Abstract
Of the five herpes simplex virus type 1 immediate early (IE) proteins, the least is known about the function of ICP22 during productive infection and latency. Research characterizing the physical and functional properties of the protein has been limited because ICP22 has proven to be difficult to express in transient assays. In addition, genetic analysis of ICP22 has been complicated by the fact that the C terminus of ICP22 is expressed as a discrete protein product. In order to characterize properties of mutant and wild-type ICP22, we developed a transient expression system. We found that ICP22 can be expressed at detectable levels when placed under the control of the cytomegalovirus IE promoter, confirming recent observations by K. A. Fraser and S. A. Rice (J. Virol. 81:5091-5101, 2007). We extended this analysis to show that ICP22 can also be expressed from its own promoter in the presence of other viral factors, either by coexpression with ICP0 or by infection with an ICP22 null virus. Notably, infection of cells transfected with an ICP22 expression vector yielded ICP22 protein that was modified in a manner similar to that of ICP22 protein detected in wild-type-infected cells. We go on to demonstrate that the failure of ICP22 protein to be expressed in transiently transfected cells was not due to inactivity of the ICP22 promoter, but rather to the ability of ICP22 to inhibit expression of reporter gene activity, including its own, in transient assays. Of special note was the observation that expression of ICP22 was sufficient to prevent transactivation of reporter genes by ICP0. Finally, transient expression of ICP22 was sufficient to complement replication of an ICP22 null virus, demonstrating that this system can be used to study functional properties of ICP22. Collectively, this transient expression system facilitates tests of the physical and functional properties of ICP22 and ICP22 mutants prior to introduction of mutant genes into the viral genome.
Upon infection of the host cell by herpes simplex virus type 1 (HSV-1), a highly ordered transcriptional cascade is initiated from the viral genome. The first viral proteins to be expressed, the immediate early (IE) proteins infected cell protein 0 (ICP0), ICP4, ICP22, and ICP27, prime the cell to support efficient viral replication. While the IE proteins have been studied extensively over the past 3 decades, a precise picture of how they function to promote viral replication is not yet available. At present, the least is known about ICP22, as functional characterization of this protein has been hindered by difficulty in expressing ICP22 protein in transient assays. Since it is desirable to know the physical and functional properties of wild-type (WT) and mutant forms of the protein before introduction of specific mutant genes into the viral genome, genetic analysis of ICP22 has also been hindered.
Although study of ICP22 in isolation has been limited, experiments comparing infections with existing ICP22 null viruses to infection with WT HSV-1 have revealed several interesting mutant properties. First, ICP22 is not required for replication of HSV-1 in cell culture; however; the absence of ICP22 reduces the efficiency of viral replication (8, 11, 12, 16). While this phenotype is modest (a <10-fold reduction in viral replication efficiency) in some immortalized cell lines, including Vero, Hep-2, and HeLa cells (permissive cells), replication in the majority of primary cells and rodent cells is reduced by several orders of magnitude (restrictive cells). It is unclear at this time what cellular factors distinguish permissive cells from restrictive cells; however, it is unlikely that the difference is due solely to transformation, as many restrictive cell types are transformed (e.g., Rab-9 cells) (8, 16). Second, cells infected with ICP22− viruses produce viral RNAs at reduced levels and fail to accumulate WT levels of several late viral proteins (8, 11, 16). These phenotypic properties are believed to be due, at least in part, to ICP22-induced alteration of the large subunit of RNA Pol II (15). Third, virions produced by cells infected with ICP22− viruses sediment at densities that differ from the density of WT virions in sucrose gradients and contain altered levels of tegument proteins (9). Finally, the phenotype of ICP22− viruses is most striking in the mouse eye model. Replication of ICP22− viruses is greatly restricted in the eye, trigeminal ganglion, and cerebellum, and viral DNA loads in latently infected trigeminal ganglion are markedly reduced (9). Not surprisingly, the virulence of ICP22− viruses is also greatly reduced compared to that of the WT virus (10, 17, 9).
In an effort to understand the mechanisms that underlie the phenotypes of ICP22− viruses, several groups have attempted to characterize ICP22 when expressed in isolation. Initial studies by Prod'hon et al. (13) examined the effect of expression of ICP22 on HSV promoter activity. In contrast to its role in upregulating late viral gene expression during infection, cotransfection of ICP22 alone with a panel of HSV-1 promoters repressed transcription of reporter genes. This study further demonstrated that the repressive function of ICP22 was partially abrogated by coexpression of the UL13 gene, which encodes a viral Ser/Thr kinase. The observed repression of viral gene expression by ICP22 first noted by Prod'hon et al. has been corroborated by several groups (2, 5). To date, however, the mechanism underlying this repression remains unclear. An unfortunate aspect of many of these studies is that they failed to show that ICP22 was even expressed in the transient reporter assays, making it difficult to conclude that ICP22 expression resulted in repression of the reporter gene.
Transient expression of ICP22 has also been used to examine the subcellular localization of ICP22 (18). In this study, researchers fused full-length ICP22 or fragments thereof to green fluorescent protein (GFP) and were able to identify and map two nuclear localization signals within the ICP22 open reading frame (ORF). Although the data used to identify the nuclear localization signals were convincing, expression of full-length ICP22 fused to GFP was reported only in a single cell, again reflecting the difficulty in expressing the protein transiently. Most recently, Fraser and Rice reported expression of ICP22 at readily detectable levels by Western blotting from the cytomegalovirus (CMV) promoter in the plasmid pCDNA3 (4). Despite this recent breakthrough, it is interesting to note that the authors were unable to detect ICP22 when expression was driven from the CMV promoter in an alternative plasmid. Using the pCDNA3:ICP22 expression cassette, Fraser and Rice demonstrated that ICP22 alone is sufficient to alter the phosphorylation state of the C-terminal domain of RNA Pol II. A similar strategy using the pCDNA3 vector was employed by Cun et al. to demonstrate transcriptional repression by ICP22 alone (2). The use of the CMV promoter in pCDNA3 as an engine to drive ICP22 expression presents numerous opportunities for screening and characterizing mutant forms of ICP22 without the need to introduce mutations directly into the viral genome.
In the present study, we attempted to determine whether ICP22 can be expressed from its own promoter. To this end, we first transfected cells with an ICP22 expression cassette containing the ICP22 ORF under the control of its own promoter and then infected these cells with an ICP22− virus. We show that ICP22 can be expressed efficiently from its own regulatory elements when transfected cells are either (i) infected with an ICP22 null virus or (ii) cotransfected with an ICP0 expression vector. This is the first demonstration showing that ICP22 can be expressed transiently from its own regulatory elements. We further demonstrate that the inability to detect ICP22 expressed from its own promoter in the absence of ICP0 is not specific to the ICP22 promoter but, rather, is a consequence of the ability of ICP22 to downregulate transcription of all promoters in transient assays. In our hands, the repressive activity of ICP22 in transient assays is sufficiently strong to inhibit activation of gene expression by ICP0. Finally, the efficiency of expression of ICP22 from either its own promoter or the CMV promoter is sufficient to complement ICP22− virus replication in restrictive cells, providing a transient assay with which to characterize physical and functional properties of mutant forms of ICP22.
