Abstract
Highly active antiretroviral therapy (HAART) can reduce human immunodeficiency virus type 1 (HIV-1) viremia to clinically undetectable levels. Despite this dramatic reduction, some virus is present in the blood. In addition, a long-lived latent reservoir for HIV-1 exists in resting memory CD4+ T cells. This reservoir is believed to be a source of the residual viremia and is the focus of eradication efforts. Here, we use two measures of population structure—analysis of molecular variance and the Slatkin-Maddison test—to demonstrate that the residual viremia is genetically distinct from proviruses in resting CD4+ T cells but that proviruses in resting and activated CD4+ T cells belong to a single population. Residual viremia is genetically distinct from proviruses in activated CD4+ T cells, monocytes, and unfractionated peripheral blood mononuclear cells. The finding that some of the residual viremia in patients on HAART stems from an unidentified cellular source other than CD4+ T cells has implications for eradication efforts.
Successful treatment of human immunodeficiency virus type 1 (HIV-1) infection with highly active antiretroviral therapy (HAART) reduces free virus in the blood to levels undetectable by the most sensitive clinical assays (18, 36). However, HIV-1 persists as a latent provirus in resting, memory CD4+ T lymphocytes (6, 9, 12, 16, 48) and perhaps in other cell types (45, 52). The latent reservoir in resting CD4+ T cells represents a barrier to eradication because of its long half-life (15, 37, 40-42) and because specifically targeting and purging this reservoir is inherently difficult (8, 25, 27).
In addition to the latent reservoir in resting CD4+ T cells, patients on HAART also have a low amount of free virus in the plasma, typically at levels below the limit of detection of current clinical assays (13, 19, 35, 37). Because free virus has a short half-life (20, 47), residual viremia is indicative of active virus production. The continued presence of free virus in the plasma of patients on HAART indicates either ongoing replication (10, 13, 17, 19), release of virus after reactivation of latently infected CD4+ T cells (22, 24, 31, 50), release from other cellular reservoirs (7, 45, 52), or some combination of these mechanisms. Finding the cellular source of residual viremia is important because it will identify the cells that are still capable of producing virus in patients on HAART, cells that must be targeted in any eradication effort.
Detailed analysis of this residual viremia has been hindered by technical challenges involved in working with very low concentrations of virus (13, 19, 35). Recently, new insights into the nature of residual viremia have been obtained through intensive patient sampling and enhanced ultrasensitive sequencing methods (1). In a subset of patients, most of the residual viremia consisted of a small number of viral clones (1, 46) produced by a cell type severely underrepresented in the peripheral circulation (1). These unique viral clones, termed predominant plasma clones (PPCs), persist unchanged for extended periods of time (1). The persistence of PPCs indicates that in some patients there may be another major cellular source of residual viremia (1). However, PPCs were observed in a small group of patients who started HAART with very low CD4 counts, and it has been unclear whether the PPC phenomenon extends beyond this group of patients. More importantly, it has been unclear whether the residual viremia generally consists of distinct virus populations produced by different cell types.
Since the HIV-1 infection in most patients is initially established by a single viral clone (23, 51), with subsequent diversification (29), the presence of genetically distinct populations of virus in a single individual can reflect entry of viruses into compartments where replication occurs with limited subsequent intercompartmental mixing (32). Sophisticated genetic tests can detect such population structure in a sample of viral sequences (4, 39, 49). Using two complementary tests of population structure (14, 43), we analyzed viral sequences from multiple sources within individual patients in order to determine whether a source other than circulating resting CD4+ T cells contributes to residual viremia and viral persistence. Our results have important clinical implications for understanding HIV-1 persistence and treatment failure and for improving eradication strategies, which are currently focusing only on the latent CD4+ T-cell reservoir.
MATERIALS AND METHODS
Patient populations.
