the wild-type CcmCDE complex. This indicates that axial ligands to the heme iron have not yet switched over to the CcmE tyrosine 134 in the intermediate complexes, and thus all heme is still liganded to the CcmC H60 and H184.
(iii) Release of holoCcmE via the CcmABCD complex.
We conclude that the stable intermediate holoCcmCDE complexes described above require the release function provided by CcmAB and that in the release process axial ligands must switch from CcmC (H60 and H184) to CcmE (Y134) (125). Only when the CcmA and CcmB proteins are coexpressed is the released holoCcmE free from CcmC (56). We envision that major conformational movements are needed to accomplish this switching and holoCcmE release, thus explaining the requirement for an ABC transporter complex (125). Note that a CcmABCD complex has been suggested for Rhodobacter capsulatus (62, 63, 65) and E. coli (56), but others in the field offer alternative views proposing that a CcmAB transporter operates independently of CcmC by exporting an unknown substrate such as a reductant (37, 141, 146).
We believe that all data are consistent with the ABC transporter release function for CcmABCD (Fig. 6). Conceptually, the release is similar to that for the Lol ABC transporter (LolCDE), designated the lol system, that has been characterized by Tokuda and colleagues (Fig. 6) (81, 153, 161). We note that both the LolCDE proteins and the CcmABC proteins are not typical ABC transporters in that they do not translocate a substrate but instead are involved in recognition and release of periplasmic molecules. The release function of each has been shown to require the ABC component (LolD or CcmA) for a proposed conformation change and “release” of the target molecule, lipoproteins for LolCDE and holoCcmE for CcmABC (56). The lol system is dedicated toward recognizing lipoproteins that will be sent from the cytoplasmic membrane to the outer membrane of gram-negative bacteria. Lipoproteins have a fatty acid attached to an N-terminal cysteine residue. Some lipoproteins function tethered to the cytoplasmic membrane, but most require tethering to the inner surface of the outer membrane. LolCDE in E. coli recognizes lipoproteins that do not have an aspartate next to the cysteinyl attachment (97). The aspartate is an exclusion feature, and therefore lipoproteins with the aspartate remain as cytoplasmic membrane-anchored lipoproteins. Outer membrane-targeted lipoproteins are tightly bound to LolCDE in the absence of ATP hydrolysis (by LolD) or in the absence of the periplasmic LolA chaperone. Upon binding of LolA, ATP hydrolysis releases the lipoprotein to LolA, which transfers it to the outer membrane LolB. LolB and LolA are related, and this energy-independent exchange has been discussed previously (110). Thus, the holoCcmE release is analogous to the lipoprotein-LolA release. LolA is the lipoprotein chaperone to the outer membrane, and CcmE is the heme chaperone to what we refer here as “step 2,” the synthetase component of system I (Fig. 6).
FIG. 6.
Comparative models of the system I CcmABCD holoCcmE release pathway and the Lol lipoprotein release system (LolABCDE).
It was previously determined that the small polypeptide CcmD is part of a CcmABCD complex in R. capsulatus (63) and in E. coli (127). Recently we have shown that CcmD is essential for the holoCcmE release but not for the heme attachment to CcmE or binding of CcmE to CcmC (127). As we have described for the R. capsulatus CcmD, the E. coli CcmD has a single TMD with an N terminus in the periplasm (Fig. 4). The short N terminus contains a completely conserved tyrosine of unknown function, although it was proven not to be an axial ligand in the intermediate CcmCDE complexes (125).
In summary, three recent experimental results lead us to suggest that the released holoCcmE is in the oxidized Fe3+ state rather than Fe2+: (i) all heme in the purified, trapped holoCcmCDE complex and holoCcmE is Fe3+ (125); (ii) all heme in the purified WWD protein CcsBA from system II is in the reduced (Fe2+) state, as one would predict, and thus even purification under ambient air yields a reduced WWD holoprotein (59); and (iii) a highly conserved mechanism to rereduce the heme iron of holoCcmE appears to be present in CcmF [see “Step 2: heme attachment to apocytochrome c (from holoCcmE), and CcmFH as a holoCcmE reductase and cytochrome c synthetase” below] (125). (Daltrop et al. have observed heme transfer from holoCcmE* [truncated] to an apocytochrome c in vitro [albeit without CcmF/H present and after overnight incubation] [42]. The heme for this transfer from holoCcmE* required reduction by dithionite. Although the authors concluded that one function of holoCcmE may be to retain the heme in the reduced state, unlike our proposal here, their data are consistent with a need for iron reduction in holoCcmE.)
Step 2: heme attachment to apocytochrome c (from holoCcmE), and CcmFH as a holoCcmE reductase and cytochrome c synthetase.
Released holoCcmE is free to interact with the CcmF/H complex, but a key question is the mechanism by which the heme is reduced for covalent bond formation to the CXXCH motif. We have recently purified CcmF in milligram quantities from E. coli (125). Similar to the results of Sanders et al. with R. capsulatus CcmF (138), the E. coli CcmF and CcmH proteins form a complex. Surprisingly, the preparations were tinted red, which is suggestive of heme (125). Initially, we thought that this might originate from holoCcmE, since it has previously been shown using coimmunoprecipitation experiments that CcmE and CcmF interact (2, 123). However, spectral analysis, heme assays, and pyridine hemochromagens indicated that this is a b-type heme and that (holo)CcmE was not copurifying (125). Even in the absence of the ccmE or ccmH gene, CcmF has a b heme in a 1:1 stoichiometry. Thus, the data indicate that the CcmF/H complex is itself a cytochrome b that has the cytochrome c synthetase function. This result is conceptually reminiscent of the heme a synthase encoded by ctaA described above. Recall that CtaA has a heme b which is involved in electron transfer and the heme o substrate for modification to heme a (76). Based on experimentally determined topological models for CcmF (62, 129) and new phoA fusion results (125), a unified topological diagram for CcmF is shown in Fig. 7. The completely conserved residues (in red in Fig. 7) are used to discuss the following three functions for the CcmF/H complex.
FIG. 7.
Topologies of the CcmF and CcmH integral membrane proteins required for system I cytochrome c biogenesis for step 2, i.e., heme attachment to apocytochrome c by CcmFH. The indicated sequences are from Escherichia coli. The topologies of CcmH and CcmF are experimentally established, as described in the text (see, e.g., reference 125). Completely conserved (i.e., identical) amino acid residues (red) were identified by individual protein alignments with bacterial sequences. The potential histidine axial ligands to the b heme and heme in holoCcmE are starred. The highly conserved WWD domain in CcmF is shaded, as are the hydrophobic patches. The following organisms were used for determining conserved residues: the alphaproteobacteria, Agrobacterium tumefaciens C58, R. capsulatus, Caulobacter crescentus CB15, and Bradyrhizobium japonicum (blr 0467); the betaproteobacteria, Nitrospira multiformis ATCC 25196 and Nitrosomonas europaea ATCC 19718; the gammaproteobacteria, E. coli K-12 MG1655, Pseudomonas fluorescens Pf01, Shewanella oneidensis MR-1, and Vibrio parahaemolyticus RIMD 2210633; the deltaproteobacteria, Myxococcus xanthus and Desulfovibrio desulfuricans; and the deinococci, Deinococcus geothermalis and Thermus thermophilus.
First, at least one histidine (CcmF H261) acts as an axial ligand to the b heme to yield the cytochrome b function. A CcmF (H261A) mutant protein has one-quarter the amount of b heme in the wild type and a perturbed absorption spectra (125). The other heme b axial ligand could be H491, but this has not yet been tested. The b heme of CcmF is reduced in vitro by quinols, suggesting that its cytochrome b functions as a quinol:heme oxidoreductase. We note a partially conserved motif, (“Q” in Fig. 7) with the amino acid sequence NPF at the outer surface of the second periplasmic loop, which is analogous to the quinone binding motif of RegB (GGXXNPF) in a comparable topological position (152). We suggest that the CcmF protein accepts electrons from quinols to reduce the b heme, which subsequently reduces the heme from holoCcmE (125). Note also that the b heme is positioned topologically directly below the WWD domain, the predicted location for the heme of holoCcmE (Fig. 7). Although we cannot rule out that the cytochrome b is directly involved in the thioether bond formation, the simplest interpretation is that it acts as a quinol:heme oxidoreductase in vivo.