MATERIALS AND METHODS
Cells and viruses.
Vero (ATCC CCL-81) and Rab-9 (ATCC CRL-1414) cells were propagated in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 100 mM penicillin-streptomycin (Invitrogen), and 2 mM l-glutamine (Invitrogen) at 37°C in 5% CO2. The WT virus used in these studies was HSV-1 strain KOS in passage 8 from initial isolation. All viruses used in the study were propagated in Vero cells by standard protocols.
Plasmids. (i) p22:eGFP.
p22:eGFP was constructed to replace the ICP22 ORF with the enhanced GFP (eGFP) cassette. The eGFP-N1 vector (Clontech, Mountain View, CA) was cut with BamHI and BglII and religated to remove the multiple cloning site (MCS) (eGFP:−MCS). eGFP:−MCS was cut with AscI and AflII, and the ends were filled to produce a blunt-ended fragment containing both the CMV promoter and the full-length eGFP ORF. The p22 vector was cut with AseI, the ends were filled in with Klenow, and the vector was religated to remove the AseI site within the vector sequences (p22A). p22A was digested with EagI and EcoN1 to remove the ICP22 ORF. The ends were filled in, and the eGFP fragment was ligated into the p22A vector to produce p22:eGFP. The orientation of the insert was determined by digestion with NotI and verified by sequencing.
(ii) ICP22 expression vectors.
In order to maximize the chance of identifying a plasmid that expressed ICP22, we constructed three additional ICP22 expression vectors.
(iii) pAlter22.
The 3.2-kb EcoRI-KpnI fragment containing the entire KOS US1 gene was subcloned from the p22 plasmid into pAlter (Promega, Madison, WI).
(iv) pCDNA3:ICP22.
pCDNA3:ICP22 was kindly provided by David Davido. The EcoRI-to-KpnI fragment of the ICP22 gene from p22 was subcloned into the same sites in pGem3zf(+) to make the plasmid pGem3zf(+)-ICP22. pGem3zf(+)-ICP22 was cut with EcoNI, filled in with Klenow and digested with XbaI. The 2.2-kb fragment containing the ICP22 ORF was isolated and cloned into the EcoRV and XbaI sites of pCDNA3.1.
(v) pCDNA3:22ORF.
The ICP22 ORF was PCR amplified with Platinum PFX (Invitrogen) from KOS infectious DNA, using primers to the 5′ (ATGGCCGACATTTCCCC) and 3′ (TCACGGCCGGAGAAACG) boundaries of the ICP22 ORF. The PCR product was cloned into the pTOPO:blunt vector per the manufacturer's protocol (Invitrogen). The ICP22 ORF was excised from the TOPO vector by digestion of pTOPO:ICP22 with EcoRI and subcloned into pCDNA3.1 (Invitrogen). The orientation of the insert was determined by restriction enzyme digestion and confirmed by sequencing.
(vi) Luciferase reporter plasmids.
The ICP0 promoter luciferase construct has been previously described (3). The ICP22, ICP4, TK, UL9, gC, and L42 promoter constructs were provided by Anna Kushnir and David Davido (submitted for publication). pGL3:CMV was constructed by cutting the 700-bp BglII-to-HindIII fragment containing the CMV IE promoter from pCDNA3.1 and ligating it into the same sites in the MCS in pGL3:basic.
Construction of d22:GFP.
In order to produce a 3.5-kb fragment containing the GFP ORF, the plasmid p22:eGFP was digested with EcoRI and KpnI. The fragment was purified and cotransfected with KOS infectious DNA (d22:GFP). Forty-eight hours posttransfection, total virus was harvested and plated onto Vero cells. Introduction of eGFP into the viral genome by homologous recombination was determined by the formation of green plaques on Vero cell monolayers. GFP-positive isolates were plaque purified three times. A final plaque isolate was amplified, and the integrity of the genome was confirmed by Southern blotting. Specifically, DNA isolated from KOS and d22:GFP-infected cells was digested with EcoRI and NotI, producing a 15-kb fragment for KOS and 2.3- and 12.9-kb fragments for d22:GFP (Fig. 1A to C). The digested DNA was separated in a 1% agarose gel and transferred to nitrocellulose. The blots were UV-cross-linked and blocked with ExpressHyb (Clontech) at 60°C. Probes specific for either ICP22 or eGFP were hybridized overnight at 60°C. The following day, blots were washed twice with 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS) at room temperature and twice for 1 h with 0.1× SSC and 0.1% SDS at 60°C. Blots were exposed to a phosphorimager screen (GE Healthcare, Sunnydale, CA), and images were analyzed by Imagequant software (Molecular Dynamics). Digestion of KOS DNA yielded a single band larger than 10 kb when probed for ICP22-specific sequences (Fig. 1D). Bands specific to the eGFP probe were not detected in the KOS sample. In contrast, ICP22-specific sequences were not detected in DNA isolated from d22:GFP-infected cells; however, a band migrating at 2.3 kb was detected with the eGFP probe. The larger 12.9-kb fragment was not detected in the d22:GFP-infected samples, as the probe was specific to sequences 5′ to the NotI site (Fig. 1C). Neither ICP22 nor the eGFP-specific bands were detected in mock-infected samples.
FIG. 1.
Construction and characterization of d22:GFP. (A) Diagram of the KOS genome. The positions of the unique (lines) and repeat (boxes) regions of the genome are indicated. The position of the US1 gene is noted. TRL, terminal repeat long; TRS, terminal repeat short; IRL, internal repeat long; IRS, internal repeat short. (B) The EcoRI-EcoRI fragment of the KOS genome containing US1 is shown. The positions of the US1 and US2 genes are represented by the gray boxes. The positions of the ICP22-specific probe is marked by the hatched bar. (C) Map of the d22:GFP genome. The ICP22 ORF was deleted (section in parentheses) and replaced with an eGFP cassette. Introduction of GFP introduced a novel fragment containing a NotI site, which is shown. The probe specific to eGFP is represented by the hatched bar. (D) Southern blot of KOS and d22:GFP. KOS and d22:GFP DNAs were digested with EcoRI and NotI and probed for ICP22- and eGFP-specific sequences by Southern blotting. (E) Replication of d22:GFP in Rab-9 cells. Replicate cultures of Rab-9 cells were infected with 2.5 PFU/cell KOS, 22/n199 (N199), or d22:GFP. At 24 hours postinfection, total virus was harvested and assayed on Vero cells by standard plaque assays. The experiment was repeated three times, and the error bars indicate the standard deviations. (F) Viral gene expression in d22:GFP-infected Rab-9 cells. Replicate cultures of Rab-9 cells were mock infected or infected with 2.5 PFU/cell KOS, 22/n199, or d22:GFP. At 6 hours postinfection, total cell lysates were prepared, and levels of ICP22, ICP4, and gC were determined by Western blotting.
