Abstract
SIN2V is an engineered mutant Sindbis virus (SIN) that is unable to process the P23 cleavage site in polyproteins P123 and P1234 that are translated from the genome after its entry into cells. Unlike wild-type (wt) SIN, it caused minus strands to be made continuously and replication-transcription complex (RTC) activity to be unstable (R. Gorchakov, E. Frolova, S. Sawicki, S. Atasheva, D. Sawicki, and I. Frolov, J. Virol. 82:6218-6231, 2008). We examined further the effects of P23 on SIN RNA replication and RTC activity. Continuous minus-strand synthesis by SIN2V produced 250% of wt levels of minus strands but accumulated only 110% of wt levels (0.39 pg, or 2.7 × 104 molecules of double-stranded RNA per cell). Because SIN2V-infected cells accumulated only 40% of the minus strands that were made, cells must possess some process to limit RTC accumulation. The loss of activity by SIN2V RTC after translation was inhibited was stochastic and not due to their inherent instability, based on finding that activity was lost without the degradation of the minus-strand templates. In addition to their normal functions, P23 RTCs exhibited the novel phenotype of being unable to switch from making less to making more genomes than subgenomic 26S mRNA at late times during infections. Our results lend credence to the hypothesis that nsP2 (and possibly nsP3) possesses functions other than those needed solely for RTC activity and that it may also act with the host to regulate minus-strand synthesis and the stability of the RTC.
Alphaviruses are enveloped plus-strand RNA viruses, members of the family Togaviridae, that replicate in and cycle between vertebrate hosts and mosquito vectors. Sindbis virus (SIN) is a prototype alphavirus whose genome is an mRNA of 11,703 nucleotides with a 5′ type 0 cap structure and a 3′ poly(A) tail (40). After entry, the wild-type (wt) SIN genome is translated into nonstructural proteins (nsP) P123 and P1234, which are autoproteolytically and sequentially cleaved to form mature nsP1, nsP2, nsP3, and nsP4 (reviewed in references 35 and 40). Sequential cleavage of the nonstructural polyproteins was proposed to play a role in the regulation of viral minus-strand and plus-strand syntheses and to be essential to form three different polymerase activities from the P1234 precursors (14, 20, 21, 37, 44). This model suggested that release of nsP4 activated the polymerase subunit, first for minus-strand synthesis (P123 plus nsP4), followed by nsP1 cleavage that formed enzymes active in both genome plus-strand and minus-strand syntheses (nsP1, P23, and nsP4) but not active, or less active, in 26S mRNA synthesis and, finally, for P23 cleavage to release nsP2 and nsP3, a mature (nsP1 to -4) polymerase or replication-transcription complex (RTC) that efficiently recognizes the internal promoter for 26S mRNA synthesis while retaining the ability to produce genomes. Such a scheme provided explanations both for the short-lived nature of the minus-strand polymerase (33, 34) as being due to loss of promoter recognition with cleavage of P23 and for the long-lived nature of the plus-strand polymerases (30) as being due to a lack of further cleavages that would alter or block its activity.
Even when nonstructural polyproteins are synthesized throughout infection, alphavirus minus-strand synthesis normally is restricted to the early phase, within the first 4 to 7 h postinfection (p.i.), depending on the cell type, and plus-strand synthesis is a stable property of the viral RTC replicative structures. Two exceptions were identified recently (28, 29). We found that a failure to cease minus-strand synthesis and to stabilize RTC activity occurred when wt-SIN-infected RNase L-deficient mouse embryo fibroblasts (29) and when wt BHK21 cells were infected with one of two different kinds of SIN nsP2 mutants (28). The first were SIN or Semliki Forest virus single-residue nsP2 loss-of-function mutants (28), and the second were SIN polyprotein cleavage-defective mutants that produced P123 (SIN-1V2V) or P23 (SIN2V) and not mature forms of the nsPs (12). These exceptions suggested nsP2 functions were necessary for both phenotypes in wt-SIN infections and could involve a known activity of the protein or a previously unknown one. The N domain of nsP2 has RNA helicase and NTPase activities (2, 10, 17, 25) and encodes a nucleoside triphosphatase (42) that provides the first enzymatic reaction for capping the 5′ ends of both viral genomic and subgenomic RNAs (17). The C domain of nsP2 contains a papain-like protease activity responsible for autocatalytic processing of P1234 and P123 polyproteins (4, 6, 16, 43), influences both minus-strand and 26S mRNA promoter recognition (18, 31, 32, 39), and encodes nuclear, as well as nucleolar, localization signals and a 2-0-methyltransferase-like domain (2, 7, 24-26). Of the four nsPs, only nsP2 is found in the nucleus. SIN cleavage-defective mutants successfully replicated. They produced minus and plus strands, continued plus-strand synthesis after minus-strand synthesis was inhibited, and recognized the internal 26S promoter (12). This result was unexpected and meant that alphavirus polymerase activities may be enhanced by polyprotein processing but do not require it.
Finding that SIN2V was defective in two features characteristic of wt SIN suggested that each required function was expressed only by mature nsP2 (and possibly nsP3) protein. Having a mutant that continuously produced minus strands allowed us to determine if those made late had a different fate than those made early and if the P23-containing RTCs were inherently unstable, which would readily explain their activity loss. In order to do this, we developed an ethidium bromide-based assay to quantify the amount of viral replicative RNA (replication intermediates [RI]) (isolated as an RNase-resistant replicative form [RF] or cores) in SIN-infected cells independent of their activity in transcription and compared total wt-SIN RTC amounts to those in SIN2V-infected cells. In addition to these findings, we detected a novel and subtle defect in SIN2V RTC function.
MATERIALS AND METHODS
Cell cultures and viruses.
BHK21 cells (CCL 10), obtained from the American Type Culture Collection (Manassas, VA), and BHK-21 F8 cells selected in the Frolov laboratory (12) were grown in Dulbecco's minimum essential medium supplemented with penicillin (50 U/ml), streptomycin (100 U/ml), tryptose phosphate broth (5% [vol/vol]), and fetal bovine serum (6% [vol/vol]) at 37°C.
The heat-resistant strain of SIN (SIN HR), a prototype SIN, was described previously and will be referred to as wt SIN in this report (13). Virus stocks were grown in DF-1 cells that were obtained from the American Type Culture Collection (Manassas, VA). The SIN2V mutant, described previously (1, 12), has a replacement of Gly at the P2 position of nsP2 by Val and is unable to process the nsP2/3 site of the nonstructural polyproteins P23, P123, and P1234. SIN2V had the UGA (opal) codon that is located between the coding sequences for nsP3 and nsP4 changed to UGU, encoding Cys, to increase replication efficiency and the production of nsP4 (12). The same change is present in SIN/GFP, the parental virus, which does not share the phenotypes of SIN2V in minus-strand synthesis and RTC instability and replicates identically to wt SIN (12). In addition, SIN2V and SIN/GFP contain a copy of the gene encoding the green fluorescent protein (GFP) reporter between the nucleotides coding for amino acids 389 and 390 in the nsP3 sequence and in frame with the nsP3 sequence.
Infection, viral-RNA labeling, and analysis.
BHK21 cells were cultured in 35- or 60-mm-diameter petri dishes and infected with wt SIN or SIN2V at a multiplicity of infection (MOI) of 30 to 50 PFU/cell or as indicated. Infected cells were labeled with 1 ml of 5′ [3H]uridine (50 μCi/ml or 200 μCi/ml as indicated [the stock solution was 1.0 mCi/ml or 37 MBq/ml]; PE Life Sciences, Shelton, CT) in Dulbecco's minimal essential medium containing 10% fetal bovine serum, 20 mM HEPES (pH 7.4), and 20 μg/ml of actinomycin D. At the end of each pulse period, the cells were harvested with 5% lithium dodecyl sulfate in LET buffer (0.1 M LiCl, 1 mM EDTA, and 10 mM Tris-HCl, pH 7.4) containing 200 μg/ml proteinase K, and the DNA in the cell lysates was sheared by repeated passage through a 27-gauge needle (3). The [3H]uridine incorporation was determined by trichloroacetic acid precipitation. The acid-insoluble precipitates were collected, dried, and immersed in toluene-containing OmniFluor (Perkin Elmer, Boston, MA), and the radioactivity was determined with a Beckman LS 3801 (Fremont, CA). Viral and host RNAs were identified by size after separation by electrophoresis on agarose gels as described previously (3). Amounts of mock and infected cell lysates equivalent to 5 × 104 cells were electrophoresed on 1.2% agarose gels in TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA) running buffer for 400 V-hours, after which viral and cell RNAs were visualized by staining them with ethidium bromide. For fluorography, the gels were dehydrated with methanol and soaked in methanol containing 1% 2,5-diphenyloxazole and then in water, vacuum dried, and exposed to X-ray film at −80°C. Where indicated, the gel sections corresponding to the viral RNA species were excised, and the [3H]uridine incorporation in each gel section was determined by liquid scintillation counting.