A second predicted function of CcmF is binding to holoCcmE as part of its synthetase activity, using the holoCcmE heme for attachment to apocytochrome c (57, 123, 125). The CcmF (H173A) protein has wild-type cytochrome b function (125) yet is completely nonfunctional (123, 125). In analogy to CcmC and CcsA, as discussed above, the H173 and H303 of CcmF are periplasmically located, adjacent to the WWD domain, and are likely ligands to the heme from holoCcmE, which is used for the synthetase reaction (Fig. 7). How does axial ligand switching of this heme (from holoCcmE to CcmF to CXXCH) occur? Possibly, the energy from cytochrome electron transfer is used for this, and/or the covalent attachment to CXXCH induces this switching. Uchida et al. used Raman spectroscopy to indicate that one axial ligand in holoCcmE* (truncated with no TMD) is a histidine and the other is the tyrosine 134 discussed above (Fig. 4) (158), but it is not necessary to envision two heme ligands from CcmE immediately switched to two from CcmF and then to the two from cytochrome c. Garcia-Rubio et al. used various spectral approaches on holoCcmE* indicating that tyrosine is one axial ligand, but an unknown sixth ligand may exhibit conformational flexibility (61). In any event, it is likely that the WWD domain binds holoCcmE heme, precisely positioning the vinyl side chains (one with the CcmE adduct) to accept the CXXCH motif of the apocytochrome c (see below). The synthetase reaction is likely spontaneous, as long as the iron of heme is reduced (see Fig. 5B for chemical reaction of one thiol of CXXCH to the α carbon of the heme vinyl group). At this site, the CcmE H130 imidazole side chain must be ejected from the β carbon of the 2-vinyl group. As shown in Fig. 5A (large blue arrow), iron reduc-tion of the heme in holoCcmE would also favor ejection of the imidazole by the reverse Michael reaction.
The third function of the CcmF/H complex is recognition of the CXXCH motif for the synthetase activity. The apocytochrome c CXXCH motif may be recognized by CcmH, potentially chaperoning the motif to the WWD active site containing heme (from holoCcmE). Although the large periplasmic N- and C-terminal loops of CcmF (Fig. 7) have no designated functions as of now, possibly these are involved in interaction with CcmH. Both the R. capsulatus (138) and E. coli CcmF proteins are easily dissociated from CcmH, in the latter case simply by using high-salt conditions (125). These results suggest that interaction between TMDs of CcmF and -H are minimal and that electrostatic interactions via soluble domains may predominate, hence leading to the suggestion that the large periplasmic domains of CcmF are involved. CcmH has an RCXXC motif (Fig. 7), which was previously shown to reduce the CXXCH of apocytochrome c (see, e.g., reference 103). The structure of the first CcmH periplasmic domain (from Pseudomonas) containing the RCXXC motif has been shown to be a three-helix bundle, unlike the case for other thioredoxin-like proteins (45). The recently described analogous E. coli CcmH domain suggests that it is a dimer (1). Note that in most organisms (e.g., R. capsulatus) ccmH is split into two ORFs (Fig. 7). The first was previously called ccl2 and has a single TMD and the RCXXC periplasmic domain (25, 103). The second ORF is called cycH (or ccmI), which had been topologically dissected and studied in alphaproteobacteria such as R. capsulatus and is potentially involved in the chaperone function (of the apocytochrome c), using a tetratricopeptide repeat (38, 44, 91, 136, 137). Employing two-hybrid analysis in yeast, Meyer et al. have shown that the Arabidopsis CcmH interacts directly with apocytochrome c (102). Because only the N-terminal periplasmic domain (Ccl2) appears to be essential for cytochrome c synthesis under certain conditions (see, e.g., references 44 and 132), it is important to key in on this domain (Fig. 7). It clearly has both chaperonin and thiol reduction functions. After attachment to the CXXCH motif, the c heme must be released from CcmF/H. Potentially, the reactions of the two vinyl groups with CXXCH (including ejection of the histidine-vinyl from CcmE) and/or the subsequent folding of the apocytochrome c around the heme provides the energy for release from the CcmF/H active site.
When we first presented the sequence of CcmF (called Ccl1) in 1992 (25) and subsequently performed a topological and residue consensus profile in 1998 (62), we wondered why CcmF was so large and why there were so many completely conserved histidines. We believe that multiple functions described here provide the rationale for these conservations and observations. Interestingly, in plant mitochondrial genomes, CcmF is separated as two (72) and sometimes three individual ORFs, called CcmFN1, CcmFN2, and CcmFC, which form a complex with CcmH (70, 121). Has genomic reorganization of the mitochondrial endosymbiont naturally separated multiple functions of the CcmF protein, albeit while still forming a complex? Finally, we must note that the detailed mechanism of formation of the two thioether bonds at the CcmF/H active site remains a very significant open question in the field. With the likely addition now of a mechanism to reduce holoCcmE at the active site prior to ligation, the molecular mechanisms and timing of reactions become even more intricate. Below we address why such a complicated system has been maintained for billions of years through evolution.
System IV and Others
System IV.
Kuras et al. identified four integral membrane proteins (called Ccb1 to -4) that are necessary to attach heme covalently to a single cysteine in cytochrome b6 of the cytochrome b6f complex (hence the designation ccb genes) (87, 88). The ccb genes are present only in oxygenic phototrophs (95), where the cytochrome b6 has been known for some time to have this spectrally unique heme. The cysteine is located on the opposite side of the membrane from the cytochrome f (c type), and so this putative Ccb complex functions on the opposite side (n side) from that of the three systems for cytochrome c synthesis (p side) just described. The authors suggested designating Ccb “system IV,” which seems appropriate. Like systems I to III, it is widely distributed, and clearly the mechanism of attachment and the functions of the Ccb proteins are of tremendous interest and may provide parallels with and clues to other systems down the road.
Others.
The apocytochrome c recognition motif for system I has been much studied and appears to be only the CXXCH motif (6-8, 13, 14, 29), with the caveat that CXXXCH may also be used (130). Some variants of system I (e.g., NrfEFG [51], which is related to CcmFGH) and system II (CcsBA) recognize slightly altered apocytochrome c signature motifs such as CXXCK (51, 74, 117) or CX15CH (73). These variants are likely to play major roles in the future in understanding of the apocytochrome c recognition determinants and the features of the substrate and synthetases that are important for such.
Some variations of system I exist in archaea and a few bacterial outliers (e.g., Desulfovibrio) in that the CcmE protein has a cysteine instead of the histidine (H130) for covalent bond formation (11). Note that a cysteine (thiol) can carry out reactions similar to those of the imidazole (histidine), forming a covalent bond to the β vinyl group of heme (Fig. 5A); likewise, this thiol adduct would be ejected preferentially by reduction of the heme iron (Fig. 5A). Interestingly, the CcmH protein is missing in the organisms that have a cysteine in CcmE. The significance of this should become clearer as we understand the chemical mechanisms underlying holoCcmE adduct formation and the CcmF synthetase reactions (on the holoCcmE and CXXCH motif of the apocytochromes). One speculation might be that the ejected thiol (of CcmE) reduces the CXXCH (rather than the CcmH RCXXC), and thus CcmH is no longer required for reduction, possibly with CcmF taking over the CXXCH recognition function.
Certain rare protozoans (Euglenozoa such as trypanosomes and Leishmania) have a cytochrome c in their mitochondria with a single cysteine in a CH motif (rather than the classic CXXCH) (10, 12). Surprisingly, they do not appear to have any proteins from system I, II, or III. The structure of their cytochrome c is similar to that of other mitochondrial cytochromes c, so Fulop et al. have speculated that a new, yet-unknown system is still to be discovered for these protozoa (60). It also remains possible that a diverged CCHL that cannot be detected by BLAST searches is present yet recognizes more than just the CH in the apocytochromes. Recall that there clearly are determinants on apocytochromes c and c1 (other than the CXXCH) recognized by the CCHLs. Possibly both the recognition determinants and CCHLs have coevolved, drifting so that they are not detectable by homology searches.
EVIDENCE FOR WHY THREE SYSTEMS EXIST: EVOLUTION AND ECOLOGY
Why are there cytochromes c? Studies in the last decade have suggested that the covalent linkage provides added stability to the cytochrome c, and so they are much less likely to lose the heme than the globins or other cytochromes (see, e.g., reference 16; reviewed in reference 3). The idea that a wider range of redox potentials result from attachment (28) has been discussed as being unlikely (3, 18). As noted earlier, in prokaryotes there are many different types of cytochromes c in hundreds of different electron transport chains, and efficient mechanisms to recognize a CXXCH motif are critical (12). Remember also that cytochromes c in prokaryotes function outside the protected cytoplasm. Using cytochromes c may be one way to “hard-wire” a chain or conduit for electron transfer that will function under diverse and sometimes hazardous environmental conditions.