Transfection.
Rab-9 and Vero cells were seeded in six-well plates at a density of 3 × 105 cells/well. Twenty-four hours postseeding, DMEM was replaced with 2 ml Opti-MEM (Invitrogen). Cells were transfected with Lipofectamine 2000 (6 μl Lipo/3 μg total DNA; Invitrogen) or Fugene 6 (3 μl Fugene/2 μg total DNA; Roche, Indianapolis, IN) per the manufacturer's instructions. When necessary, the total DNA in each reaction was normalized with carrier DNA. Six hours after addition of the transfection reagent, Opti-MEM was replaced with 2 ml of DMEM and incubated until cells were processed further as described below.
Western blotting.
ICP0, ICP4, ICP22, and gC protein levels were measured by Western blotting. At the time of harvest, Vero and Rab-9 cells were washed with phosphate-buffered saline and lysed in 250 μl radioimmunoprecipitation assay buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) by rocking for 30 min at 4°C. Lysates were transferred to Eppendorf tubes and clarified by centrifugation at 14,000 × g for 10 min. The supernatant fluid was transferred to a new tube containing 4× sample buffer (0.25 M Tris-HCl [pH 6.8], 8% SDS, 40% glycerol, 0.02% bromophenol blue, 10% beta-mercaptoethanol) and boiled for 3 min. Proteins in lysates equivalent to 2 × 104 cells were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane. Membranes were blocked for 1 h at room temperature in TBST buffer (25 mM Tris [pH 7.4], 3.0 mM KCl, 140 mM NaCl, and 0.1% Tween 20) with 5% nonfat milk. Blots were incubated with antibody (Ab) specific for the N terminus of ICP22 (8), ICP4 (Virusys Sykesville, MD), or ICP0 (Virusys) or gC-specific Ab (Virusys) overnight in TBST with 5% nonfat milk at 4°C at dilutions of 1:250, 1:5,000, 1:1,000, and 1:1,000, respectively. The next morning, the membranes were washed three times for 10 min with TBST buffer at room temperature. Goat anti-rabbit or mouse horseradish peroxidase-conjugated secondary Abs (Jackson Laboratories, Bar Harbor, ME) were diluted 1:25,000 in TBST buffer containing 5% nonfat milk and incubated with the blots for 45 min at room temperature. Blots were subsequently washed six times with TBST buffer for a minimum of 30 min per wash, treated with Millipore ECL reagent (Millipore, Bedford, MA) and exposed to X-ray film (Pierce, Rockford, IL).
Northern blotting.
In order to measure ICP22 and ICP0 transcript levels, Northern blot assays were performed essentially as described by Lee and Schaffer (6). Briefly, 3 × 105 Rab-9 cells per well were seeded into six-well plates. Twenty-four hours later, cells were transfected with the plasmids indicated, using Fugene 6. Total RNA was harvested 24 h later using Trizol (Invitrogen, Carlsbad, CA) per the manufacturer's instructions. Ten micrograms of RNA per sample was separated in 1% agarose-formaldehyde gels and transferred to nitrocellulose. Blots were UV-cross-linked, dried, and blocked for 1 h at 60°C. ICP22 (1)- and ICP0 (6)-specific riboprobes were synthesized using the Promega T7/SP6 in vitro transcription kit (Promega, Madison, WI) per the manufacturer's instructions. The probes were hybridized to blots overnight at 68°C and washed as previously described (1). Blots were exposed to a phosphorimager screen (GE Healthcare) and data analyzed using ImageQuant 3.3 software (GE Healthcare).
Luciferase assays.
Luciferase assays were performed in order to measure viral-promoter activity. Replicate cultures of Vero or Rab-9 cells were transfected with the plasmids as indicated in the figure legend. Twenty-four hours posttransfection, whole-cell lysates were prepared using passive lysis buffer (Promega) per the manufacturer's instructions. The levels of luciferase activity in each sample were measured using the luciferase reporter assay system (Promega, Madison WI) on a Microlumat LB 96 luminometer (Berthhold Technologies, Oak Ridge, TN).
Complementation assays.
In order to access the ability of ICP22-expressing plasmids to complement the replication of ICP22− virus, Rab-9 cells were seeded into replicate six-well plates at 3 × 105 cells/well. Twenty-four hours postplating, cells were transfected with the plasmids indicated in the figure legend, using Lipofectamine 2000. Twenty-four hours posttransfection, cells were infected with 2.5 PFU per cell of d22:GFP and harvested 24 h later. Production of new infectious virus was determined by a standard plaque assay on Vero cell monolayers. The complementation index was determined by dividing the amount of virus produced in cells transfected with the ICP22 expression vector by the amount of virus produced in cells transfected with a GFP expression vector. Introduction of ICP22 sequences into the viral genome by homologous recombination was monitored by looking for the presence of white plaques. In no case were white plaques detected.
RESULTS
Transfection efficiency in restrictive cell lines.
In order to develop a suitable complementation assay for functional studies of mutant forms of ICP22, it was necessary to use a cell line that is both highly transfectable and restrictive for replication of ICP22− viruses. To this end, we tested a panel of cell lines for their ability to express GFP in transient transfection assays and support ICP22− virus replication. Most of the restrictive cell lines tested, including human embryonic lung cells and human foreskin fibroblasts, were unable to achieve expression of GFP in more than 10% of the cells, making them unsuitable for our studies. In contrast, cell lines in which transfection was efficient (i.e., in which more than 70% of the cells expressed GFP), including Vero and ARPE-19 cells, were permissive for replication of the ICP22− viruses, supporting replication to 90% of the WT levels (data not shown). Rab-9 cells, an immortalized rabbit skin cell line, however, proved to be both restrictive for ICP22− virus replication (8) and moderately transfectable (between 30 and 40%). Consequently, Rab-9 cells were used in this study.
Construction and characterization of d22:GFP.
Although an ICP22 mutant virus, 22/n199, had previously been constructed in strain KOS, this virus expresses the first 198 amino acids of ICP22 which may exhibit function. In order to eliminate possible complications from expression of the N terminus of ICP22, a complete ICP22 deletion virus in KOS was necessary. To this end, the ICP22 ORF of strain KOS was deleted and replaced with an eGFP cassette containing the CMV promoter and full-length eGFP gene as described in Materials and Methods (Fig. 1B and C). Recombination of eGFP sequences into the viral genome and loss of ICP22 were documented by Southern blotting as described in Materials and Methods (Fig. 1D).