Quantification of RTC RNA.
The total infected-cell RNA was obtained by extracting cell lysates first with low-pH (pH 4.3) phenol (Fisher Scientific) containing 0.4% lithium dodecyl sulfate, followed by chloroform/isoamyl alcohol (95:5 [vol/vol]), and then by ethanol precipitation. The RNA pellet was resuspended in water and any remaining DNA was eliminated by digestion with Turbo DNase (Ambion, Austin, TX) in 40 mM Tris, pH 7.9, 10 mM NaCl, 6 mM MgCl2, and 1 mM CaCl2 buffer or 1× DNase buffer provided by the manufacturer at 30°C for 30 min, after which the RNA was extracted with water-saturated phenol, followed by chloroform/isoamyl alcohol (95:5), and then ethanol precipitated. The RNA concentration was determined using a NanoDrop spectrophotometer (ND-1000; NanoDrop Technology, Inc., Wilmington, DE). The double-stranded cores (RFs) of the viral RI were obtained by limited digestion with RNase T1 (10 units/μg total cell RNA) in 0.3 M NaCl, 0.01 M Tris-HCl, pH 7.4, and 0.001 M EDTA buffer at 30°C for 30 min or with RNase A (10 ng/μg total cell RNA) in 0.3 M NaCl, 50 mM Tris, pH 7, 1 mM EDTA buffer at 30°C for 15 min, after which the RFs were electrophoresed on 1.2% agarose-TBE gels for 600 V-hours and then stained with ethidium bromide (0.5 μg/ml; 100 ml per gel) for 20 min and destained in excess water for 20 min. The optical density of the bound ethidium bromide was imaged using the Omega Molecular Imaging System. Bound ethidium bromide was quantified as image pixels within the gel area containing each RF band, and the sum of these optical densities gave the integrated optical density (IOD). The amounts of RF RNA were quantified from the IOD signal using UltraQuant version 6.0 1-D-Gels Tool software (UltraLum, Inc., CA). The resulting IOD values were plotted using KaleidaGraph version 4.0 graphing and analysis software.
Isolation of viral RF RNA species.
The RF RNA was obtained from total infected-cell RNA samples. The resuspended total RNA was treated with RNase T1 (20 U/μg RNA at 30°C for 30 min) and Turbo DNase (described above) as used previously (12). To purify the RF RNA, the samples were subjected to electrophoresis on 0.8% low-melting-temperature (LMT) agarose gels (BioWhittaker Molecular Applications, Rockland, ME) in TBE buffer for 400 V-hours, and the gels were stained with ethidium bromide. The areas of the gels containing the three individual RF species were excised, 5 volumes of LET buffer was added to each sample, and they were heated to 66°C for 5 min, followed by extraction with phenol (pH 5 to 6) and chloroform and alcohol precipitation in the presence of 100 μg of carrier yeast tRNA. Alcohol precipitation was performed in the absence of any carrier RNA when studies to determine the ratio of RF RNA to the IOD were done.
Quantifying minus strands in SIN2V- and wt SIN-infected cells.
Cultures infected with SIN were incubated in medium containing 200 μCi of 5′ [3H]uridine/ml and 20 μg of actinomycin D/ml for the times indicated. The viral RF RNA was obtained as described above or by limited digestion with RNase A (0.1 μg/ml) at 25°C for 15 min in 0.15 M NaCl, followed by chromatography on CF-11 cellulose (Whatman, Clifton, NJ), as described previously (3). Minus-strand RNA synthesis was determined in a nuclease protection assay. Briefly, the 5′-[3H]uridine-labeled RF RNA was heat denatured in 1 mM EDTA at 100°C for 5 min and fast cooled at 0°C, after which samples of the denatured RF were allowed to anneal in the presence of a 100 molar excess of purified virion plus strands and 0.4 M NaCl for 30 min at 68 to 70°C, followed by 30 min at 25°C. The controls were equivalent amounts of the same heated and fast-cooled RF samples that were either left on ice (heat-denatured control) or put through the annealing procedure in the absence of added plus strands (self-annealed control). The percentage of the total radioactivity in the [3H]uridine-labeled RF RNA that was in minus strands was determined as described previously (3).
RESULTS
Optimizing infection with SIN2V.
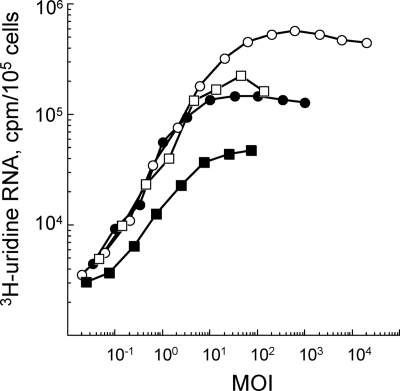
We first verified that all susceptible BHK21 and DF-1 cells were infected during the initial infection cycle to exclude the possibility that there could be a second round of infection. We used the incorporation of [3H]uridine into viral RNA to determine the MOI (PFU/cell) that gave the maximum incorporation during the first 5 hours of the infection cycle. An MOI of 25 was required to ensure that all (93 to 100%) BHK21 and DF-1 cells were infected with SIN2V (Fig. 1); this was similar to the results found for wt-SIN-infected cultures. The experiments, therefore, used MOIs of 30 to 50 for each virus to ensure that all cells were infected in the first round. Also, it was found that SIN2V-infected BHK21 cells incorporated 40% and DF-1 cells only ∼20% as much [3H]uridine as did wt-SIN-infected BHK21 or DF-1 cells, respectively (Fig. 1).
FIG. 1.
The MOI (or the number of PFU/cell) of wt SIN and SIN2V required to infect cultures of BHK21 and chicken embryo fibroblasts (DF-1 cells) in the initial round of infection. Cultures (35-mm petri dishes) of cells were infected at MOIs that ranged from 0.02 PFU/cell to 2 × 104 PFU/cell at 25°C for 1 h and then were maintained at 37°C. The cultures were labeled continuously with [3H]uridine (50 μCi/ml) in the presence of actinomycin D (20 μg/ml) beginning at 1 h p.i. and were harvested at 5 h p.i. The acid-insoluble incorporation in viral RNA per 105 cells was determined. Shown are wt-SIN-infected BHK21 cells (○), wt-SIN-infected DF-1 cells (•), SIN2V-infected BHK21 cells (□), and SIN2V-infected DF-1 cells (▪).
Quantitative assay to monitor SIN RI/native RF levels.
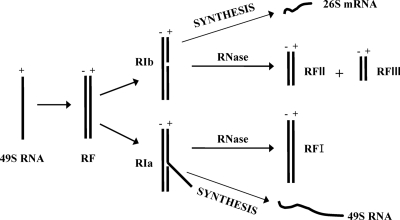
Our goal was to study activities that might be altered in SIN RTCs containing uncleaved P23. We first developed a method to determine the number and the stability of wt SIN RI RNAs. When extracted free of proteins and lipids, SIN minus strands that are active in RNA synthesis are recovered as (i) partially double-stranded, high-salt-insoluble RI possessing multiple nascent strands or (ii) more fully double-stranded, high-salt-soluble native RFs that have one or only a few polymerases and nascent strands (reviewed in references 32 and 40). For simplicity, we refer to both as RI in this report. Alphavirus RI exhibit two functional states, called RIa and RIb, that are distinguished by the kinds of double-stranded cores or RF RNA left after RNase A or T1 digestion (Fig. 2) (38). The multistranded RIa yields a 20S double-stranded RNA core structure (RFI) and is associated with efficient production of genomes. RIb is associated with efficient 26S mRNA synthesis by the SIN polymerase that recognizes an internal promoter and copies the downstream 3′ one-third of the minus-strand template into 26S mRNA. RIb produces two RFs (RFII and RFIII). While the 26S encoding sequence is present in RFIII, the 5′ two-thirds of the same 49S minus-strand template, which is occupied by incomplete 49S genome strands, is present in RFII (Fig. 2) (38).