Why are there three predominant systems for assembly rather than one? Let us start with system III, the simplest, which uses the CCHL enzyme. Clearly the CCHL evolved after mitochondrial endosymbiosis (which occurred with an alphaproteobacterium possessing the system I pathway) (68, 89, 90). Recall that all alphaproteobacteria possess system I, as do the mitochondria of plants and some primitive protozoa (Fig. 2). One case for the need for systems I and II in bacteria and plants is that they must ultimately end with a reduced heme iron for ligation to occur, even under environmental conditions that may be oxidizing. What would make the more complicated system I obsolete in certain mitochondria? For some eukaryotes, it is likely that the internal milieu requiring synthesis by system I changed. We speculate that the heme was largely protected against oxidation, and additionally that reduced iron, and therefore reduced heme, was not limiting for growth (see below for a discussion of the benefits of system I). For example, one could envision that the levels of iron needed for Fe/S proteins, hemoglobins, and/or other cytochromes significantly surpassed that needed for cytochromes c and c1. A simpler mechanism that did not involve the synthesis of at least eight integral membrane proteins and the use of ATP (as with system I) became more economical. Since only two cytochromes c were used (cytochromes c and c1), a specialized enzyme that recognizes these two became possible. Circumstantial proof for these ideas is the retention of system I in plant and some protozoal mitochondria, which likely face more oxidizing and varying environments. Possibly, some protozoa in the future with both systems I and III, in the process of losing system I and/or using each under different conditions, will be discovered.
At present, because of the significant amount of lateral transfer that has occurred with systems I and II, it is not possible to say which came first (Fig. 2) (66). Because of its simplicity, one would guess that system II evolved initially. Its presence in cyanobacteria and plant chloroplasts indicates that it was also part of the cyanobacterial endosymbiotic events leading to the algal chloroplast. Its retention might well be due to the fact that the heme is well protected from oxidation (59). Still, some accessory players in chloroplasts, possibly for maintaining reduced thiols of the CXXCH in cytochrome f, may have evolved subsequent to the endosymbiosis. These have been discussed previously (116).
What possible advantages might system I confer that are not features of system II? Why is system I so complicated, as it clearly takes more energy to produce the system I proteins than system II and we know that ATP is required for system I function? We have evidence from the comparison of these two recombinant systems in E. coli that system I can use heme at potentially five-times-lower concentrations than system II (126). Iron is the limiting factor in heme biosynthesis (109). Thus, in certain iron-limiting environments an organism with system I can make a cytochrome c while those with system II cannot. Moreover, in experiments where holoCcmE was synthesized and then heme synthesis shut off completely (either with heme synthesis inhibitors or by genetically knocking out heme biosynthesis), E. coli with system I but not with system II was still able to synthesize cytochromes c de novo (57). This means that holoCcmE can function as a heme reservoir when no iron is available. (These experiments also prove that the heme attached to the cytochrome c comes from holoCcmE.) Together, these advantages might provide for a fitter organism in particular habitats. Interestingly, a very few species of bacteria have both systems I and II, such as Bordetella parapertussis. Possibly these are in the process of lateral transfer, awaiting selection against the system that provides less advantage. (For example, B. pertussis has only system II.) Alternatively, B. parapertussis may use system I under one growth condition and system II under a different condition. Studying the use of each system in these organisms and their ecology may further address the advantages and disadvantages of each in nature. We have attempted to evaluate the types of organisms that possess each system and to analyze the habitats in which these are typically found (B. San Francisco and R. G. Kranz, unpublished data). No clear patterns have emerged in this analysis, in part because the microhabitats that microorganisms use and their ecological distributions are poorly understood.
FOR GENOME ANNOTATORS: WHAT DEFINES A SYSTEM I (ccm GENES), A SYSTEM II (ccs GENES), OR A SYSTEM III (CCHL)
Given the fact that some proteins from both systems I and II (CcsA, CcmC, and CcmF) possess the highly conserved WWD domain, it can sometimes be confusing and time-consuming to properly annotate the sequences of new genomes for cytochrome c synthesis proteins. What signature genes and sequences can identify a system when the picture is murky? Here we provide some brief guidance for proper annotation and how to quickly establish whether the organism has system I (ccm), system II (ccs), both systems I and II (rarely), or system III. Table 1 presents some signature motifs found in each of the subfamilies of the WWD proteins (heme-handling proteins) and other proteins. The signature genes for system I that should emerge from a BLAST search are ccmC, ccmF, and ccmE. In outlying organisms with system I, the other ccm genes are most likely present, with the exception of ccmH, as noted above. For system II, the signature gene is ccsA with the indicated WWD motif. As shown for the few conserved residues in CcsB (Fig. 3, in red), the ccsB ORF can be troublesome to detect by homology searches in outliers, although it is typically in an operon with ccsA. For system III in mitochondria, all CCHLs are related, but specifically conserved residues in outliers also might be difficult to detect by BLAST searches (as discussed above for Euglenozoa). Finally, a few organisms can have multiple copies of a system. For example, Salmonella enterica serovar Typhimurium has two complete sets of ccm genes (98). Remember that the variations that recognize rarer apocytochrome substrates may be additionally present (e.g., in Wolinella two more CcsBA proteins, one for a CXXCK and one for a CX14CH motif, and in E. coli the nrf genes, which encode CcmFGH paralogs that recognize CXXCK).
TABLE 1.
System identification information for genome annotators
| System | Gene product | Conserved motif(s) | Additional key residues | Location of N terminus | No. of TMDs | Comments |
|---|---|---|---|---|---|---|
| I | CcmC | -WGX(F/W/Y)WXW(D/E)XR- | Two conserved His residues flanking the WWD domain | Inside | 6 | |
| CcmD | -(G/A)XY- | Outside | 1 | CcmD is always downstream of CcmC if it is present in bacteria; CcmD often shows as a small hypothetical protein of 40-80 amino acids; most organisms have the conserved -GXY- motif, with the exception of some alphaproteobacteria which have -AXY- | ||
| CcmE | -(C/H)XXXY- | Inside | 1 | Most organisms have a His in the conserved motif; some archaea and deinococci have a Cys instead. | ||
| CcmF | -W(S/A)YXXLGWGG(F/W/Y) WXWDPVENAS(F/L)(L/M/I)WL- | Four conserved His residues | Outside | 13 | Has up to 15 predicted TMDs (see Fig. 3B legend) | |
| CcmH/CcmI | -LRCXXC- (thioredox domain), TPR repeat domains | Outside | 2 | In most organisms, the E. coli CcmH is divided into two proteins, CcmH (Ccl2) and CcmI (CycH); CcmH is amino acids 1-127 of the E. coli protein and has one TMD and contains the -LRCXXC- motif; CcmI is a TPR repeat protein with one TMD; inability to identify CcmH does not automatically exclude an organism from being classified as system I | ||
| II | CcsA | -WAXX(A/S)WGX(F/Y)WXWDXKEXX- | Three conserved His residues, two flanking the WWD domain and one in a TMD on the n side | Outside | 6 | CcsA proteins may have 8 predicted TMDs (see text); they are also preceded or followed by CcsB in bacterial genomes |
| CcsB | Large soluble domain between TMDs 3 and 4 | -VNXP- in soluble domain, conserved His in third TMD | Inside | 4 | CcsB proteins may have 5-6 predicted TMDs, due to hydrophobic regions in the extracellular domain; CcsB proteins have little sequence conservation, though the protein secondary structure is conserved, and they may proceed or follow CcsA in the genome. | |
| CcsBA | -WAXX(A/S)WGX(F/Y)WXWDXKEXX- | -VNXXP- in CcsB soluble domain, four conserved His residues (His-77, -716, -858, and -897; -see Fig. 5) | Inside | 10 | CcsBA proteins may have up to 11-14 predicted TMDs (see text); CcsA-like proteins with 11-14 predicted TMDs that are over 2.5 kb in length may be candidate CcsBA fusions, particularly if a superfamily search indicates the presence of a ResB superfamily domain; a homology search against a known CcsBA fusion sequence (i.e., Helicobacter) can provide additional confirmation | |
| III | CCHL | -CPX- (in N-terminal part), -PFDRHDW- (in C-terminal part) | See Pfam 01265 | Mitochondrial intermembrane space | 0 | CCHL proteins are soluble proteins that lack a typical mitochondrial targeting sequence; The N termini are poorly conserved but contain several -CPX- heme-responsive motifs (with two known CC1HL exceptions); proteins that group to Pfam 01265 and contain -CPX- motifs are likely CCHL |
FUTURE STUDIES
Throughout this review we have noted some particularly important studies that are needed in the field. Recall that the structures for only a few proteins in systems I and II have been determined, and then only soluble, truncated versions that are typically nonfunctional (CcmE*, CcmG*, CcsX* [ResA], and CcmH* [periplasmic domains] [see, e.g., references 52 and 53] and others cited above). The crystal structure of a WWD domain (heme-handling) protein (CcmC, CcmF, or CcsA), with and without heme, remains a critical need. Although some in the field have proposed that the WWD domain interacts with the CcmE polypeptide component (70, 124), the presence of the WWD domain in CcsA (system II), where no CcmE operates, suggests otherwise. We suggest that the fundamental purpose of the WWD domain is as an external scaffold for heme, whereby the vinyl groups are surface exposed for interaction with various proteins, yielding a heme environment (including the flanking histidines as axial ligands to the iron) that is ideal for the necessary vinyl reactivity. The three-dimensional structures of the heme o and a synthases are important in and of themselves but also for comparison with the cytochrome c synthetases (CcmF and CcsA) that are discussed in this review. A CCHL structure, with and without substrates, will be very interesting.