In order to determine whether the phenotypes of d22:GFP are similar to the phenotypes previously reported for other ICP22 null viruses, Vero and Rab-9 cells were infected with 2.5 PFU/cell of KOS, 22/n199, or d22:GFP. Twenty-four hours postinfection, total virus was harvested and quantified by standard plaque assays on Vero cell monolayers. Replication of d22:GFP was slightly reduced in permissive Vero cells (data not shown) but was restricted 30-fold in Rab-9 cells relative to replication of wild-type KOS (Fig. 1E), similar to levels observed in 22/n199-infected Rab-9 cells. Replication of d22:GFP was similarly restricted in several other known restrictive cell lines, including human foreskin fibroblasts and human embryonic lung cells (data not shown).
To further characterize the phenotype of d22:GFP in cell culture, levels of the viral IE proteins ICP22 and ICP4 and the late protein gC were compared in KOS-, 22/n199-, and d22:GFP-infected cells (Fig. 1F). To this end, Rab-9 cells were infected with 10 PFU/cell of each virus. At 8 hours postinfection, whole-cell lysates were prepared and levels of viral proteins analyzed by Western blotting. As expected, ICP22 was detected only in WT-infected cell lysates but not in lysates of 22/n199- or d22:GFP-infected Rab-9 cells. Consistent with previous reports, levels of ICP4 were not altered in Rab-9 cells infected with either 22/n199 or d22:GFP compared to those in KOS-infected Rab-9 cells. In contrast, levels of gC were greatly reduced in cells infected with d22:GFP and 22/n199. The reduction in gC levels was not observed in cells infected with a rescued virus (data not shown). It is important to note that while we observed a very large difference in the expression of gC in WT-infected cells versus that in ICP22 null virus-infected cells, it has been reported previously that the levels of gC were not affected by the absence of ICP22 (14). It is unclear from this previous study what conditions were used in these tests, but our data indicate that gC is an excellent marker for defects in KOS late gene expression observed in ICP22 null virus-infected cells. Collectively, the restriction in viral replication and the reduced levels of late viral gene expression make d22:GFP a suitable ICP22− virus for studies of ICP22 function.
Transient expression of ICP22.
When this project was initiated, expression of ICP22 in transient assays was poorly defined, and ICP22 expression in the absence of other viral proteins had been demonstrated only at or near the limit of detection. More recently, ICP22 was expressed at detectable levels when transcription was driven from the CMV promoter in the expression vector pCDNA3 (2, 4). To further characterize expression of ICP22 in isolation, a panel of ICP22 expression vectors (Fig. 2A) driven by the ICP22 or CMV promoters was tested for levels of ICP22 expression following transfection by Western blotting (Fig. 2B). The first vector, p22 (8), contains a KOS EcoRI-KpnI fragment that encompasses the entire US1 gene, including the ICP22 promoter and 5′ and 3′ untranslated region (UTR) sequences, cloned into pBR322. The same KOS fragment was subcloned into pAlter to yield the plasmid pAlter22. In order to express ICP22 from a heterologous promoter, the ICP22 ORF was subcloned from the p22 vector into pCDNA3 to generate plasmid pCDNA3:ICP22. This plasmid contains the full-length ICP22 ORF as well as 5′ and 3′ UTR sequences specific to ICP22 driven by the CMV promoter. Finally, in order to reproduce the findings of Fraser and Rice (4), the ICP22 ORF was subcloned into pCDNA3 via PCR to generate pCDNA3:22ORF in which ICP22 is also driven by the CMV promoter.
FIG. 2.
Transient expression of ICP22. (A) Maps of ICP22 expression vectors. HSV-1-specific sequences are denoted by solid lines. Dotted lines indicate vector sequences. The promoter and polyadenylation sequences utilized in each vector are noted. (B) Expression of ICP22 from plasmid vectors in transfected mock-infected and transfected d22:GFP-infected Vero and Rab-9 cells. The position of the ICP22-specific bands is marked with a bracket. A nonspecific band routinely detected with the anti-ICP22 Ab is marked with an arrow.
(i) Transfection.
To test for ICP22 expression from these vectors, replicate cultures of Vero and Rab-9 cells were transfected with the plasmids indicated in Fig. 2A. Twenty-four hours later, whole-cell lysates were prepared and ICP22 levels determined by Western blot analysis. The Ab used in these studies is specific for the N terminus of ICP22 and is therefore unable to detect the smaller, C-terminal protein US1.5.
ICP22 was below the limit of detection in cells transfected with p22, pAlter22, or pCDNA3:ICP22 in both Vero cells and Rab-9 cells (Fig. 2B). In contrast, a single band migrating at approximately 68 kDa, the expected size of ICP22, specific for ICP22 Ab was detected in Vero and Rab-9 cells transfected with pCDNA3:22ORF (Fig. 2B). This finding is consistent with the data published by Fraser and Rice (4) that ICP22 is expressed efficiently from the CMV promoter in pCDNA3. It is unclear why ICP22 was below the limit of detection for pCDNA3:ICP22, as this plasmid utilizes the same promoter as pCDNA3:22ORF. We hypothesized that regulatory elements located within the 5′ and 3′ UTR sequences might regulate ICP22 expression; however, transfer of the 5′ and 3′ UTR elements from pCDNA3:ICP22 to pCDNA:22ORF did not result in loss of ICP22 protein expression (data not shown). Though puzzling, these data seem consistent with other observations discussed above regarding the expression of ICP22 in transient assays, specifically the failure to express ICP22 from a pCI vector, which also utilizes the CMV promoter (4).
(ii) Transfection/infection.
Although ICP22 was expressed when the ICP22 ORF was subcloned into the CMV expression cassette, one concern was whether the other ICP22 expression vectors were competent to express ICP22. To address this question, Vero and Rab-9 cells were transfected with the ICP22 expression vectors or a GFP control and infected 24 h later with d22:GFP (Fig. 2B). At 6 hours postinfection, whole-cell lysates were prepared and levels of ICP22 assayed by Western blotting. ICP22 was not detected in either Vero or Rab-9 cells transfected with the GFP reporter vector, demonstrating that, as expected, ICP22 is not expressed from the incoming virus (Fig. 2B). In contrast, Vero and Rab-9 cells transfected with all four of the ICP22 expression vectors and infected with d22:GFP expressed ICP22 at high levels. Notably, ICP22 expressed in cells infected with d22:GFP migrated as multiple bands, consistent with the high degree of posttranslation modification of this protein detected in WT-infected samples. These data demonstrate that all four vectors are competent to express ICP22. In addition, they show that ICP22 can be expressed and modified in transient assays when the cells are infected with an ICP22− virus, raising the possibility that mutant forms of ICP22 can be characterized in transient assays without first introducing the mutation into the viral genome.