FIG. 2.
Schematic of the RNA structures that function in SIN-infected cells. The viral 49S plus-strand genome is used as a template by the viral-RNA-dependent RNA polymerase to produce a complementary minus-strand RNA (shown here as a double-stranded or RF structure; the actual template structures are not known). The nascent minus strand is then used as a template for plus-strand synthesis and is found in two types of structures (38). The replicative structures (RI and native RF) making genomes (RIa-type structures) contain plus-strand sequences that are complementary to all of the template sequences and therefore are resistant to mild RNase treatment. Their nuclease-resistant core structure is RFI, which is 20S in size and represents the double-stranded form of the genome RNA. Replicative structures active in subgenomic 26S mRNA synthesis (RIb-type structures) have an internal promoter site that is efficiently recognized by the viral polymerase and that creates a nuclease-sensitive gap in the template strand of RI/native RF when purified from infected cell lysates. Cleavage by RNase yields RFII and RFIII cores from such replicative structures.
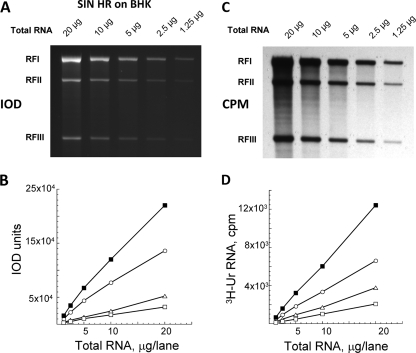
We monitored the viral RI levels hourly by measuring the relative amounts of RFI, RFII, and RFIII. We chose the fluorescent dye ethidium bromide, which binds to both single-stranded and double-stranded RNA in gels, and verified that its use would provide a quantitative and sensitive method to determine the amounts of RF. The three SIN RFs were separated by electrophoresis on gels and stained with sufficient ethidium bromide (0.5 μg/ml; 100 ml per gel) to saturate the amounts of RNA in the gel (data not shown) (increased concentrations of ethidium bromide led to greater background intensity but did not increase the staining in specific RF bands). The intensity of the bound ethidium bromide was imaged using the Omega Molecular Imaging System, and the numbers of RFs were quantified from the IOD values for the fluorescence associated with each RF. In pilot studies, we verified that there was a linear relationship between IOD values and numbers of RFs obtained from 1.25 to 20 μg of total infected-cell RNA when such numbers were analyzed per gel lane (Fig. 3A and B) (RFs from 40 μg of total infected-cell RNA/lane exceeded the linear limit [data not shown]) based on finding that the R values (Pearson correlation coefficients) that reflected the degree of linear relationship between two variables were all higher than 0.99 and close to 1. This result was confirmed using [3H]uridine incorporation into the same RF bands that were cut from the dried gels and gave similar sets of linear values for each RF species (Fig. 3C and D). Thus, ethidium bromide staining of purified RFs within an agarose gel was a sensitive means to both quantify viral RI during infection and assess whether they were making either genomes (RFI generated from RIa) or 26S mRNA (RFII and RFIII generated from RIb).
FIG. 3.
A linear correlation existed between the RF concentration and the level of bound ethidium bromide fluorescence (IOD values) or of [3H]uridine incorporated by wt SIN RF RNA. Total wt-SIN-infected cell RNA was obtained as described in Materials and Methods, and various amounts (1 to 20 μg/lane) were electrophoresed on 1.2% agarose-TBE gels. (A) Gels were stained with ethidium bromide and destained, and the bound ethidium bromide associated with RFI, RFII, and RFIII RNAs was imaged. (B) The IOD values obtained for RFI (○), RFII (▵), and RFIII (□) cores and for the sum of the RF core RNA (▪) shown in panel A. (C) A fluorogram of the same gel after it was processed for fluorography, dried, and exposed to X-ray film. (D) The individual areas of the dried gel that corresponded to RFI (○), RFII (▵), and RFIII (□) cores and the sum of the RF core RNA (▪) shown in panel C were cut out and counted in a Beckman scintillation counter.
The fate of wt SIN HR minus strands in BHK21 cells.
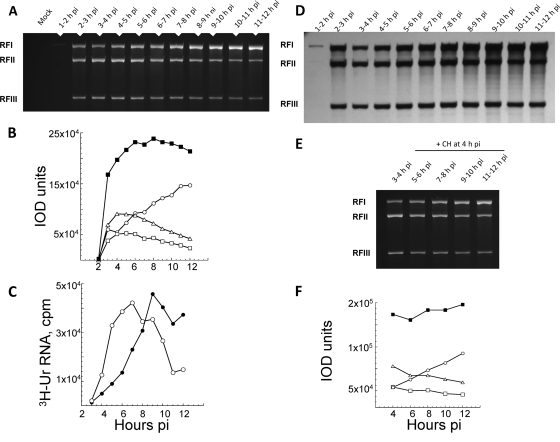
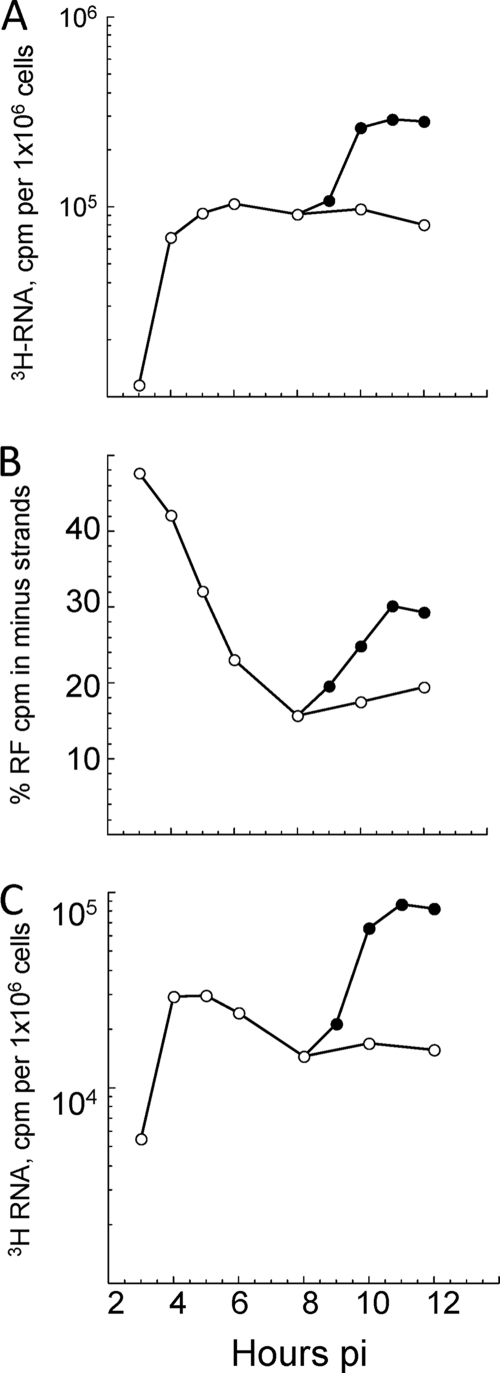
The rate of alphavirus plus-strand synthesis, which is determined by the amount of [3H]uridine incorporated into viral RNA per 30-min or 60-min labeling period, increases early and reaches a maximum at ∼5 or 6 h p.i.; it then remains maximal and constant until ∼12 h p.i., when the cell starts to die. In contrast, minus strands normally are only made early. They are recovered as RI and not in single-stranded form, and their number determines the rate of plus-strand synthesis (35). Thus, as shown using SIN and Semliki Forest virus, alphavirus minus strands are stable templates, i.e., once made they are not degraded or lost. Using IOD and [3H]uridine incorporation measurements, we quantified the RFs in wt-SIN-infected cells that were incubated in the absence or presence of translation inhibitors (i.e., cycloheximide), which block minus-strand synthesis. By IOD analysis, SIN RFs were detectable at ∼2 h p.i. in wt-SIN-infected BHK21 cells (Fig. 4), reached their maximal level by 6 h p.i., and remained at this level until 12 h p.i. We verified that the IOD values obtained for 20 μg of wt-SIN-infected cell samples were within the linear range by also testing 1:2, 1:5, and 1:10 dilutions of each of the late-time-p.i. samples (data not shown). As originally observed by Simmons and Strauss (38) from radioactive uridine incorporation, we observed changes in the ratios of RFI to RFII plus RFIII over time by IOD assay. For simplicity, we used the RFI/RFIII ratios. Figure 4 shows the results from one of three replicate experiments in which BHK cells were infected with SIN2V and pulse-labeled for 1 h with [3H]uridine. The RFs obtained from the cells at different times after infection were separated by gel electrophoresis and are shown in Fig. 4A (ethidium bromide stained) and D (fluorogram). There were smaller amounts of RFI than of RFIII early (2 to 4 h p.i.), but RFI increased steadily whereas RFIII (and RFII) peaked and decreased steadily after 5 to 6 h p.i. (Fig. 4B). The ratio of RFI to RFIII at 10 h p.i., obtained by averaging the three replicate