It would be invaluable to reconstitute in vitro the holoCcmE release in step 1 and the step 2 synthetase reaction for system I and purified system II (CcsBA) and III (CCHL) synthetases. We need to learn much about the synthetase mechanisms for all three systems. Understanding the details of heme movement into or through the membrane, through channels or by other mechanisms, has been discussed above. Finally, more ecological and evolutionary studies are needed for further speculation on the advantages of each system.
Acknowledgments
We thank Brian San Francisco for analysis of the natural distribution of systems, Kevin Moeller for discussions on imidazole reactivities, and Patrick Lane for illustrations.
R.G.K. was supported by NIH grant GM 47909.
REFERENCES
- 1.Ahuja, U., A. Rozhkova, R. Glockshuber, L. Thony-Meyer, and O. Einsle. 2008. Helix swapping leads to dimerization of the N-terminal domain of the c-type cytochrome maturation protein CcmH from Escherichia coli. FEBS Lett. 5822779-2786. [DOI] [PubMed] [Google Scholar]
- 2.Ahuja, U., and L. Thony-Meyer. 2003. Dynamic features of a heme delivery system for cytochrome C maturation. J. Biol. Chem. 27852061-52070. [DOI] [PubMed] [Google Scholar]
- 3.Allen, J. W., P. D. Barker, O. Daltrop, J. M. Stevens, E. J. Tomlinson, N. Sinha, Y. Sambongi, and S. J. Ferguson. 2005. Why isn't ‘standard’ heme good enough for c-type and d1-type cytochromes? Dalton Trans. 20053410-3418. [DOI] [PubMed] [Google Scholar]
- 4.Allen, J. W., P. D. Barker, and S. J. Ferguson. 2003. A cytochrome b562 variant with a c-type cytochrome CXXCH heme-binding motif as a probe of the Escherichia coli cytochrome c maturation system. J. Biol. Chem. 27852075-52083. [DOI] [PubMed] [Google Scholar]
- 5.Allen, J. W., O. Daltrop, J. M. Stevens, and S. J. Ferguson. 2003. C-type cytochromes: diverse structures and biogenesis systems pose evolutionary problems. Philos. Trans. R. Soc. London B 358255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen, J. W., and S. J. Ferguson. 2006. The Escherichia coli cytochrome c maturation (Ccm) apparatus can mature cytochromes with an extra cysteine within or adjacent to the CXXCH motif. Biochem. Soc. Trans. 3491-93. [DOI] [PubMed] [Google Scholar]
- 7.Allen, J. W., and S. J. Ferguson. 2003. Variation of the axial haem ligands and haem-binding motif as a probe of the Escherichia coli c-type cytochrome maturation (Ccm) system. Biochem. J. 375721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen, J. W., and S. J. Ferguson. 2006. What is the substrate specificity of the System I cytochrome c biogenesis apparatus? Biochem. Soc. Trans. 34150-151. [DOI] [PubMed] [Google Scholar]
- 9.Allen, J. W., M. L. Ginger, and S. J. Ferguson. 2005. Complexity and diversity in c-type cytochrome biogenesis systems. Biochem. Soc. Trans. 33145-146. [DOI] [PubMed] [Google Scholar]
- 10.Allen, J. W., M. L. Ginger, and S. J. Ferguson. 2004. Maturation of the unusual single-cysteine (XXXCH) mitochondrial c-type cytochromes found in trypanosomatids must occur through a novel biogenesis pathway. Biochem. J. 383537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen, J. W., E. M. Harvat, J. M. Stevens, and S. J. Ferguson. 2006. A variant System I for cytochrome c biogenesis in archaea and some bacteria has a novel CcmE and no CcmH. FEBS Lett. 5804827-4834. [DOI] [PubMed] [Google Scholar]
- 12.Allen, J. W., A. P. Jackson, D. J. Rigden, A. C. Willis, S. J. Ferguson, and M. L. Ginger. 2008. Order within a mosaic distribution of mitochondrial c-type cytochrome biogenesis systems? FEBS J. 2752385-2402. [DOI] [PubMed] [Google Scholar]
- 13.Allen, J. W., N. Leach, and S. J. Ferguson. 2005. The histidine of the c-type cytochrome CXXCH haem-binding motif is essential for haem attachment by the Escherichia coli cytochrome c maturation (Ccm) apparatus. Biochem. J. 389587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen, J. W., E. J. Tomlinson, L. Hong, and S. J. Ferguson. 2002. The Escherichia coli cytochrome c maturation (Ccm) system does not detectably attach heme to single cysteine variants of an apocytochrome c. J. Biol. Chem. 27733559-33563. [DOI] [PubMed] [Google Scholar]
- 15.Arnesano, F., L. Banci, P. D. Barker, I. Bertini, A. Rosato, X. C. Su, and M. S. Viezzoli. 2002. Solution structure and characterization of the heme chaperone CcmE. Biochemistry 4113587-13594. [DOI] [PubMed] [Google Scholar]
- 16.Arnesano, F., L. Banci, I. Bertini, S. Ciofi-Baffoni, T. L. Woodyear, C. M. Johnson, and P. D. Barker. 2000. Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562. Biochemistry 391499-1514. [DOI] [PubMed] [Google Scholar]
- 17.Banci, L., I. Bertini, C. Cefaro, S. Ciofi-Baffoni, A. Gallo, M. Martinelli, D. P. Sideris, N. Katrakili, and K. Tokatlidis. 2009. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 16198-206. [DOI] [PubMed] [Google Scholar]
- 18.Barker, P. D., and S. J. Ferguson. 1999. Still a puzzle: why is haem covalently attached in c-type cytochromes? Structure 7R281-R290. [DOI] [PubMed] [Google Scholar]
- 19.Barker, P. D., J. C. Ferrer, M. Mylrajan, T. M. Loehr, R. Feng, Y. Konishi, W. D. Funk, R. T. MacGillivray, and A. G. Mauk. 1993. Transmutation of a heme protein. Proc. Natl. Acad. Sci. USA 906542-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barker, P. D., E. P. Nerou, S. M. Freund, and I. M. Fearnley. 1995. Conversion of cytochrome b562 to c-type cytochromes. Biochemistry 3415191-15203. [DOI] [PubMed] [Google Scholar]
- 21.Barrick, D. 1995. Depletion and replacement of protein metal ligands. Curr. Opin. Biotechnol. 6411-418. [DOI] [PubMed] [Google Scholar]
- 22.Barrick, D. 1994. Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93→Gly. Biochemistry 336546-6554. [DOI] [PubMed] [Google Scholar]
- 23.Beckett, C. S., J. A. Loughman, K. A. Karberg, G. M. Donato, W. E. Goldman, and R. G. Kranz. 2000. Four genes are required for the system II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol. Microbiol. 38465-481. [DOI] [PubMed] [Google Scholar]
- 24.Beckman, D. L., and R. G. Kranz. 1993. Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc. Natl. Acad. Sci. USA 902179-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beckman, D. L., D. R. Trawick, and R. G. Kranz. 1992. Bacterial cytochromes c biogenesis. Genes Dev. 6268-283. [DOI] [PubMed] [Google Scholar]
- 26.Bernard, D. G., S. T. Gabilly, G. Dujardin, S. Merchant, and P. P. Hamel. 2003. Overlapping specificities of the mitochondrial cytochrome c and c1 heme lyases. J. Biol. Chem. 27849732-49742. [DOI] [PubMed] [Google Scholar]
- 27.Bernard, D. G., S. Quevillon-Cheruel, S. Merchant, B. Guiard, and P. P. Hamel. 2005. Cyc2p, a membrane-bound flavoprotein involved in the maturation of mitochondrial c-type cytochromes. J. Biol. Chem. 28039852-39859. [DOI] [PubMed] [Google Scholar]
- 28.Bowman, S. E., and K. L. Bren. 2008. The chemistry and biochemistry of heme c: functional bases for covalent attachment. Nat. Prod. Rep. 251118-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braun, M., and L. Thony-Meyer. 2004. Biosynthesis of artificial microperoxidases by exploiting the secretion and cytochrome c maturation apparatuses of Escherichia coli. Proc. Natl. Acad. Sci. USA 10112830-12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown, B. M., Z. Wang, K. R. Brown, J. A. Cricco, and E. L. Hegg. 2004. Heme O synthase and heme A synthase from Bacillus subtilis and Rhodobacter sphaeroides interact in Escherichia coli. Biochemistry 4313541-13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown, K. R., B. M. Allan, P. Do, and E. L. Hegg. 2002. Identification of novel hemes generated by heme A synthase: evidence for two successive monooxygenase reactions. Biochemistry 4110906-10913. [DOI] [PubMed] [Google Scholar]