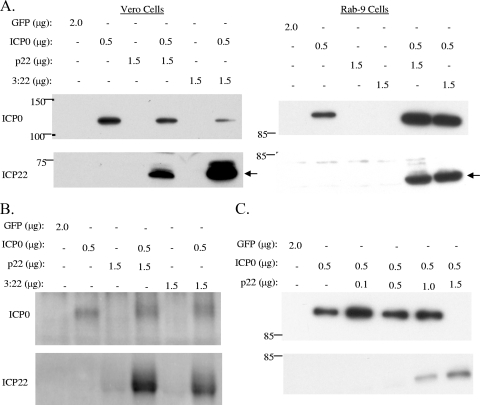
Coexpression of ICP22 with ICP0.
Because ICP22 was expressed transiently in the presence of other viral factors, we wanted to determine if expression of specific viral factors facilitated efficient expression of ICP22. Given the role of ICP0 as a global transcriptional activator, we hypothesized that it may induce ICP22 expression. To test this hypothesis, Vero cells and Rab-9 cells were transfected with the ICP22 expression plasmids indicated in Fig. 3A. Replicate cultures were cotransfected with the ICP0 expression vector, pSH, which drives expression of ICP0 from its own promoter. Twenty-four hours later, whole-cell lysates were prepared, and levels of ICP22 and ICP0 were measured by Western blotting. As previously described, ICP22 was not detected in cells transfected with p22 or pCDNA3:ICP22 alone. In contrast, ICP22, evident as an approximately 68-kDa band, was readily detected when Vero and Rab-9 cells were cotransfected with the ICP0 expression vector, demonstrating that ICP0 expression is sufficient to induce the expression of ICP22. ICP0 was detected in all samples transfected with the ICP0 expression vector. Expression of ICP22 was not detected when the p22 plasmid was cotransfected with an ICP4 expression vector (data not shown).
FIG. 3.
Cotransfection of ICP0 and ICP22. (A) Western blots of Vero and Rab-9 cells cotransfected with plasmids expressing ICP22 and ICP0. The ICP22-specific band is marked with an arrow. (B) ICP0 and ICP22 RNA levels in transfected Rab-9 cells as determined by Northern blotting. (C) ICP22 protein expression in Rab-9 cells transfected with increasing amounts of an ICP0 expression vector.
In order to determine if the enhanced ICP22 expression was due to an increase in ICP22 transcript levels, the experiment was repeated, and levels of ICP22 RNA were examined. To this end, Rab-9 cells were transfected with the plasmids shown in Fig. 3B. Twenty-four hours later, total RNA was harvested, and levels of ICP0 and ICP22 RNAs were assayed by Northern blotting. As expected, ICP0 RNA was detected in cells transfected with the ICP0 expression vector, pSH. ICP22 RNA was detected at very low levels in cells transfected with the ICP22 expression vectors alone. In contrast, ICP22 RNA levels were greatly increased in the presence of ICP0, consistent with ICP0's recognized function as a global transcriptional activator.
Since ICP0 was able to induce expression of ICP22 in trans, we next wanted to determine if ICP0-induced ICP22 expression was dose dependent, in order to be able to vary the amount of ICP22 for functional studies. To this end, Rab-9 cells were transfected with increasing amounts of the ICP22 expression vector, p22, and a constant amount of the ICP0 expression vector, pSH (Fig. 3C). Twenty-four hours later, whole-cell lysates were prepared, and levels of ICP22 and ICP0 were determined by Western blotting. As described previously, ICP22 was not detected in cells transfected with p22 alone but was readily detected when cotransfected with ICP0. The levels of ICP22 protein increased with increasing amounts of ICP22 expression vector transfected, demonstrating that the amount of ICP22 expressed can be regulated. ICP0 was detected in all samples transfected with the ICP0 expression vector, with one notable exception. At the highest concentration of ICP22 expression vector tested (1.5 μg), ICP0 levels reproducibly fell below the limit of detection. This dominance of ICP22 expression over ICP0 occurred repeatedly as levels of ICP22 increased and will be discussed further below.
ICP22 promoter activity in Vero cells.
Since both the ICP22 transcript and protein were not detected when expressed from the ICP22 promoter in the absence of other viral factors, we wanted to determine whether the ICP22 promoter is active in cell culture. To test this possibility, Vero cells were transfected with a panel of viral promoter-luciferase reporter constructs (Fig. 4). Plasmid p36, a minimal, cellular promoter, was included as a negative control. Twenty-four hours later, cells were lysed, and the levels of firefly luciferase activity were measured. As expected, p36 failed to drive high levels of luciferase expression. In contrast, and as expected, the strong ICP0 promoter induced high levels of luciferase expression. Similarly, the ICP4 promoter also induced high levels of luciferase expression. The early UL9 and late L42 promoters induced little luciferase expression, yielding levels as low as those of p36, the basal promoter control. The gC and TK promoters both induced luciferase expression but not as efficiently as the ICP0 and ICP4 promoters. The ICP22/47 promoter induced high levels of luciferase activity, approximately equal to the levels produced by ICP4. Results similar to those observed in Vero cells were also observed in Rab-9 and HeLa cells (data not shown). Since the ICP22 promoter was highly active in all cell types tested, the failure to demonstrate ICP22 protein expression from its own promoter (Fig. 2 and 3) must be explained by an alternative hypothesis.
FIG. 4.
ICP22 promoter activity in Rab-9 cells. Vero cells were transfected with the luciferase reporter constructs indicated. Data presented are the averages of the results for three experiments. Error bars indicate the standard deviations.
Repression of luciferase reporter activity by ICP22.