experiments, was 2.6 by IOD assay and 2.2 by cpm of [3H]uridine when the RF RNA bands were cut from the gel and counted. Figure 4C shows the levels of 49S genomes and 26S subgenomic mRNA synthesis over time and demonstrates the increase of genome synthesis and decrease of 26S mRNA synthesis, which is similar to the RF pattern. A similar shift was observed in the ratio of genome to 26S mRNA and in the ratio of RFI to RFIII in SIN/GFP-infected BHK21 cells between 4 h p.i. and 9 h p.i. (data not shown). Therefore, SIN/GFP RTC composed of mature nsP2 proteins and nsP3-GFP fusion proteins retained this property, as well as other wt phenotypes previously described (12). Stopping the synthesis of both host and viral proteins at 4 h p.i. with cycloheximide did not alter the stability of RF using the IOD assay (Fig. 4E). The same number of RFs (the sum of the IODs of RFI, RFII, and RFIII) that were present at 4 h p.i. were present 8 h later. As expected, cycloheximide added at 4 h p.i. stopped the synthesis of minus strands, and no more RFs accumulated. However, RFI increased and RFII and RFIII decreased over time (Fig. 4F), as they did in untreated cells (Fig. 4B). This means the slow loss from early to late times p.i. of internal initiation of subgenomic mRNA synthesis that caused the RIb structure to convert to the RIa structure was a property of preformed components of the RTC.
FIG. 4.
Analysis of RI (RF core) numbers; genome and 26S mRNA levels; and RFI, RFII, and RFIII ratios during the infectious cycle for wt-SIN-infected BHK21 cells. Cultures of BHK21 cells were infected with wt SIN (MOI, 30) and maintained at 37°C. Beginning at 1 h p.i., the cultures were labeled with [3H]uridine (3H-Ur) for 1-h periods and harvested at the end of each labeling period. Total infected-cell RNA was obtained, and aliquots were analyzed by electrophoresis on 1.2% agarose-TBE gels for the [3H]uridine incorporation in 49S and 26S plus strands. The RF cores of wt SIN RI were prepared from the remaining lysates, as described in Materials and Methods. Samples of RF applied to each lane were derived from 20 μg of total infected-cell RNA and were electrophoresed on 1.2% agarose-TBE gels, stained with ethidium bromide, and monitored for fluorescence (IOD values) before the gels were fluorographed. (A) Relative fluorescence of ethidium bromide-stained RF cores from infected cell lysates harvested between 2 and 12 h p.i. (B) IOD value for ethidium bromide-bound RF cores shown in panel A. Shown are RFI (○), RFII (▵), and RFIII (□) cores and the sum of the RF core RNA (▪). (C) Analysis of the levels of [3H]uridine incorporation in 49S genome (•) and 26S mRNA (○) during infection. (D) Fluorogram of the same wt SIN RNA agarose gel, showing the level of incorporation of [3H]uridine into individual RF species over time p.i. (E) Fluorescence image of an agarose gel containing wt SIN RF from infected BHK21 cells untreated (harvested at 4 h p.i.) or treated with cycloheximide (+ CH) starting at 4 h p.i. (F) Analysis of the IOD values obtained for each RF species and their sum in the gel shown in panel E. Shown are RFI (○), RFII (▵), and RFIII (□) cores and the sum of the RF core RNA (▪).
The RFs detected by IOD assays were from RI active in transcription (Fig. 4D). They incorporated [3H]uridine in a pattern of accumulation that paralleled the IOD patterns for the same RF samples, averaging at maximum 1.1 × 104 cpm by 2.3 × 105 IOD units of wt SIN RF under the conditions used. Thus, the results in Fig. 3 and 4 verified the ability of IOD analysis to monitor viral RI levels and confirmed that in wt-SIN-infected cells, minus strands (as RF cores of the RTC) were physically stable during infection and in the presence of translation inhibitors.
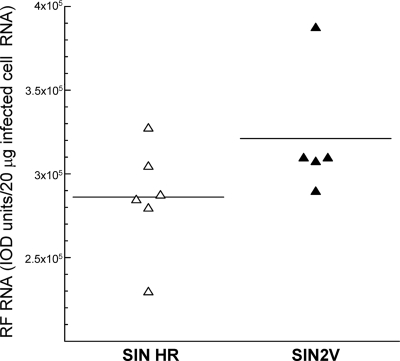
We also used the IOD assay to determine the amount of RF RNA in wt-SIN-infected BHK21 cells and the ratio of RF RNA to IOD units (Table 1) . For this purpose, viral RF cores were purified from ∼2 × 107 SIN HR-infected BHK21 cells as described in Materials and Methods. Briefly, RNase T1-resistant RNA recovered from infected cells was electrophoresed on two lanes of an LMT agarose gel. The areas of the gel containing each of the RF species were identified by ethidium bromide staining and cut out, and the RNA was recovered after phenol-chloroform extraction and ethanol precipitation in the absence of any added RNA carrier. Recoveries of the RFs from the LMT gel ranged from 10% (RFI) to 55% (RFIII). This was determined from the IOD value for 10% of each purified RF preparation and by comparing this to the IOD obtained for a known concentration of the original pre-LMT RF RNA when all samples were subjected to electrophoresis on the same gel (Table 1) and the analysis was performed in duplicate. When corrected for recoveries and using the molecular masses obtained by Simmons and Strauss (38) for RFI (8.8 × 106 daltons [1.46 × 10−17 g]), RFII (5.6 × 106 daltons [9.3 × 10−18 g]), and RFIII (2.9 × 106 daltons [4.8 × 10−18 g]), there was about 0.39 pg of total RF RNA/cell, of which ∼58% was RIa, and at late times after infection, RF RNA accounted for ∼0.6% of the total infected-cell RNA (Table 1). From these numbers, we estimated there were ∼2.7 × 104 RI molecules per infected BHK21 cell, using RFI as the average size for the core of an RI molecule. Finally, when the three RF values were averaged, 1 ng of RF RNA was equivalent to 1,774 IOD units (Table 1).
TABLE 1.
Analysis of viral RNA species in wt-SIN-infected BHK21 cellsa
| RNA species | Total (ng) | % Recovery from LMT gels | Avg total (ng) | % of total infected-cell RNA | IOD/ng of RF RNA |
|---|---|---|---|---|---|
| RFI | 437 | 10.1/9.8 | 4,326 | 1,864 | |
| RFII | 399 | 17.8/12.4 | 2,242 | 1,471 | |
| RFIII | 523 | 55.3/40.2 | 947 | 1,988 | |
| RFI + RFII + RFIII | 7,515 | 0.6 | 1,774 |
The RNA amounts shown were from 19.2 × 106 BHK21 cells (1,245 μg). The analysis represents the results from two experimental determinations, with the recovered RF levels averaged for the total values. RNA concentrations were determined using a Nanodrop spectrophotometer.