- 32.Brown, K. R., B. M. Brown, E. Hoagland, C. L. Mayne, and E. L. Hegg. 2004. Heme A synthase does not incorporate molecular oxygen into the formyl group of heme A. Biochemistry 438616-8624. [DOI] [PubMed] [Google Scholar]
- 33.Chang, G. 2003. Structure of MsbA from Vibrio cholerae: a multidrug resistance ABC transporter homolog in a closed conformation. J. Mol. Biol. 330419-430. [DOI] [PubMed] [Google Scholar]
- 34.Chepuri, V., and R. B. Gennis. 1990. The use of gene fusions to determine the topology of all of the subunits of the cytochrome o terminal oxidase complex of Escherichia coli. J. Biol. Chem. 26512978-12986. [PubMed] [Google Scholar]
- 35.Chepuri, V., L. Lemieux, D. C. Au, and R. B. Gennis. 1990. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J. Biol. Chem. 26511185-11192. [PubMed] [Google Scholar]
- 36.Chepuri, V., L. Lemieux, J. Hill, J. O. Alben, and R. B. Gennis. 1990. Recent studies of the cytochrome o terminal oxidase complex of Escherichia coli. Biochim. Biophys. Acta 1018124-127. [DOI] [PubMed] [Google Scholar]
- 37.Christensen, O., E. M. Harvat, L. Thony-Meyer, S. J. Ferguson, and J. M. Stevens. 2007. Loss of ATP hydrolysis activity by CcmAB results in loss of c-type cytochrome synthesis and incomplete processing of CcmE. FEBS J. 2742322-2332. [DOI] [PubMed] [Google Scholar]
- 38.Cinege, G., A. Kereszt, S. Kertesz, G. Balogh, and I. Dusha. 2004. The roles of different regions of the CycH protein in c-type cytochrome biogenesis in Sinorhizobium meliloti. Mol. Genet. Genomics 271171-179. [DOI] [PubMed] [Google Scholar]
- 39.Daltrop, O., J. W. Allen, A. C. Willis, and S. J. Ferguson. 2002. In vitro formation of a c-type cytochrome. Proc. Natl. Acad. Sci. USA 997872-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daltrop, O., and S. J. Ferguson. 2004. In vitro studies on thioether bond formation between Hydrogenobacter thermophilus apocytochrome c(552) with metalloprotoporphyrin derivatives. J. Biol. Chem. 27945347-45353. [DOI] [PubMed] [Google Scholar]
- 41.Daltrop, O., K. M. Smith, and S. J. Ferguson. 2003. Stereoselective in vitro formation of c-type cytochrome variants from Hydrogenobacter thermophilus containing only a single thioether bond. J. Biol. Chem. 27824308-24313. [DOI] [PubMed] [Google Scholar]
- 42.Daltrop, O., J. M. Stevens, C. W. Higham, and S. J. Ferguson. 2002. The CcmE protein of the c-type cytochrome biogenesis system: unusual in vitro heme incorporation into apo-CcmE and transfer from holo-CcmE to apocytochrome. Proc. Natl. Acad. Sci. USA 999703-9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deshmukh, M., G. Brasseur, and F. Daldal. 2000. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol. Microbiol. 35123-138. [DOI] [PubMed] [Google Scholar]
- 44.Deshmukh, M., M. May, Y. Zhang, K. K. Gabbert, K. A. Karberg, R. G. Kranz, and F. Daldal. 2002. Overexpression of ccl1-2 can bypass the need for the putative apocytochrome chaperone CycH during the biogenesis of c-type cytochromes. Mol. Microbiol. 461069-1080. [DOI] [PubMed] [Google Scholar]
- 45.Di Matteo, A., S. Gianni, M. E. Schinina, A. Giorgi, F. Altieri, N. Calosci, M. Brunori, and C. Travaglini-Allocatelli. 2007. A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c. J. Biol. Chem. 28227012-27019. [DOI] [PubMed] [Google Scholar]
- 46.Doerrler, W. T., and C. R. Raetz. 2002. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 27736697-36705. [DOI] [PubMed] [Google Scholar]
- 47.Dreyfuss, B. W., P. P. Hamel, S. S. Nakamoto, and S. Merchant. 2003. Functional analysis of a divergent system II protein, Ccs1, involved in c-type cytochrome biogenesis. J. Biol. Chem. 2782604-2613. [DOI] [PubMed] [Google Scholar]
- 48.Drygas, M. E., A. M. Lambowitz, and F. E. Nargang. 1989. Cloning and analysis of the Neurospora crassa gene for cytochrome c heme lyase. J. Biol. Chem. 26417897-17906. [PubMed] [Google Scholar]
- 49.Dumont, M. E., J. F. Ernst, D. M. Hampsey, and F. Sherman. 1987. Identification and sequence of the gene encoding cytochrome c heme lyase in the yeast Saccharomyces cerevisiae. EMBO J. 6235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dumont, M. E., J. F. Ernst, and F. Sherman. 1988. Coupling of heme attachment to import of cytochrome c into yeast mitochondria. Studies with heme lyase-deficient mitochondria and altered apocytochromes c. J. Biol. Chem. 26315928-15937. [PubMed] [Google Scholar]
- 51.Eaves, D. J., J. Grove, W. Staudenmann, P. James, R. K. Poole, S. A. White, I. Griffiths, and J. A. Cole. 1998. Involvement of products of the nrfEFG genes in the covalent attachment of haem c to a novel cysteine-lysine motif in the cytochrome c552 nitrite reductase from Escherichia coli. Mol. Microbiol. 28205-216. [DOI] [PubMed] [Google Scholar]
- 52.Edeling, M. A., L. W. Guddat, R. A. Fabianek, J. A. Halliday, A. Jones, L. Thony-Meyer, and J. L. Martin. 2001. Crystallization and preliminary diffraction studies of native and selenomethionine CcmG (CycY, DsbE). Acta Crystallogr. D 571293-1295. [DOI] [PubMed] [Google Scholar]
- 53.Edeling, M. A., L. W. Guddat, R. A. Fabianek, L. Thony-Meyer, and J. L. Martin. 2002. Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment. Structure 10973-979. [DOI] [PubMed] [Google Scholar]
- 54.Enggist, E., M. J. Schneider, H. Schulz, and L. Thony-Meyer. 2003. Biochemical and mutational characterization of the heme chaperone CcmE reveals a heme binding site. J. Bacteriol. 185175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feissner, R. E., C. S. Beckett, J. A. Loughman, and R. G. Kranz. 2005. Mutations in cytochrome assembly and periplasmic redox pathways in Bordetella pertussis. J. Bacteriol. 1873941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feissner, R. E., C. L. Richard-Fogal, E. R. Frawley, and R. G. Kranz. 2006. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol. Microbiol. 61219-231. [DOI] [PubMed] [Google Scholar]
- 57.Feissner, R. E., C. L. Richard-Fogal, E. R. Frawley, J. A. Loughman, K. W. Earley, and R. G. Kranz. 2006. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol. Microbiol. 60563-577. [DOI] [PubMed] [Google Scholar]
- 58.Ferguson, S. J., J. M. Stevens, J. W. Allen, and I. B. Robertson. 2008. Cytochrome c assembly: a tale of ever increasing variation and mystery? Biochim. Biophys. Acta 1777980-984. [DOI] [PubMed] [Google Scholar]
- 59.Frawley, E. R., and R. G. Kranz. 2009. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc. Natl. Acad. Sci. USA 10610201-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fulop, V., K. A. Sam, S. J. Ferguson, M. L. Ginger, and J. W. Allen. 2009. Structure of a trypanosomatid mitochondrial cytochrome c with heme attached via only one thioether bond and implications for the substrate recognition requirements of heme lyase. FEBS J. 2762822-2832. [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Rubio, I., M. Braun, I. Gromov, L. Thony-Meyer, and A. Schweiger. 2007. Axial coordination of heme in ferric CcmE chaperone characterized by EPR spectroscopy. Biophys. J. 921361-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldman, B. S., D. L. Beck, E. M. Monika, and R. G. Kranz. 1998. Transmembrane heme delivery systems. Proc. Natl. Acad. Sci. USA 955003-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldman, B. S., D. L. Beckman, A. Bali, E. M. Monika, K. K. Gabbert, and R. G. Kranz. 1997. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J. Mol. Biol. 268724-738. [DOI] [PubMed] [Google Scholar]