Since previous studies have suggested that ICP22 can repress transcription from a large number of promoters in transient reporter assays, we wanted to determine if ICP22 can repress expression from its own promoter (13). To this end, Vero cells were transfected with the ICP22 promoter-luciferase construct and increasing amounts of pCDNA3:22ORF. Twenty-four hours posttransfection, total cell lysates were prepared and luciferase activity measured (Fig. 5A). Transfection of the ICP22 reporter plasmid alone yielded high levels of firefly luciferase expression. Luciferase activity was reduced with increasing amounts of the ICP22 expression vector to 20% at the highest concentration. Notably, ICP22 protein was detected in replicate samples (Fig. 5B), suggesting that the repressive effect was due to expression of ICP22. These data extend observations presented by Prod'hon et al. (13) and demonstrate that ICP22 can repress its own promoter. In addition, similar results were observed when cells were transfected with other ICP22 expression vectors but not with plasmids expressing only the 200 N-terminal amino acids of ICP22 (data not shown). ICP22 also repressed luciferase activity of other HSV-1 promoter-reporter constructs, including ICP0, VP16, TK, and VP5 (data not shown), consistent with previous reports. Although the mechanism of repression of the reporter activity by ICP22 is unclear, we believe it most likely acts on promoters. While we present data generated using a luciferase reporter assay, Prod'hon et al. observed similar results utilizing a CAT reporter gene, which strongly argues that ICP22 does not affect reporter enzyme activity. In addition, the general defect in reporter activity observed in the presence of transiently expressed ICP22 is consistent with the observation that ICP22 expression is sufficient to eliminate phosphorylation of serine 2 of the RNA Pol II CTD heptad repeat. Phosphorylation of Ser2 within the heptad repeat is thought to be necessary for efficient elongation of the Pol II complex, and thus, loss of Ser2 phosphorylation should have a global effect on transcription. Taken together, these observations indicate that ICP22 expressed in isolation is capable of inhibiting its own expression as well as expression of other HSV proteins in transient assays.
FIG. 5.
Repression of promoter activity by ICP22. (A) ICP22 represses its own promoter. Vero cells were transfected with the ICP22 luciferase reporter construct and increasing amounts of the ICP22 expression vector. Firefly luciferase activity was measured and normalized to the “no pCDNA3:22ORF” control. (B) Western blots of ICP22 expression. MT represents mock-transfected cells, and the plus sign indicates a positive cell lysate control. (C) ICP22 expression represses expression from the CMV IE promoter. (D) ICP22 prevents transactivation by ICP0. The ICP22 promoter construct was cotransfected with the ICP0 expression vector, pSH, and increasing concentrations of pCDNA3:22ORF as indicated. The level of luciferase activity was measured 24 hours posttransfection. Data presented are the averages of the results for three experiments. Error bars indicate the standard deviations.
While this hypothesis is intriguing, it is unclear if ICP22 acts as a universal repressor of all reporters in transient assays or is specific to HSV promoters. Alteration of the Pol II CTD suggests a global inhibition of transcription; however, ICP22 protein was detected when expressed from the CMV IE promoter, suggesting that the CMV promoter is not affected by ICP22 protein expression. Similarly, data presented by Prod'hon et al. reported that the CMV promoter was not downregulated by expression of ICP22 in CAT assays. In order to confirm this observation, we tested the CMV IE promoter activity in the presence of ICP22 in our promoter-luciferase assay. Results of initial experiments using large amounts of the CMV reporter construct (>1 μg) argued that expression of ICP22 had little to no effect on reporter expression (data not shown), consistent with our Western blots and the reporter data presented by Prod'hon et al. However, given the high levels of signal produced in these experiments, we were concerned that we were outside the linear range of the assay. We therefore reduced the amount of the CMV IE promoter construct to 0.1 μg and tested the ability of ICP22 to downregulate reporter activity. We found that, like the ICP22 promoter, increasing levels of ICP22 reduced expression from the CMV IE promoter to approximately 10% of the activity at the highest level of ICP22 tested (Fig. 5C). Given this observation, we also tested the SV40 early promoter and the adenovirus major late promoter and found that reporter activity was reduced in all cases in the presence of the ICP22 expression vector.
Since ICP0 facilitates efficient expression of ICP22 in transient assays, we hypothesized that ICP0 may prevent repression by ICP22. To test this hypothesis, the ICP22 promoter-luciferase reporter constructs were cotransfected with the ICP0 expression vector, pSH (which drives ICP0 expression from its own promoter), and increasing amounts of the ICP22 expression vector, pCDNA3:ICP22ORF (Fig. 5D). As expected, ICP0 greatly enhanced expression of the ICP22 promoter as well as that of a large panel of representative viral genes, consistent with ICP0's well-known ability to activate gene expression (data not shown). Unexpectedly, ICP0 did not prevent repression by ICP22; in fact, the opposite effect was observed. Cotransfection of ICP22 with ICP0 resulted in reduction (to less than 5%) in ICP22 promoter activity compared to cells transfected with ICP0 alone. Similar results were obtained when ICP0 was expressed from the CMV IE promoter. This observation poses an interesting conundrum, as ICP0 can increase the expression of ICP22, but at the same time, repression of the promoter by ICP22 can prevent or overcome transactivation by ICP0. It is unclear if ICP22 is interfering directly with activation by ICP0 or, alternatively, if expression of ICP0 is reduced in the presence of ICP22 and is therefore not able to transactivate to maximum levels. Unfortunately, the levels of ICP0 expressed in these assays were below the limit of detection.
Complementation of d22:GFP replication in restrictive Rab-9 cells.
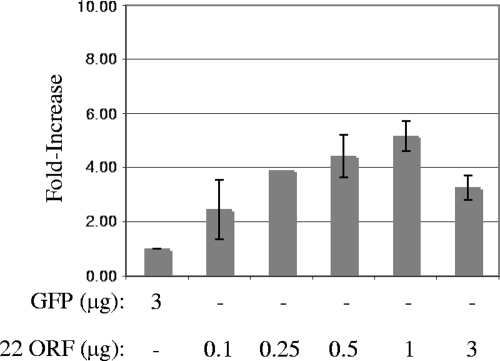
Having demonstrated multiple approaches to expressing ICP22 transiently, we next asked if expression of ICP22 can complement the replication of an ICP22− virus. For this test, Rab-9 cells were transfected with increasing amounts of pCDNA3:22ORF or a GFP control vector, pCMV:GFP. Twenty-four hours later, cells were infected with 2.5 PFU per cell of d22:GFP, and total virus replication was measured 24 hours postinfection by a standard plaque assay on Vero cell monolayers. Infection of Rab-9 cells transfected with pCMV:GFP yielded no increase in d22:GFP replication over input (approximately 1 PFU/cell) in cells transfected with GFP alone, as shown in Fig. 6. In contrast, transfection of cells with the ICP22 expression vector increased the total amount of virus produced (fivefold with 1 μg of DNA). Similar results were obtained when cells were transfected with p22 or pAlter22 (data not shown). These findings demonstrate that ICP22− viruses can be complemented, albeit modestly, in transfection-infection assays.
FIG. 6.
Complementation of d22:GFP replication in Rab-9 cells. Rab-9 cells were transfected with the amounts of plasmid indicated. Total virus replication was scored by a plaque assay on Vero cells and reported as an n-fold increase over that on pCMV:GFP-transfected cells. Bars represent the averages of the results for three experiments (except for at 0.25 μg, for which there were two experiments), and error bars represent the standard deviation.