Accumulation of minus strands in SIN2V-infected cells.
Because minus-strand synthesis and RTC formation did not cease with SIN2V (12), we determined if SIN2V-infected cells accumulated significantly larger numbers of RI than wt-SIN-infected cells. We analyzed the IOD values for these samples when diluted 1:2, 1:5, and 1:10 to ensure the observed IOD values represented the actual levels of RF cores and were within the linear range of the assay (data not shown). The maximal IOD units detected for SIN2V RF cores were, on average, ∼110% of the maximal levels detected in wt-SIN-infected cells, with a range of 100% to 174% of wt levels, from the results of five or six independent experiments (Fig. 5). This meant continuous minus-strand synthesis by SIN2V-infected cells did not result in a correspondingly greater accumulation of RI. Rather, accumulation stopped and SIN2V RI in excess of this must have been lost, either actively degraded or excreted. Either the minus strands made after the maximum is reached are lost, or there is a stochastic process in which both newly made and old minus strands are lost after the maximum numbers are reached.
FIG. 5.
Steady-state levels of viral RI RNA (RF cores) in wt-SIN- and SIN2V-infected BHK21 cells. Cultures of BHK21 cells were infected with wt SIN (▵) or SIN2V (▴) at an MOI of 30 at 37°C. The infected cultures were harvested at hourly intervals from 6 to 12 h p.i., when the rates of viral RNA synthesis were maximal. The RNase-resistant RF cores of the viral RI were obtained and electrophoresed on 1.2% agarose-TBE gels and stained with ethidium bromide. The IODs of the bound ethidium in RFI, RFII, and RFIII cores were summed to give the total IOD/RF cores present in 20 μg of total infected-cell RNA. The results shown are the average values for one to four time points per experiment and for five or six independent experiments. The mean value for wt SIN RF was 286,000, and that for SIN2V RF was 321,200, or 112% of the wt value.
SIN2V minus strands made late are recruited into the RTC and function as templates for plus-strand synthesis.
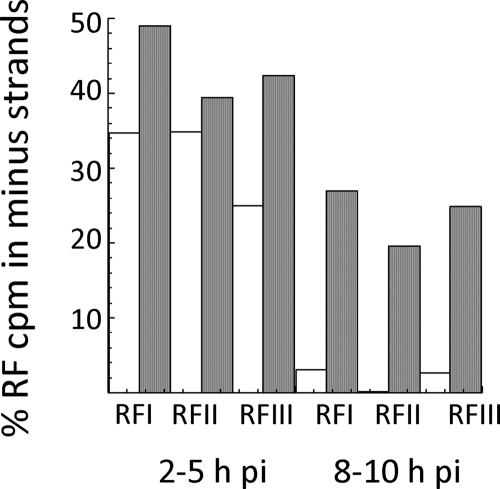
To determine if the minus strands made after SIN2V plus-strand synthesis and RI stopped increasing were dead end and not assembled into an RTC as RI, we directly determined if they were used as templates to make subgenomic 26S mRNA late in infection. Continuous labeling with [3H]uridine beginning at 8 h p.i. was used to examine the population of minus strands made at late times, and the results were compared to 1-h pulse-labels given between 2 and 12 h p.i. that monitored the total, RI, and minus-strand populations over time. The minus strands made after 8 h p.i. in SIN2V-infected cells were in RI that could be converted with RNase to RFI, RFII, and RFIII. When the label was added at 8 h p.i., RI accumulated to levels that were greater than the 1-h pulse-labeled (active RF core) population, which remained relatively constant over the same period (Fig. 6A). When pulse-labeled for 1-h periods between 8 and 12 h p.i., ∼20% of the total radiolabel in SIN2V RF RNA was in minus strands and ∼80% of the incorporation was in plus strands (Fig. 6B and C). Since minus strands make up half of the total RNA molecules in double-stranded RF structures, a value of 20% means that 40% of the total minus-strand molecules were newly made each hour between 8 and 12 h p.i. This value increased with time under continuous-labeling conditions, with 60% of the minus strands recovered from transcriptional active RF cores being made after 8 h p.i. The findings meant the minus strands made late were not released as single strands and were not selectively turned over. There also was not a selective increase in only RFI but a proportional increase in all three RF species (IOD assay; data not shown), as expected if they became part of the active RI population. To show directly that the minus strands made after 8 h p.i. functioned as templates, we isolated the individual RFI to RFIII species and determined whether labeled minus strands were present in each, and specifically in RFIII cores. While RFI is derived from RI making genomes, structures of the same size would be expected for the RI engaged in minus strands, since both RNA products are 49S in size. However, RFIII functions solely in 26S mRNA synthesis, and any radiolabeled minus strands found there would be serving as templates for 26S mRNA. BHK21 cells were infected with wt SIN or SIN2V and labeled with [3H]uridine from 2 to 5 h p.i. to monitor minus strands made in the early phase of infection and from 8 to 10 h p.i. for those made in the late phase of infection, after which the RF cores were isolated (Fig. 7). Similar to minus strands made early in infection, SIN2V late minus strands were present in all three RF species, including RFIII. The wt-SIN-infected cells made few, if any, minus strands late in infection (Fig. 7).
FIG. 6.
Fate of SIN2V minus strands made late in infection. Cultures of SIN2V-infected BHK21 cells were pulsed with [3H]uridine (200 μCi/ml) in the presence of actinomcyin D for 1-h periods starting at 2 h p.i. (○) or were labeled continuously only late in infection, starting at 8 h p.i. (•). The RFs were obtained, and their contents of labeled minus strands were determined as described in Materials and Methods. (A) SIN2V-infected cells accumulate RTCs (RF cores) late in infection and to levels in excess of those active in 1-h pulse periods. (B) The percentage of the total incorporation in SIN2V RF cores that is in minus strands newly made during each pulse period. (C) Minus strands made late accumulated and were present in the RTC (RF cores) in numbers that exceeded those present in the transcription-active RTC population.
FIG. 7.
SIN2V minus strands made late in infection are functional templates for plus-strand synthesis. Wt-SIN- and SIN2V-infected cells were pulse-labeled from 2 to 5 h p.i. (early in infection) or from 8 to 10 h p.i. (late in infection) with [3H]uridine (200 μCi/ml in the presence of actinomycin D). The viral RF species were isolated after electrophoresis of nuclease-treated total infected-cell RNA (20 μg total cell RNA/lane) on 0.8% LMT agarose-TBE gels as described in Materials and Methods. The area containing each RF was identified by ethidium bromide staining and cut out, and the RNA was isolated following phenol extraction and ethanol precipitation. The amount of radiolabeled minus-strand RNA in each RF was determined using nuclease protection assays. Open bars, wt-SIN minus strands; closed bars, SIN2V minus strands.
As shown above, SIN2V-infected cells accumulated only slightly more RI than wt-SIN-infected cells (Fig. 5). Using data derived from 1-hour pulse-labels with [3H]uridine, as shown in Fig. 6, we determined that SIN2V- infected cells actually produced ∼250% more minus strands than did wt-SIN-infected cells. This was calculated by determining the cpm of [3H]uridine that was incorporated into RI for each hour, determining the fraction of the cpm that was in minus strands to give the amount in minus strands, and adding these together to obtain the total cpm in minus strands made during 12 h. Thus, even though minus-strand synthesis does not shut off, i.e., it is continuous, with SIN2V, the infected cells do not accumulate minus strands much above the level found with wt SIN. Since the minus strands made late are also assembled into the RTC and used for transcription, the cell must somehow limit the number of RTCs. The extra RTCs must be lost by some mechanism that we do not yet know. It is interesting that when translation is inhibited in SIN2V-infected cells, the activity of the RTC is diminished over time but the RI is not diminished, suggesting that only excess RTC is lost and the wt SIN stops minus-strand synthesis just when more minus strands would not result in more RTCs.