- 64.Goldman, B. S., K. K. Gabbert, and R. G. Kranz. 1996. Use of heme reporters for studies of cytochrome biosynthesis and heme transport. J. Bacteriol. 1786338-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goldman, B. S., and R. G. Kranz. 2001. ABC transporters associated with cytochrome c biogenesis. Res. Microbiol. 152323-329. [DOI] [PubMed] [Google Scholar]
- 66.Goldman, B. S., and R. G. Kranz. 1998. Evolution and horizontal transfer of an entire biosynthetic pathway for cytochrome c biogenesis: Helicobacter, Deinococcus, Archae and more. Mol. Microbiol. 27871-873. [DOI] [PubMed] [Google Scholar]
- 67.Gralnick, J. A., and D. K. Newman. 2007. Extracellular respiration. Mol. Microbiol. 651-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gray, M. W. 1993. Origin and evolution of organelle genomes. Curr. Opin. Genet. Dev. 3884-890. [DOI] [PubMed] [Google Scholar]
- 69.Green, G. N., H. Fang, R. J. Lin, G. Newton, M. Mather, C. D. Georgiou, and R. B. Gennis. 1988. The nucleotide sequence of the cyd locus encoding the two subunits of the cytochrome d terminal oxidase complex of Escherichia coli. J. Biol. Chem. 26313138-13143. [PubMed] [Google Scholar]
- 70.Hamel, P., V. Corvest, P. Giege, and G. Bonnard. 2009. Biochemical requirements for the maturation of mitochondrial c-type cytochromes. Biochim. Biophys. Acta 1793125-138. [DOI] [PubMed] [Google Scholar]
- 71.Hamel, P. P., B. W. Dreyfuss, Z. Xie, S. T. Gabilly, and S. Merchant. 2003. Essential histidine and tryptophan residues in CcsA, a system II polytopic cytochrome c biogenesis protein. J. Biol. Chem. 2782593-2603. [DOI] [PubMed] [Google Scholar]
- 72.Handa, H., G. Bonnard, and J. M. Grienenberger. 1996. The rapeseed mitochondrial gene encoding a homologue of the bacterial protein Ccl1 is divided into two independently transcribed reading frames. Mol. Gen. Genet. 252292-302. [DOI] [PubMed] [Google Scholar]
- 73.Hartshorne, R. S., M. Kern, B. Meyer, T. A. Clarke, M. Karas, D. J. Richardson, and J. Simon. 2007. A dedicated haem lyase is required for the maturation of a novel bacterial cytochrome c with unconventional covalent haem binding. Mol. Microbiol. 641049-1060. [DOI] [PubMed] [Google Scholar]
- 74.Hartshorne, S., D. J. Richardson, and J. Simon. 2006. Multiple haem lyase genes indicate substrate specificity in cytochrome c biogenesis. Biochem. Soc. Trans. 34146-149. [DOI] [PubMed] [Google Scholar]
- 75.Harvat, E. M., C. Redfield, J. M. Stevens, and S. J. Ferguson. 2009. Probing the heme-binding site of the cytochrome c maturation protein CcmE. Biochemistry 481820-1828. [DOI] [PubMed] [Google Scholar]
- 76.Hederstedt, L., A. Lewin, and M. Throne-Holst. 2005. Heme A synthase enzyme functions dissected by mutagenesis of Bacillus subtilis CtaA. J. Bacteriol. 1878361-8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Higgins, C. F., and M. M. Gottesman. 1992. Is the multidrug transporter a flippase? Trends Biochem. Sci. 1718-21. [DOI] [PubMed] [Google Scholar]
- 78.Huang, L., G. Wojciechowski, and P. R. Ortiz de Montellano. 2006. Role of heme-protein covalent bonds in mammalian peroxidases. Protection of the heme by a single engineered heme-protein link in horseradish peroxidase. J. Biol. Chem. 28118983-18988. [DOI] [PubMed] [Google Scholar]
- 79.Hurd, T. R., T. A. Prime, M. E. Harbour, K. S. Lilley, and M. P. Murphy. 2007. Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J. Biol. Chem. 28222040-22051. [DOI] [PubMed] [Google Scholar]
- 80.Inoue, K., B. W. Dreyfuss, K. L. Kindle, D. B. Stern, S. Merchant, and O. A. Sodeinde. 1997. Ccs1, a nuclear gene required for the post-translational assembly of chloroplast c-type cytochromes. J. Biol. Chem. 27231747-31754. [DOI] [PubMed] [Google Scholar]
- 81.Ito, Y., K. Kanamaru, N. Taniguchi, S. Miyamoto, and H. Tokuda. 2006. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Mol. Microbiol. 621064-1075. [DOI] [PubMed] [Google Scholar]
- 82.Johansson, C., C. H. Lillig, and A. Holmgren. 2004. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J. Biol. Chem. 2797537-7543. [DOI] [PubMed] [Google Scholar]
- 83.Kol, M. A., A. van Dalen, A. I. de Kroon, and B. de Kruijff. 2003. Translocation of phospholipids is facilitated by a subset of membrane-spanning proteins of the bacterial cytoplasmic membrane. J. Biol. Chem. 27824586-24593. [DOI] [PubMed] [Google Scholar]
- 84.Kranz, R., R. Lill, B. Goldman, G. Bonnard, and S. Merchant. 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29383-396. [DOI] [PubMed] [Google Scholar]
- 85.Kranz, R. G. 1989. Isolation of mutants and genes involved in cytochromes c biosynthesis in Rhodobacter capsulatus. J. Bacteriol. 171456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kranz, R. G., C. S. Beckett, and B. S. Goldman. 2002. Genomic analyses of bacterial respiratory and cytochrome c assembly systems: Bordetella as a model for the system II cytochrome c biogenesis pathway. Res. Microbiol. 1531-6. [DOI] [PubMed] [Google Scholar]
- 87.Kuras, R., C. de Vitry, Y. Choquet, J. Girard-Bascou, D. Culler, S. Buschlen, S. Merchant, and F. A. Wollman. 1997. Molecular genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 27232427-32435. [DOI] [PubMed] [Google Scholar]
- 88.Kuras, R., D. Saint-Marcoux, F. A. Wollman, and C. de Vitry. 2007. A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 1049906-9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kurland, C. G., and S. G. Andersson. 2000. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 64786-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lang, B. F., E. Seif, M. W. Gray, C. J. O'Kelly, and G. Burger. 1999. A comparative genomics approach to the evolution of eukaryotes and their mitochondria. J. Eukaryot. Microbiol. 46320-326. [DOI] [PubMed] [Google Scholar]
- 91.Lang, S. E., F. E. Jenney, Jr., and F. Daldal. 1996. Rhodobacter capsulatus CycH: a bipartite gene product with pleiotropic effects on the biogenesis of structurally different c-type cytochromes. J. Bacteriol. 1785279-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Le Brun, N. E., J. Bengtsson, and L. Hederstedt. 2000. Genes required for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol. 36638-650. [DOI] [PubMed] [Google Scholar]
- 93.Lee, D., K. Pervushin, D. Bischof, M. Braun, and L. Thony-Meyer. 2005. Unusual heme-histidine bond in the active site of a chaperone. J. Am. Chem. Soc. 1273716-3717. [DOI] [PubMed] [Google Scholar]
- 94.Lee, J. H., E. M. Harvat, J. M. Stevens, S. J. Ferguson, and M. H. Saier, Jr. 2007. Evolutionary origins of members of a superfamily of integral membrane cytochrome c biogenesis proteins. Biochim. Biophys. Acta 17682164-2181. [DOI] [PubMed] [Google Scholar]
- 95.Lezhneva, L., R. Kuras, G. Ephritikhine, and C. de Vitry. 2008. A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 28324608-24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lundberg, M., C. Johansson, J. Chandra, M. Enoksson, G. Jacobsson, J. Ljung, M. Johansson, and A. Holmgren. 2001. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J. Biol. Chem. 27626269-26275. [DOI] [PubMed] [Google Scholar]