DISCUSSION
In this study, we demonstrate multiple means of expressing ICP22 in transient assays: (i) by driving expression from the CMV promoter; (ii) by cotransfection of an ICP22 expression vector with the viral regulatory protein, ICP0; and (iii) by infection of cells transfected with an ICP22 expression vector with an ICP22 null virus. We also show that the difficulty in achieving expression of ICP22 in transfected cells is not a consequence of ICP22 promoter inactivity but, rather, a consequence of the ICP22 protein repressing expression from its own promoter. Notably, in our hands, this repressive activity was strong enough to inhibit the potent CMV IE promoter or ICP0-mediated transactivation of viral promoters. Finally, ICP22 expressed in cells infected with d22:GFP migrates as multiple bands by SDS-PAGE, suggesting that the protein is modified as it is in WT-infected cells and is able to complement replication of an ICP22 null virus, demonstrating that both physical and functional properties of the protein can be studied in transient assays prior to construction of mutant viruses.
Transient expression of ICP22.
Difficulty in achieving and detecting transient expression of ICP22 has been a road block to the elucidation of ICP22 function. The original report demonstrating transient expression of ICP22 utilized a construct with the SV40 promoter driving ICP22 expression, but the conditions used to detect expression of the ICP22 protein were different from those used to assay its functional properties (13). Recent studies demonstrating detectable levels of ICP22 using CMV promoter-driven vectors should greatly facilitate further study of this gene (4). Interestingly, not all CMV promoter:ICP22 constructs express ICP22 as subcloning of the ICP22 ORF into pCI did not allow for efficient expression of ICP22 but pCDNA3 did. In addition, we show here that different means of cloning ICP22 into pCDNA3 yielded two vectors that were both competent to express ICP22 following infection with an ICP22− virus, but only one, pCDNA3:22ORF, was able to express the protein in isolation. This observation suggested that sequences 5′ and/or 3′ of the ICP22 ORF may downregulate ICP22 expression in the absence of other viral factors; however, transfer of these elements to pCDNA3:ICP22ORF did not affect ICP22 protein expression (data not shown). It is unclear why these similar vectors differ in the ability to express ICP22 and why transfer of unique ICP22-specific sequences within pCDNA3:ICP22 to pCDNA3:22ORF did not alter ICP22 expression. At this time it appears necessary to generate multiple ICP22 expression vectors and determine empirically which are able to express in the absence of other viral factors.
Functional and physical characterization of ICP22 expressed in isolation.
Transfection of ICP22 expression vectors, under the control of either the ICP22 promoter or a heterologous promoter, led to efficient expression of ICP22 following infection with an ICP22− virus. Notably, while expression of ICP22 in isolation led to a single band in Western blots, infection with an ICP22− virus led to extensive modification of ICP22, similar to that observed in WT virus-infected cells. Moreover, transient expression of ICP22 was sufficient to complement replication of an ICP22− virus in restrictive Rab-9 cells. The level of complementation was modest, as it was limited by the transfection efficiency of Rab-9 cells and the ability of Rab-9 cells to support low levels of ICP22− viral replication. We believe this system could be further optimized by increasing the efficiency of transfection in cells that are more restrictive to ICP22− virus replication. Efforts to develop such a system are under way. Clearly, the ability to complement ICP22 null virus replication with transfected ICP22 will make screening of the functional properties of ICP22 mutants much more time efficient than will generating and testing ICP22 mutant viruses.
Regulation of gene expression by ICP22.
It had been reported previously, and confirmed in this report, that cotransfection of ICP22 with a panel of HSV reporter genes resulted in the repression of reporter protein activity (13). We have extended upon this observation and shown that the ICP22 promoter, as well as several other HSV promoters, is also inhibited by expression of ICP22. We go on to demonstrate that the CMV IE promoter is also downregulated in the presence of ICP22, suggesting that ICP22 acts as a general repressor of transcription. This observation contradicts data reported by Prod'hon et al. (13), and we believe that the difference in results is a consequence of the assays used. In this report, we have taken advantage of the luciferase assay system, which has a much greater linear range than do the CAT assays previously used by Prod'hon et al. We think that the data presented by Prod'hon et al. fell outside the linear range of the CAT assay and that little to no effect of ICP22 on CMV promoter activity was observed due to the system being overwhelmed. In support of this, we found that transfection of large amounts of the CMV promoter:luciferase construct overcomes inhibition by ICP22 (data not shown). This might also explain why ICP22 protein can be detected in some cases when expressed from the CMV promoter. In effect, providing the CMV promoter in excess can outcompete the repressive activity of ICP22.
In addition to inhibiting promoters in isolation, we also found that ICP22 expression was sufficient to prevent transactivation by ICP0 (Fig. 5D). It is unclear how ICP22 is able to reduce gene expression in the presence of ICP0; however, we propose that the inhibition is not specific but, rather, is the result of two general mechanisms. First, as ICP22 has been shown to efficiently downregulate expression from all HSV promoters in reporter assays, it seems likely that ICP22 would downregulate expression of the ICP0 protein in cotransfection assays. Evidence for this is seen in Fig. 3C where increasing amounts of the ICP22 expression vector, and ICP22 protein, resulted in loss of ICP0 expression. Similarly, increased amounts of the ICP0 expression vector in transient reporter assays were able to overcome repression by ICP22 (data not shown). Unfortunately, we have been unable to show that ICP0 is expressed in the transient reporter assays, as ICP0 levels are below the limit of detection. Efforts to improve sensitivity are ongoing to confirm this hypothesis. Second, expression of ICP22 is sufficient for alteration of the RNA Pol II CTD, suggesting that ICP22 is inhibiting transcriptional initiation and/or elongation. As ICP0 is believed to work upstream of transcriptional initiation by preventing association of heterochromatin with viral promoters, blocking downstream steps, such as transcriptional initiation, should result in reduced reporter activity.
Since all the promoters we have tested have been inhibited by expression of ICP22, we believe ICP22 expressed in isolation globally shuts down transcription; however, these assays were all performed using transient expression systems. It is unclear if ICP22 expression alters transcription from the host cell genome. Given that ICP22 expression is sufficient to alter the phosphorylation of the RNA Pol II CTD, we believe it is likely that host cell transcription will be disrupted, and experiments to address this hypothesis are under way.