The loss of activity by the SIN2V RTC after translation was inhibited was stochastic and occurred without the degradation of the minus-strand templates.
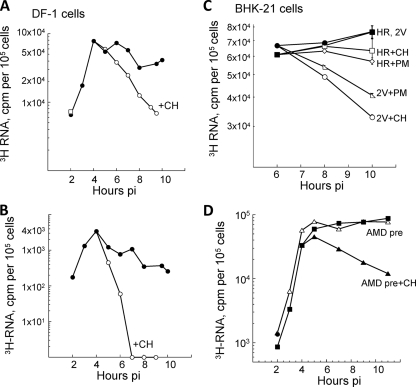
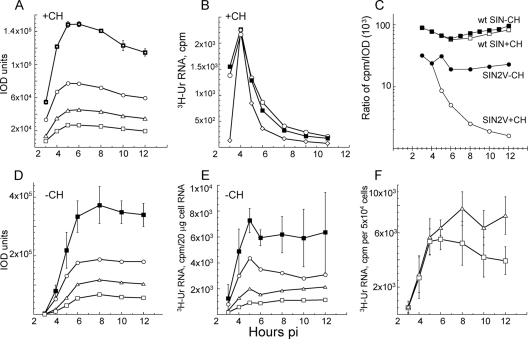
Although the P23-containing RTCs were fully functional and produced minus-strand templates, genomes, and subgenomic mRNA, their transcription activity was short-lived; however, new RTCs were assembled continuously throughout infection in DF-1 chicken embryo cells (Fig. 8A and B), not just in BHK21 (12). We verified the short-lived nature of the RTC activity in SIN2V-infected cells by using puromycin, a translation inhibitor whose mode of action differs from that of cycloheximide, which we used previously (12, 33, 34). Puromycin resembles the terminal aminoacyl-adenosine moiety of aminoacyl-tRNA, leading to its covalent attachment to and then release (termination) of the growing peptide chain, in contrast to cycloheximide, which blocks ribosome elongation and might affect other RNA-protein interactions. Wt SIN plus-strand synthesis was unaffected by either inhibitor, whereas SIN2V plus-strand synthesis decreased after the addition of either drug (Fig. 8C). Thus, the activity of RTCs assembled in SIN2V-infected cells is lost over time after translation is inhibited. This loss of RTC activity was not caused by SIN2V infection inducing expression of host genes because the loss still occurred in SIN2V-infected cells that were treated with actinomycin D just before infection (Fig. 8D).
FIG. 8.
Effects of translation inhibition on wt SIN and SIN2V transcription in chicken embryo fibroblasts and BHK21 cells. (A and B) Cultures of DF-1 chicken embryo fibroblasts were infected with SIN2V at an MOI of 30 PFU/cell at 37°C, and RNA synthesis was monitored with [3H]uridine in 1-h pulse-labels between 1 and 10 h p.i. Duplicate cultures were incubated in cycloheximide (+CH) (100 μg/ml) starting at 4 h p.i. and radiolabeled in the presence of the inhibitor. (A) SIN2V plus-strand synthesis. (B) SIN2V minus-strand synthesis. •, untreated SIN2V-infected cells; ○, cycloheximide-treated SIN2V-infected cells. (C) BHK21 cultures in 35-mm petri dishes were infected at an MOI of 30 with either wt SIN HR (▪, □, ⋄) or SIN2V (•, ○, ▵) and maintained at 37°C. Individual cultures were pulsed with [3H]uridine in medium also containing 20 μg actinomycin D/ml for 1-h periods and harvested at the end of the pulse period. Duplicate cultures were untreated (▪, •) or treated with cycloheximide (□, ○) or puromycin (+PM) (50 μg/ml) (⋄, ▵) beginning at 4 h p.i. Cultures treated with translation inhibitors were pulse-labeled in the presence of the inhibitor. (D) Effect of actinomcyin D pretreatment. BHK21 cultures in 35-mm petri dishes were incubated with 2 μg of actinomcyin D/ml starting 30 min before infection with SIN2V at an MOI of 30 (▪) or were untreated (▵). A duplicate set of actinomcyin D-pretreated (AMD pre) cultures was incubated in medium containing 100 μg/ml of cycloheximide starting at 4 h p.i. (▴). Individual cultures were labeled with [3H]uridine (50 μCi/ml in the presence of actinomcyin D) and harvested at the end of each pulse period. Incorporation into acid-insoluble RNA by 105 cells is shown.
The presence of P23 proteins might cause the SIN2V RTC to be inherently unstable and more susceptible to falling apart. If so, the RI or its minus-strand template would be released and susceptible to degradation, in which case its loss would parallel the loss of SIN2V RTC activity. BHK21 cells were infected with SIN2V and labeled with [3H]uridine (200 μCi/ml) for 1-h periods in the presence of cycloheximide and the incorporation of [3H]uridine into viral RNA, and their IOD values were measured. Cycloheximide treatment prevents the synthesis of any new minus strands and thus the creation of new RTCs. This allowed us to follow the activity of previously assembled RTCs. While overall [3H]uridine incorporation into the SIN2V RF cores decreased 15-fold on average (range, 10- to 30-fold) after 8 h of translation inhibition (Fig. 9B) and decreased similarly in all three RF species, the physical numbers of SIN2V RI decreased less than 1.5-fold (the average decrease was 31%, with a range of 22% to 46%, in three separate analyses) over the same 8-h period (Fig. 9A). The 10-fold difference in radioactive incorporation relative to physical RF RNA amounts showed that the RI was retained but its activity in transcription was lost. The RI was unchanged and not degraded. This meant SIN2V-infected cells would have two populations of RI: those derived from RTCs active in viral plus-strand synthesis and those from RTCs that were no longer active. Alternatively, it remains possible that the activity of the RTC declines with time. In this case, the newly created ones would be more active than older ones, and they would lose activity over time. We favor the first explanation, because as described above, we found evidence that the process is stochastic.
FIG. 9.
Analysis of SIN2V replicative structures. Total infected-cell RNA was treated with DNase I and RNase T1, and then amounts equivalent to 20 μg of original total RNA per lane were electrophoresed on 1.2% agarose-TBE gels to separate the three viral RF species. The RFs were stained with ethidium bromide and destained, and the IOD of their bound ethidium bromide fluorescence was measured. After fluorography, the area of the dried gel containing each RF was cut out and counted in a scintillation counter. (A and B) Loss of SIN2V transcription occurred without significant loss of RTC cores. (A) IOD analysis of the number of RTC cores in SIN2V-infected BHK21 cells treated with cycloheximide (+CH) starting at 3.5 h p.i. Shown are RFI (○), RFII (▵), RFIII (□), and the sum of the RF RNAs (▪), with error bars showing the range of values in three experimental repetitions. (B) Transcription activity of RTC in SIN2V-infected cells treated with cycloheximide starting at 3.5 h p.i. Individual results for three experiments are shown. 3H-Ur, [3H]uridine. (C) Only SIN2V showed decreasing ratios of total RNA synthesis (cpm) per RF RNA (IOD) when translation was inhibited in BHK21 cells. Cells infected with wt SIN (squares) or SIN2V (circles) were incubated in the absence (closed symbols) or presence (open symbols) of cycloheximide starting at 3.5 h p.i. (D to F) Untreated cells. (D) IOD numbers of SIN2V RF cores. Shown are RFI (○), RFII (▵), RFIII (□), and the RF sum (▪), with error bars showing the range of values obtained in three experiments. (E) Incorporation of [3H]uridine by SIN2V RF. Shown are RFI (○), RFII (▵), RFIII (□), and the sum of the RF RNA (▪), with error bars showing the range of values from three repetitions of the experiment. (F) Rates of synthesis of SIN2V 49S genome RNA (□) and 26S mRNA (▵). The error bars show the range of values from three experiments.