In the analysis of population structure between circulating activated and resting CD4+ T cells, three distinct sources of patient sequences were used. These datasets included a set of patient sequences obtained from Chun et al. (10) (patients 1, 2, 4, 5, 7, and 8), Bailey et al. (1) (patient 154), and Chun et al. (11) (patients J2 and J7). We failed to obtain complete datasets from either of the two Chun et al. (10, 11) studies. In addition, patient identifiers used in our study do not match those used in the two Chun et al. (10, 11) studies, due primarily to randomization of patient data. We had complete access to all patients and sequences used in the Bailey et al. (1) study, of which patient 154 was used in the analysis of circulating activated versus resting CD4+ T cells.
In the analysis of population structure between proviruses in circulating resting CD4+ T cells and free plasma virus, we utilized two distinct sources of patient data. The first set of patient sequences was obtained from the Bailey et al. study (1). The second source of patient data was derived in our lab, through the sequence analysis of newly enrolled patients. We enrolled asymptomatic HIV-1-infected adults who maintained suppression of viremia on antiretroviral drugs to below the limit of clinical detection (<50 copies/ml). These patients had all maintained stable suppression for at least 6 months prior to enrolling in our study, and most had maintained suppression for much longer. A summary of all patient characteristics can be found in Table S2 in the supplemental material. Sampling from these newly enrolled patients occurred at study entry, as well as at subsequent time points. Patients who volunteered donated 180 ml of blood/visit. Some patients returned periodically over the course of 3 years to provide additional blood samples. Table S3 in the supplemental material contains the periodic sampling data for all newly enrolled patients. Obtaining sequences from the plasma of patients with clinically undetectable viral loads is technically challenging, and it is often not possible to recover many, if any, sequences. Therefore, we included in the present study only patients from whom we were able to obtain a sufficient number of sequences. We chose this sufficient number to mean no less than 20 plasma and 20 proviral sequences from any single patient. Our protocol was approved by a Johns Hopkins institutional review board, and informed consent was obtained from all study participants.
Amplification, cloning, and sequencing of the env gene from free plasma virus and proviruses in circulating resting CD4+ T cells.
A 180-ml sample of blood was collected at each study visit using an acid-citrate-dextrose anticoagulant and separated by using a Ficoll density gradient. After gradient separation, the plasma layer was quickly removed, centrifuged to remove any contaminating cells, and immediately frozen and stored at −80°C until further use. The buffy coat layer was subsequently removed from the Ficoll tubes, and resting CD4+ T cells were purified from total peripheral blood mononuclear cells (PBMC) via magnetic bead depletion, as previously described (16). Purified resting CD4+ T cells were lysed with a commercial detergent cell lysis solution (Gentra), and the lysate was frozen at −80°C until further use.
To analyze free virus in the plasma, 6-ml aliquots of plasma were thawed and subjected to ultracentrifugation at 170,000 × g for 30 min at 4°C. Pelleted virus was subsequently resuspended in 400 μl of phosphate-buffered saline (Invitrogen) and lysed, and the RNA was extracted via a silica bead-based RNA isolation protocol, implemented on an EZ1 Biorobot (Qiagen). The RNA was eluted in 60 μl of elution buffer and subsequently treated with amplification grade DNase I (Invitrogen) in accordance with the manufacturer's instructions. To amplify region C2-V4 of the env gene from RNA isolated from free plasma virus, the RNA was subjected to a one-step reverse transcriptase PCR (RT-PCR) using a Superscript III RT/Platinum Taq High-Fidelity DNA polymerase one-step RT-PCR kit (Invitrogen), followed by a nested PCR, using Platinum Taq High-Fidelity DNA polymerase and 2.5 μl of the outer reaction as a template. Control reactions were carried out for all experimental amplifications, including a no-RT control to rule out DNA contamination and a no-template control. Primers for the outer and nested reactions were as follows: outer forward, 5′-CTGTTAAATGGCAGTCTAGC-3′; outer reverse, 5′-CACTTC TCCAATTGTCCCTCA-3′; nested forward, 5′-ACAATGCTAAAACCATAATAGT-3′; and nested reverse, 5′-CATACATTGCTTTTCCTACT-3′. The PCR conditions were as follows: for one-step RT-PCR, reverse transcription at 50°C for 30 min and denaturation at 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 1 min; and for the nested reaction, denaturation at 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 1 min. The products of the nested reaction were separated on 1% agarose gels, bands of appropriate size were excised, and the corresponding amplicons were eluted by using QIAquick gel extraction kits (Qiagen). Isolated amplicons were subsequently cloned by using a PCR2.1 TOPO cloning vector (Invitrogen), and at least six clones were sequenced from each PCR by using an ABI Prism 3700 DNA analyzer (Applied Biosystems). All sequences generated for newly enrolled patients have been deposited in GenBank.