- 97.Masuda, K., S. Matsuyama, and H. Tokuda. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. USA 997390-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 99.Merchant, S. S. 2009. His protects heme as it crosses the membrane. Proc. Natl. Acad. Sci. USA 10610069-10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Metcalfe, C. L., O. Daltrop, S. J. Ferguson, and E. L. Raven. 2007. Tuning the formation of a covalent haem-protein link by selection of reductive or oxidative conditions as exemplified by ascorbate peroxidase. Biochem. J. 408355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Metheringham, R., K. L. Tyson, H. Crooke, D. Missiakas, S. Raina, and J. A. Cole. 1996. Effects of mutations in genes for proteins involved in disulphide bond formation in the periplasm on the activities of anaerobically induced electron transfer chains in Escherichia coli K12. Mol. Gen. Genet. 25395-102. [DOI] [PubMed] [Google Scholar]
- 102.Meyer, E. H., P. Giege, E. Gelhaye, N. Rayapuram, U. Ahuja, L. Thony-Meyer, J. M. Grienenberger, and G. Bonnard. 2005. AtCCMH, an essential component of the c-type cytochrome maturation pathway in Arabidopsis mitochondria, interacts with apocytochrome c. Proc. Natl. Acad. Sci. USA 10216113-16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Monika, E. M., B. S. Goldman, D. L. Beckman, and R. G. Kranz. 1997. A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis; in vitro and in vivo studies. J. Mol. Biol. 271679-692. [DOI] [PubMed] [Google Scholar]
- 104.Mueller, J. P., and H. W. Taber. 1989. Isolation and sequence of ctaA, a gene required for cytochrome aa3 biosynthesis and sporulation in Bacillus subtilis. J. Bacteriol. 1714967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nicholson, D. W., H. Kohler, and W. Neupert. 1987. Import of cytochrome c into mitochondria. Cytochrome c heme lyase. Eur. J. Biochem. 164147-157. [DOI] [PubMed] [Google Scholar]
- 106.Nicholson, D. W., and W. Neupert. 1989. Import of cytochrome c into mitochondria: reduction of heme, mediated by NADH and flavin nucleotides, is obligatory for its covalent linkage to apocytochrome c. Proc. Natl. Acad. Sci. USA 864340-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nicholson, D. W., R. A. Stuart, and W. Neupert. 1989. Biogenesis of cytochrome c1. Role of cytochrome c1 heme lyase and of the two proteolytic processing steps during import into mitochondria. J. Biol. Chem. 26410156-10168. [PubMed] [Google Scholar]
- 108.Nothnagel, H. J., N. Love, and J. T. Lecomte. 2009. The role of the heme distal ligand in the post-translational modification of Synechocystis hemoglobin. J. Inorg. Biochem. 103107-116. [DOI] [PubMed] [Google Scholar]
- 109.O'Brian, M. R., and L. Thony-Meyer. 2002. Biochemistry, regulation and genomics of haem biosynthesis in prokaryotes. Adv. Microb. Physiol. 46257-318. [DOI] [PubMed] [Google Scholar]
- 110.Oguchi, Y., K. Takeda, S. Watanabe, N. Yokota, K. Miki, and H. Tokuda. 2008. Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release. J. Biol. Chem. 28325414-25420. [DOI] [PubMed] [Google Scholar]
- 111.Page, M. D., and S. J. Ferguson. 1995. Cloning and sequence analysis of cycH gene from Paracoccus denitrificans: the cycH gene product is required for assembly of all c-type cytochromes, including cytochrome c1. Mol. Microbiol. 15307-318. [DOI] [PubMed] [Google Scholar]
- 112.Page, M. D., and S. J. Ferguson. 1999. Mutational analysis of the Paracoccus denitrificans c-type cytochrome biosynthetic genes ccmABCDG: disruption of ccmC has distinct effects suggesting a role for CcmC independent of CcmAB. Microbiology 1453047-3057. [DOI] [PubMed] [Google Scholar]
- 113.Page, M. D., and S. J. Ferguson. 1997. Paracoccus denitrificans CcmG is a periplasmic protein-disulphide oxidoreductase required for c- and aa3-type cytochrome biogenesis; evidence for a reductase role in vivo. Mol. Microbiol. 24977-990. [DOI] [PubMed] [Google Scholar]
- 114.Page, M. D., D. A. Pearce, H. A. Norris, and S. J. Ferguson. 1997. The Paracoccus denitrificans ccmA, B and C genes: cloning and sequencing, and analysis of the potential of their products to form a haem or apo-c-type cytochrome transporter. Microbiology 143563-576. [DOI] [PubMed] [Google Scholar]
- 115.Page, M. D., Y. Sambongi, and S. J. Ferguson. 1998. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem. Sci. 23103-108. [DOI] [PubMed] [Google Scholar]
- 116.Page, M. L., P. P. Hamel, S. T. Gabilly, H. Zegzouti, J. V. Perea, J. M. Alonso, J. R. Ecker, S. M. Theg, S. K. Christensen, and S. Merchant. 2004. A homolog of prokaryotic thiol disulfide transporter CcdA is required for the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 27932474-32482. [DOI] [PubMed] [Google Scholar]
- 117.Pisa, R., T. Stein, R. Eichler, R. Gross, and J. Simon. 2002. The nrfI gene is essential for the attachment of the active site haem group of Wolinella succinogenes cytochrome c nitrite reductase. Mol. Microbiol. 43763-770. [DOI] [PubMed] [Google Scholar]
- 118.Pollock, W. B., F. I. Rosell, M. B. Twitchett, M. E. Dumont, and A. G. Mauk. 1998. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry 376124-6131. [DOI] [PubMed] [Google Scholar]
- 119.Poole, R. K., and G. M. Cook. 2000. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv. Microb. Physiol. 43165-224. [DOI] [PubMed] [Google Scholar]
- 120.Ramseier, T. M., H. V. Winteler, and H. Hennecke. 1991. Discovery and sequence analysis of bacterial genes involved in the biogenesis of c-type cytochromes. J. Biol. Chem. 2667793-7803. [PubMed] [Google Scholar]
- 121.Rayapuram, N., J. Hagenmuller, J. M. Grienenberger, G. Bonnard, and P. Giege. 2008. The three mitochondrial encoded CcmF proteins form a complex that interacts with CCMH and c-type apocytochromes in Arabidopsis. J. Biol. Chem. 28325200-25208. [DOI] [PubMed] [Google Scholar]
- 122.Reid, E., D. J. Eaves, and J. A. Cole. 1998. The CcmE protein from Escherichia coli is a haem-binding protein. FEMS Microbiol. Lett. 166369-375. [DOI] [PubMed] [Google Scholar]
- 123.Ren, Q., U. Ahuja, and L. Thony-Meyer. 2002. A bacterial cytochrome c heme lyase. CcmF forms a complex with the heme chaperone CcmE and CcmH but not with apocytochrome c. J. Biol. Chem. 2777657-7663. [DOI] [PubMed] [Google Scholar]
- 124.Ren, Q., and L. Thony-Meyer. 2001. Physical interaction of CcmC with heme and the heme chaperone CcmE during cytochrome c maturation. J. Biol. Chem. 27632591-32596. [DOI] [PubMed] [Google Scholar]
- 125.Richard-Fogal, C. L., E. R. Frawley, E. R. Bonner, H. Zhu, B. San Francisco, and R. G. Kranz. 23 July 2009, posting date. A conserved heme redox and trafficking pathway for cofactor attachment. EMBO J. doi: 10.1038/emboj.2009.189. [DOI] [PMC free article] [PubMed]
- 126.Richard-Fogal, C. L., E. R. Frawley, R. E. Feissner, and R. G. Kranz. 2007. Heme concentration dependence and metalloporphyrin inhibition of the system I and II cytochrome c assembly pathways. J. Bacteriol. 189455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Richard-Fogal, C. L., E. R. Frawley, and R. G. Kranz. 2008. Topology and function of CcmD in cytochrome c maturation. J. Bacteriol. 1903489-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Richardson, D. J. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology 146551-571. [DOI] [PubMed] [Google Scholar]