With SIN2V, the amount or rate of plus-strand synthesis was only about one-half of what was seen for its RI level (plus-strand cpm/IOD) compared to wt-SIN-infected cells, and isolated RF cores had cpm (active-RF)/IOD ratios for SIN2V that were half those for wt SIN (Fig. 9C). This meant that when viral-RNA synthesis is at its maximum, the incorporation of [3H]uridine into the SIN2V RI was ∼40% of that into wt SIN RI, similar to the difference in their relative rates of plus-strand synthesis (Fig. 1). Therefore, the relationship between the rate of plus-strand synthesis and the numbers of RI observed in this study and described earlier for wt SIN (38) appeared different for SIN2V because there was less plus-strand synthesis, even though there was the same number of, or more, RI. However, SIN2V plus-strand synthesis did reflect the numbers of active RI. Analysis by IOD assay of the RI in SIN2V-infected BHK21 cells showed that they increased normally early in infection and remained relatively constant after reaching a maximal level (Fig. 9D). The findings support the conclusion that with SIN2V, there is a large population of inactive RTCs. If these are not replaced with new RTCs, as occurs after inhibiting translation, overall transcription activity is lost.
P23-containing RTCs exhibited the novel phenotype of being unable to temporally switch from making less to making more genomes than subgenomic 26S mRNA.
A comparable pattern of early increase followed by maintenance of constant levels once maximal amounts accumulate was also seen for the SIN2V RI using [3H]uridine incorporation (Fig. 9E). In this case, we observed a larger variation from one experiment to the next at late times after infection that likely reflects cytopathic effects and dying cells. One difference noted was in the RFI-RFIII pattern (functionally, the RIa/RIb ratio). With both wt SIN and SIN/GFP, this ratio increased with time after infection, while with SIN2V, it remained constant throughout infection and paralleled the stable ratios of SIN2V genome/26S mRNA syntheses (Fig. 9F). Thus, P23-containing RTCs did not temporally switch between RIb and RIa states, as did the nsP2/nsP3 RTCs (Fig. 4C). Also, this ratio did not change in the presence of cycloheximide, as it does with wt SIN (compare Fig. 9A with 4F). This was an unexpected finding.
DISCUSSION
Our findings showed that alphavirus RTCs containing P23 retained most of the normal polymerase functions of RTCs containing nsP2/nsP3: they synthesized minus-strand templates and both species of plus strands, and they contained the expected RIa and RIb template structures that yielded normal-size RNase-resistant RF cores. However, RTCs continue to be formed late in infection because minus-strand synthesis does not stop as it does with wt SIN that produces nsP2/nsP3, and the RTCs containing P23 lose activity over time if not replaced by newly made ones. This loss is observed if translation is halted (12). If minus-strand synthesis continues, as it does normally when translation occurs unabated, new RTCs are produced that replace the ones that lose activity. With SIN2V, the numbers of RI that accumulate in infected cells are slightly higher than in the wt, even though the amount of viral RNA synthesis is about half of the wt rate. This is explained by the RTCs losing activity over time and the existence of two types of RTC populations in SIN2V-infected cells, one active and one inactive. RTCs containing nsP2/nsP3 do not lose activity with time, and therefore, wt plus-strand synthesis is not affected if translation is inhibited after minus-strand synthesis normally ceases.
We analyzed the hourly rates of synthesis of new SIN2V minus-strand RNA between 1 and 12 h p.i. and estimated that 220% to 260% more minus strands were made during the infectious cycle than were made in wt-SIN-infected cells. This ability to continuously form new minus strands and RTCs was essential for SIN2V to efficiently replicate and produce close to wt levels of genomes and virions. Somewhat unexpectedly, while SIN2V minus-strand templates were made continuously, they accumulated to levels only slightly greater than those in wt-SIN-infected cells that cease minus-strand synthesis after the early phase. This meant that infected cells limit the overall number of these molecules and that numbers in excess of this level must have been degraded or excreted. It was of great interest to us to observe that wt SIN synthesized numbers of RI that were just below or at this crucial total number before wt-minus-strand synthesis ceased, raising the question of how the virus or cell knows to do this. This is also the first report of the size of the RI/RTC population in SIN-infected BHK21 cells. We found that the numbers of viral RTCs appear to be proportional to the cell size when the ∼27,000 RTCs/BHK21 cell obtained in this study is compared to the 5,000 to 7,000 RTCs/cell obtained earlier (44) for chicken embryo fibroblasts that have one-fifth the amount of RNA/cell.
We found that SIN2V minus-strand synthesis was normal, as it was dependent on newly translated proteins, and the minus strands accumulated in an RI that must have been part of the RTC and functioned as authentic templates at both early and late times in infection. The unusual aspect was that viral replication retained its early permissive phenotype in these cells and never underwent the overall loss of minus-strand synthesis that is seen at ∼5 to 6 h p.i. in wt-SIN-infected BHK21 and other cells. The observation that P23-containing RTCs lost the ability to make minus strands but continued plus-strand synthesis (Fig. 8) meant that continuous minus-strand synthesis was not due to an abnormal polymerase constitutively recognizing the minus-strand promoter on genome templates. Rather, it reflected newly translated P1234 polyproteins being assembled with genome plus strands into new RTCs continuously during infection. While P23-containing polymerases were more efficient in genome synthesis than in 26S subgenomic mRNA synthesis, as expected (21), we made the novel observation that P23-containing RTCs were unable to alter the relative amounts of the two plus-strand species during infection as nsP2/nsP3 RTC does, nor could it alter the ratio of RFI to RFIII cores that produce them. The conversion or switch of RFI to RFII plus RFIII is thought to reflect the loss or gain, respectively, of an RNase-sensitive gap in the minus-strand template that is the site where factors engage the template to activate it for internal initiation of 26S RNA synthesis (38). Genome synthesis occurs when the polymerase reads through this gap, creating its plus-strand complement and thus making this region in the RF core double stranded. It is intriguing that uncleaved P23 retained polymerase and RTC assembly functions, including assembling both RIa and RIb types of RTCs, but was unable to switch reversibly between them once the RTC had formed. Earlier studies showed that such interconversions by mature RTCs occur readily and can be blocked by as little as a 10°C temperature difference in some conditionally lethal nsP2 C-domain mutants or, more rarely, nsP3 N-domain mutants (2, 3, 8, 15, 22, 31, 40, 41, 44). How these regions function in internal initiation and whether the nsP2 methyltransferase-like motif plays a specific role are not known and are of interest.
Loss of activity by P23 RTCs did not reflect an inherent instability that might arise from the presence of P23 or the absence of nsP2/nsP3 proteins. Activity was lost without a physical loss of its RF cores, arguing that inactive RTCs were not falling apart or being degraded (Fig. 9). Rather, the silenced templates remained in close association with at least the last nascent plus strand made and also retained their RIa and RIb structural integrity, as the three RF species did not vary in their relative ratios even when over 90% had been inactivated. This suggests inactivation is a more subtle and selective process that appears to stop elongation by P23 RTCs. Finally, turnover (activity loss) of viral RTC populations was stochastic, and more recently assembled RTCs were not specifically targeted (Fig. 6). Because SIN2V-infected cells continuously assemble new RTCs, it was not possible to test if restoring translation would allow inactivated RTCs to recover activity.