While ICP22 expressed in isolation acting as a global repressor is intriguing, viruses lacking ICP22 fail to express viral late genes to WT levels, particularly in restrictive cells, suggesting that ICP22 upregulates gene expression in infected cells (16). In addition, a general defect in transcription initiation has been reported for ICP22− viruses, and these viruses fail to alter the mobility of the large subunit of RNA Pol II (15). One clear difference between the two systems is that ICP22 expressed in isolation is not modified in the same manner as is ICP22 expressed during infection. In support of this observation, it has been reported that coexpression of UL13 with ICP22 in transient assays partially alleviates repression of promoter:reporter constructs by ICP22; however, data showing expression of ICP22 and UL13 in these assays were not presented, nor was evidence that UL13 modifies ICP22 when coexpressed in isolation (13). Nevertheless, it seems reasonable to assume that modification of ICP22 by viral kinases, such as UL13, could result in altered activity, although virion-associated UL13 appears not to have this function (14). Now that we have assays to address both physical and functional properties of ICP22, we have initiated experiments addressing the role UL13 plays in regulating ICP22.
Together, these observations suggest a model, originally proposed by Prod'hon et al. (13), in which the ICP22 initially expressed would act as a repressor of viral gene expression, in direct competition with the viral activating proteins (such as ICP0). In support of this model, we have shown that ICP22 expressed to sufficient levels can repress ICP0-mediated transactivation in transient assays, and this observation extends to activation by ICP4 (T. Astor and D. Davido, unpublished observations). If enough ICP22 was expressed in relation to the transactivating genes, then the genome would be pushed toward a quiescent state. In contrast, if ICP0 and ICP4 levels were high, lytic genes would be expressed, including UL13, which can phosphorylate ICP22 and inhibit its repressive activity. In support of this concept, varying the amounts of ICP22 and ICP0 expression plasmids had competing effects on viral promoter:reporter constructs (J. S. Orlando, unpublished observations). Infection of most cells would favor this latter scenario, as ICP0, ICP4, and VP16 (but not ICP22) are all present in the viral tegument and, thus, are present in the infected cell prior to any new viral gene expression, potentially masking any repressive effects that might be observed from newly synthesized ICP22. Further support for this model comes from the study of UL13− viruses in which, like ICP22− viruses, viral late gene transcriptional initiation is reduced (7). Experiments designed to further test this model, particularly reproduction of the effect of UL13 on ICP22 expression and activity, and experiments determining if ICP22 can inhibit transactivation by VP16 are currently underway. In addition, analysis of the effect of ICP22 on viral promoter activity in neurons is being explored. While infection of epithelial cells with WT virus results almost exclusively in the production of a new infectious virus, infection of neurons may result in the establishment of latency. We hypothesize that the repressive activities of ICP22 may play an important role in the establishment of latency by suppressing viral gene expression very early after infection of primary neurons. In support of this hypothesis, a direct comparison of IE gene expression during productive infection of primary neurons and epithelial cells indicated that there is a delay in RNA synthesis in neurons (1; J. Balliet, J. Min, J. Bowman and P. Schaffer, unpublished observations).
Acknowledgments
We thank members of the Schaffer laboratory for review and discussions of the data presented in the manuscript.
This work was supported by Public Health Service grant RO1 CA20260 from the National Cancer Institute to P.A.S.
Footnotes
Published ahead of print on 17 June 2009.
REFERENCES
- 1.Balliet, J. W., J. C. Min, M. S. Cabatingan, and P. A. Schaffer. 2005. Site-directed mutagenesis of large DNA palindromes: construction and in vitro characterization of herpes simplex virus type 1 mutants containing point mutations that eliminate the oriL or oriS initiation function. J. Virol. 7912783-12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cun, W., M. Hong, L. D. Liu, C. H. Dong, J. Luo, and Q. H. Li. 2006. Structural and functional characterization of herpes simplex virus 1 immediate-early protein infected-cell protein 22. J. Biochem. 14067-73. [DOI] [PubMed] [Google Scholar]
- 3.Davido, D. J., and D. A. Leib. 1998. Analysis of the basal and inducible activities of the ICPO promoter of herpes simplex virus type 1. J. Gen. Virol. 792093-2098. [DOI] [PubMed] [Google Scholar]
- 4.Fraser, K. A., and S. A. Rice. 2007. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J. Virol. 815091-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwun, H. J., S. W. Yim, D. H. Lee, and K. L. Jang. 1999. Activation of the thymidine kinase promoter by herpes simplex virus type 1 immediate early proteins. Mol. Cells 9277-280. [PubMed] [Google Scholar]
- 6.Lee, L. Y., and P. A. Schaffer. 1998. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell-type-specific expression of γ134.5 transcripts and latency-associated transcripts. J. Virol. 724250-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 735593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlando, J. S., T. L. Astor, S. A. Rundle, and P. A. Schaffer. 2006. The products of the herpes simplex virus type 1 immediate-early US1/US1.5 genes downregulate levels of S-phase-specific cyclins and facilitate virus replication in S-phase Vero cells. J. Virol. 804005-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orlando, J. S., J. W. Balliet, A. S. Kushnir, T. L. Astor, M. Kosz-Vnenchak, S. A. Rice, D. M. Knipe, and P. A. Schaffer. 2006. ICP22 is required for wild-type composition and infectivity of herpes simplex virus type 1 virions. J. Virol. 809381-9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poffenberger, K. L., A. D. Idowu, E. B. Fraser-Smith, P. E. Raichlen, and R. C. Herman. 1994. A herpes simplex virus type 1 ICP22 deletion mutant is altered for virulence and latency in vivo. Arch. Virol. 139111-119. [DOI] [PubMed] [Google Scholar]
- 11.Poffenberger, K. L., P. E. Raichlen, and R. C. Herman. 1993. In vitro characterization of a herpes simplex virus type 1 ICP22 deletion mutant. Virus Genes 7171-186. [DOI] [PubMed] [Google Scholar]
- 12.Post, L. E., and B. Roizman. 1981. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25227-232. [DOI] [PubMed] [Google Scholar]
- 13.Prod'hon, C., I. Machuca, H. Berthomme, A. Epstein, and B. Jacquemont. 1996. Characterization of regulatory functions of the HSV-1 immediate-early protein ICP22. Virology 226393-402. [DOI] [PubMed] [Google Scholar]
- 14.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 906701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice, S. A., M. C. Long, V. Lam, P. A. Schaffer, and C. A. Spencer. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 695550-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sears, A. E., I. W. Halliburton, B. Meignier, S. Silver, and B. Roizman. 1985. Herpes simplex virus 1 mutant deleted in the alpha 22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sears, A. E., B. Meignier, and B. Roizman. 1985. Establishment of latency in mice by herpes simplex virus 1 recombinants that carry insertions affecting regulation of the thymidine kinase gene. J. Virol. 55410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stelz, G., E. Rucker, O. Rosorius, G. Meyer, R. H. Stauber, M. Spatz, M. M. Eibl, and J. Hauber. 2002. Identification of two nuclear import signals in the alpha-gene product ICP22 of herpes simplex virus 1. Virology 295360-370. [DOI] [PubMed] [Google Scholar]