- 129.Rios-Velazquez, C., R. Coller, and T. J. Donohue. 2003. Features of Rhodobacter sphaeroides CcmFH. J. Bacteriol. 185422-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rios-Velazquez, C., R. L. Cox, and T. J. Donohue. 2001. Characterization of Rhodobacter sphaeroides cytochrome c(2) proteins with altered heme attachment sites. Arch. Biochem. Biophys. 389234-244. [DOI] [PubMed] [Google Scholar]
- 131.Ritz, D., L. Thony-Meyer, and H. Hennecke. 1995. The cycHJKL gene cluster plays an essential role in the biogenesis of c-type cytochromes in Bradyrhizobium japonicum. Mol. Gen. Genet. 24727-38. [DOI] [PubMed] [Google Scholar]
- 132.Robertson, I. B., J. M. Stevens, and S. J. Ferguson. 2008. Dispensable residues in the active site of the cytochrome c biogenesis protein CcmH. FEBS Lett. 5823067-3072. [DOI] [PubMed] [Google Scholar]
- 133.Saiki, K., T. Mogi, and Y. Anraku. 1992. Heme O biosynthesis in Escherichia coli: the cyoE gene in the cytochrome bo operon encodes a protoheme IX farnesyltransferase. Biochem. Biophys. Res. Commun. 1891491-1497. [DOI] [PubMed] [Google Scholar]
- 134.Saiki, K., T. Mogi, H. Hori, M. Tsubaki, and Y. Anraku. 1993. Identification of the functional domains in heme O synthase. Site-directed mutagenesis studies on the cyoE gene of the cytochrome bo operon in Escherichia coli. J. Biol. Chem. 26826927-26934. [PubMed] [Google Scholar]
- 135.Saiki, K., T. Mogi, K. Ogura, and Y. Anraku. 1993. In vitro heme O synthesis by the cyoE gene product from Escherichia coli. J. Biol. Chem. 26826041-26044. [PubMed] [Google Scholar]
- 136.Sanders, C., C. Boulay, and F. Daldal. 2007. Membrane-spanning and periplasmic segments of CcmI have distinct functions during cytochrome c biogenesis in Rhodobacter capsulatus. J. Bacteriol. 189789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sanders, C., M. Deshmukh, D. Astor, R. G. Kranz, and F. Daldal. 2005. Overproduction of CcmG and CcmFH(Rc) fully suppresses the c-type cytochrome biogenesis defect of Rhodobacter capsulatus CcmI-null mutants. J. Bacteriol. 1874245-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sanders, C., S. Turkarslan, D. W. Lee, O. Onder, R. G. Kranz, and F. Daldal. 2008. The cytochrome c maturation components CcmF, CcmH, and CcmI form a membrane-integral multisubunit heme ligation complex. J. Biol. Chem. 28329715-29722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schiott, T., M. Throne-Holst, and L. Hederstedt. 1997. Bacillus subtilis CcdA-defective mutants are blocked in a late step of cytochrome c biogenesis. J. Bacteriol. 1794523-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schiott, T., C. von Wachenfeldt, and L. Hederstedt. 1997. Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis. J. Bacteriol. 1791962-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schulz, H., R. A. Fabianek, E. C. Pellicioli, H. Hennecke, and L. Thony-Meyer. 1999. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc. Natl. Acad. Sci. USA 966462-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schulz, H., H. Hennecke, and L. Thony-Meyer. 1998. Prototype of a heme chaperone essential for cytochrome c maturation. Science 2811197-1200. [DOI] [PubMed] [Google Scholar]
- 143.Shi, L., T. C. Squier, J. M. Zachara, and J. K. Fredrickson. 2007. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 6512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Soderdahl, T., M. Enoksson, M. Lundberg, A. Holmgren, O. P. Ottersen, S. Orrenius, G. Bolcsfoldi, and I. A. Cotgreave. 2003. Visualization of the compartmentalization of glutathione and protein-glutathione mixed disulfides in cultured cells. FASEB J. 17124-126. [DOI] [PubMed] [Google Scholar]
- 145.Steiner, H., G. Kispal, A. Zollner, A. Haid, W. Neupert, and R. Lill. 1996. Heme binding to a conserved Cys-Pro-Val motif is crucial for the catalytic function of mitochondrial heme lyases. J. Biol. Chem. 27132605-32611. [DOI] [PubMed] [Google Scholar]
- 146.Stevens, J. M., O. Daltrop, J. W. Allen, and S. J. Ferguson. 2004. C-type cytochrome formation: chemical and biological enigmas. Acc. Chem. Res. 37999-1007. [DOI] [PubMed] [Google Scholar]
- 147.Stevens, J. M., O. Daltrop, C. W. Higham, and S. J. Ferguson. 2003. Interaction of heme with variants of the heme chaperone CcmE carrying active site mutations and a cleavable N-terminal His tag. J. Biol. Chem. 27820500-20506. [DOI] [PubMed] [Google Scholar]
- 148.Stevens, J. M., T. Uchida, O. Daltrop, T. Kitagawa, and S. J. Ferguson. 2006. Dynamic ligation properties of the Escherichia coli heme chaperone CcmE to non-covalently bound heme. J. Biol. Chem. 2816144-6151. [DOI] [PubMed] [Google Scholar]
- 149.Svensson, B., K. K. Andersson, and L. Hederstedt. 1996. Low-spin heme A in the heme A biosynthetic protein CtaA from Bacillus subtilis. Eur. J. Biochem. 238287-295. [DOI] [PubMed] [Google Scholar]
- 150.Svensson, B., and L. Hederstedt. 1994. Bacillus subtilis CtaA is a heme-containing membrane protein involved in heme A biosynthesis. J. Bacteriol. 1766663-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Svensson, B., M. Lubben, and L. Hederstedt. 1993. Bacillus subtilis CtaA and CtaB function in haem A biosynthesis. Mol. Microbiol. 10193-201. [DOI] [PubMed] [Google Scholar]
- 152.Swem, L. R., X. Gong, C. A. Yu, and C. E. Bauer. 2006. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J. Biol. Chem. 2816768-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Taniguchi, N., and H. Tokuda. 2008. Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J. Biol. Chem. 2838538-8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Thony-Meyer, L. 1997. Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61337-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Thony-Meyer, L. 2000. Haem-polypeptide interactions during cytochrome c maturation. Biochim. Biophys. Acta 1459316-324. [DOI] [PubMed] [Google Scholar]
- 156.Throne-Holst, M., and L. Hederstedt. 2000. The Bacillus subtilis ctaB paralogue, yjdK, can complement the heme A synthesis deficiency of a CtaB-deficient mutant. FEMS Microbiol. Lett. 183247-251. [DOI] [PubMed] [Google Scholar]
- 157.Turkarslan, S., C. Sanders, and F. Daldal. 2006. Extracytoplasmic prosthetic group ligation to apoproteins: maturation of c-type cytochromes. Mol. Microbiol. 60537-541. [DOI] [PubMed] [Google Scholar]
- 158.Uchida, T., J. M. Stevens, O. Daltrop, E. M. Harvat, L. Hong, S. J. Ferguson, and T. Kitagawa. 2004. The interaction of covalently bound heme with the cytochrome c maturation protein CcmE. J. Biol. Chem. 27951981-51988. [DOI] [PubMed] [Google Scholar]
- 159.Xie, Z., D. Culler, B. W. Dreyfuss, R. Kuras, F. A. Wollman, J. Girard-Bascou, and S. Merchant. 1998. Genetic analysis of chloroplast c-type cytochrome assembly in Chlamydomonas reinhardtii: one chloroplast locus and at least four nuclear loci are required for heme attachment. Genetics 148681-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xie, Z., and S. Merchant. 1996. The plastid-encoded ccsA gene is required for heme attachment to chloroplast c-type cytochromes. J. Biol. Chem. 2714632-4639. [DOI] [PubMed] [Google Scholar]
- 161.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2212-218. [DOI] [PubMed] [Google Scholar]
- 162.Zollner, A., G. Rodel, and A. Haid. 1992. Molecular cloning and characterization of the Saccharomyces cerevisiae CYT2 gene encoding cytochrome-c1-heme lyase. Eur. J. Biochem. 2071093-1100. [DOI] [PubMed] [Google Scholar]