Future studies will attempt to elucidate what is inactivating the SIN2V RTCs. One possibility is proteasome targeting of a component of the RTC, as proteasomes can target ubiquitinated nsP4 proteins (5, 19) and proteasomes become activated during many virus infections and are inhibited or actually required by different viruses (23, 27, 36). Such potential SIN RTC targets may not include the P23 proteins themselves, as our earlier pulse-chase study found P23 levels were unchanged over a 4-h chase period that saw a 10-fold loss in transcription activity following cycloheximide-induced translation inhibition (12). Alternatively, inactivation may reflect an altered ability of P23 to stably interact with other viral or host factors or to recruit such factors to the RTC. Also, it is possible that cellular processes apart from the RTC play a role, based on the reported failure of SIN2V-infected cells to limit host responses and host functions. Although SIN2V reduced host translation to ∼10% of uninfected-cell levels, similar to wt SIN, it was not able to stop induction of type 1 interferon and interferon-stimulated antiviral responses by SIN2V-infected NIH 3T3 cells (8, 9, 11, 13). While BHK21 cells are deficient in aspects of the innate immune response, their use as host cells was appropriate given their traditional use (38) and because wt-SIN replication was normal and the same in BHK21 and DF-1 cells as in NIH 3T3 cells that have full innate responses to virus infection (reference 12 and references therein). Also, deficiency in type 1 interferon signaling did not prevent BHK21 cells from stopping replication by SIN2V (and the related SIN-1V2V, which forms only P123 and nsP4) or from activating cell death (12). Coincident with the inability to block all host responses was the failure of P23 polyproteins to enter the nucleus, as is normally seen for ∼50% of the mature nsP2 population (12). These findings suggested that polyprotein processing contributes to alphavirus fitness because changes needed for efficient replication in vertebrate cells require release of the nsP2 sequence in a form permitting nuclear translocation. It is thus interesting that a similar pattern of continuous minus-strand synthesis and loss of RTC activity are also seen in BHK21 cells expressing mutant mature nsP2 proteins that are efficiently transported to the nucleus (28) and in wt-SIN-infected mouse embryo fibroblasts deficient in the latent host endonuclease RNase L (29). The ability to restore the wt phenotype to these nsP2 mutants by superinfection with wt virus confirmed that their defects were due to a loss of function(s) and showed that this wt function(s) could act to halt even ongoing RTC inactivation processes and to restore efficient viral replication (28). Certain of these nsP2 mutants were altered in their abilities to affect the host environment and metabolic processes, with C-domain mutants unable to inhibit host translation and/or transcription and attenuated in vivo (8, 11, 13). The pattern of similar phenotypes suggests that both viral (nsP2 and possibly also nsP3) and host (RNase L) factors may play roles in establishing the wt-SIN-infected cell environment. We are reminded that the success of minus-strand synthesis determines the overall success of virus replication and virion production, and this in turn plays a direct role in infection outcome. This supports further study of two such unusual alphavirus phenotypes, as an understanding of them may lead to the identification of novel alphavirus-host interacting partners and potentially could provide new targets for antiviral drug discovery.
Acknowledgments
We acknowledge the gift of SIN2V from Ilya Frolov and Rodion Gorchakov (University of Alabama, Birmingham) and the technical assistance of Laci Bloomfield.
We dedicate this study to James and Ellen Strauss on their retirement and acknowledge the impact their many contributions to alphavirus research has had on our own studies of these viruses.
The studies were supported by a grant (AI057571) from NIAID, NIH, to D.S.
Footnotes
Published ahead of print on 10 June 2009.
REFERENCES
- 1.Atasheva, S., R. Gorchakov, R. English, I. Frolov, and E. Frolova. 2007. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J. Virol. 815046-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balistreri, G., J. Caldentey, L. Kaariainen, and T. Ahola. 2007. Enzymatic defects of the nsP2 proteins of Semliki Forest virus temperature-sensitive mutants. J. Virol. 812849-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De, I., C. Fata-Hartley, S. G. Sawicki, and D. L. Sawicki. 2003. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 7713106-13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Groot, R. J., W. R. Hardy, Y. Shirako, and J. H. Strauss. 1990. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 92631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot, R. J., T. Rumenapf, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1991. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. USA 888967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, M. X., and M. J. Schlesinger. 1989. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171280-284. [DOI] [PubMed] [Google Scholar]
- 7.Fazakerley, J. K., A. Boyd, M. L. Mikkola, and L. Kaariainen. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 76392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 7611254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garmashova, N., R. Gorchakov, E. Frolova, and I. Frolov. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 805686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez de Cedron, M., N. Ehsani, M. L. Mikkola, J. A. Garcia, and L. Kaariainen. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 44819-22. [DOI] [PubMed] [Google Scholar]
- 11.Gorchakov, R., E. Frolova, and I. Frolov. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 799397-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorchakov, R., E. Frolova, S. Sawicki, S. Atasheva, D. Sawicki, and I. Frolov. 2008. A new role for ns polyprotein cleavage in Sindbis virus replication. J. Virol. 826218-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorchakov, R., E. Frolova, B. R. Williams, C. M. Rice, and I. Frolov. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 788455-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, Y. S., A. Grakoui, C. M. Rice, E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J. Virol. 631194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, Y. S., E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 633142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 634653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaariainen, L., and T. Ahola. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 71187-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keranen, S., and L. Kaariainen. 1979. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J. Virol. 3219-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keranen, S., and L. Ruohonen. 1983. Nonstructural proteins of Semliki Forest virus: synthesis, processing, and stability in infected cells. J. Virol. 47505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 726546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 132925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lulla, V., D. L. Sawicki, S. G. Sawicki, A. Lulla, A. Merits, and T. Ahola. 2008. Molecular defects caused by temperature-sensitive mutations in Semliki Forest virus nsP1. J. Virol. 829236-9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neznanov, N., E. M. Dragunsky, K. M. Chumakov, L. Neznanova, R. C. Wek, A. V. Gudkov, and A. K. Banerjee. 2008. Different effect of proteasome inhibition on vesicular stomatitis virus and poliovirus replication. PLoS ONE 3e1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peranen, J., M. Rikkonen, P. Liljestrom, and L. Kaariainen. 1990. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 641888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rikkonen, M. 1996. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology 218352-361. [DOI] [PubMed] [Google Scholar]
- 26.Rikkonen, M., J. Peranen, and L. Kaariainen. 1992. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology 189462-473. [DOI] [PubMed] [Google Scholar]
- 27.Satheshkumar, P. S., L. C. Anton, P. Sanz, and B. Moss. 2009. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J. Virol. 832469-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawicki, D. L., S. Perri, J. M. Polo, and S. G. Sawicki. 2006. Role for nsP2 proteins in the cessation of alphavirus minus-strand synthesis by host cells. J. Virol. 80360-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawicki, D. L., R. H. Silverman, B. R. Williams, and S. G. Sawicki. 2003. Alphavirus minus-strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R. J. Virol. 771801-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawicki, D. L., and S. G. Sawicki. 1994. Alphavirus positive and negative strand RNA synthesis and the role of polyproteins in formation of viral replication complexes. Arch. Virol. Suppl. 9393-405. [DOI] [PubMed] [Google Scholar]
- 31.Sawicki, D. L., and S. G. Sawicki. 1985. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology 14420-34. [DOI] [PubMed] [Google Scholar]
- 32.Sawicki, D. L., and S. G. Sawicki. 1998. Role of the nonstructural polyproteins in alphavirus RNA synthesis. Adv. Exp. Med. Biol. 440187-198. [DOI] [PubMed] [Google Scholar]
- 33.Sawicki, D. L., and S. G. Sawicki. 1980. Short-lived minus-strand polymerase for Semliki Forest virus. J. Virol. 34108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawicki, D. L., S. G. Sawicki, S. Keranen, and L. Kaariainen. 1981. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J. Virol. 39348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawicki, D. L., and S. G. Sawicki. 1987. Alphavirus plus and minus strand RNA synthesis, p. 251-259. In M. Brinton and R. Ruckert (ed.), Positive strand RNA viruses. Alan R. Liss, Inc. New York, NY.
- 36.Shackelford, J., and J. S. Pagano. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41139-156. [DOI] [PubMed] [Google Scholar]
- 37.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 681874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmons, D. T., and J. H. Strauss. 1972. Replication of Sindbis virus. II. Multiple forms of double-stranded RNA isolated from infected cells. J. Mol. Biol. 71615-631. [DOI] [PubMed] [Google Scholar]
- 39.Strauss, E. G., E. M. Lenches, and J. H. Strauss. 1976. Mutants of Sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology 74154-168. [DOI] [PubMed] [Google Scholar]
- 40.Strauss, E. G., and J. H. Strauss. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol. Rev. 58491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suopanki, J., D. L. Sawicki, S. G. Sawicki, and L. Kaariainen. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J. Gen. Virol. 79309-319. [DOI] [PubMed] [Google Scholar]
- 42.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kaariainen. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of nsP2. J. Biol. Chem. 27517281-17287. [DOI] [PubMed] [Google Scholar]
- 43.Vasiljeva, L., L. Valmu, L. Kaariainen, and A. Merits. 2001. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J. Biol. Chem. 27630786-30793. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 686466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]