To analyze provirus in circulating resting CD4+ T cells, DNA from purified, lysed cells was isolated by using the Puregene method (Gentra). An outer and nested PCR designed to amplify full-length env was then carried out on the isolated DNA in a limiting dilution fashion, as previously described (1). The amplification was carried out by using AccuPrime Pfx DNA polymerase (Invitrogen) and 5 μl of template DNA. The following primers were used in these reactions: outer forward, 5′-ATGGCAGGAAGAAGCGGAGACAG-3′; outer reverse, 5′-GCTCAACTGGTACTAGCTTGAAGCACC-3′; nested forward, 5′-GATAGACGCGTAGAAAGAGCAGAAGACAGTGGCAATG-3′; and nested reverse, 5′-CCTTGTGCGGCCGCCTTAAAGGTACCTGAGGTCTGACTGG-3′. The PCR conditions were as follows: denaturation at 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 68°C for 3 min. The products of the nested reaction were then separated on 0.8% agarose gels. Bands of the appropriate size were excised, and the amplicons were eluted by using QIAquick gel extraction kits. Because the reactions were set up in a limiting dilution fashion, with >90% Poisson probability of being clonal, isolated amplicons could be directly sequenced as outlined above, without the need for a cloning step. Clonality was ensured after direct sequencing via a manual inspection of the corresponding electropherograms for doublet peaks. Only sequences that were verified as being clonal were included in our analysis.
Sequence analysis.
Several steps were taken to ensure that we were working with quality sequences, devoid of contamination, PCR error, and PCR resampling (28). We also took measures to ensure that all sequences analyzed were clonal and derived from independent reactions. All of these procedures have been previously described (1).
Phylogenetic analysis.
Sequences were subjected to multiple sequence alignment with the HXB2 reference sequence by using Gene Cutter (http://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html), preserving codon positions. The quality of the alignments was manually inspected, and adjustments were made when necessary, using the sequence editor BioEdit version 7.0.9 (Tom Hall, Ibis Biosciences, Carlsbad, CA). Gaps in multiple alignments were removed prior to estimating phylogenies in cases where there were length polymorphisms. Duplicate identical sequences derived from independent reactions were removed from the alignment for phylogenetic reconstruction but subsequently added back to the trees. Phylogenies were estimated by using both a “classical” approach and a Bayesian approach, both functioning under a maximum-likelihood (ML) optimality criterion. The classical approach was implemented using a web-based version of RAxML (44) available through the CIPRES supercomputing cluster (http://www.phylo.org/). We used the general time-reversible model of nucleotide substitution with an estimation of the proportion of invariant sites and with gamma-distributed rate variation and included an M-group ancestral sequence as an outgroup. The precision of phylogenetic reconstruction (nodal support) was assessed via bootstrap analysis, with the number of bootstrap pseudoreplicates determined empirically by the software. The Bayesian approach (21) was implemented by using a web-based version of MrBayes, also available through the CIPRES web portal. Again, we used the general time-reversible model of nucleotide substitution with an estimation of the proportion of invariant sites and with gamma distributed rate variation and included an M-group ancestral sequence used as an outgroup. For each patient, we carried out the Bayesian inference by running 10 Markov-chain-Monte-Carlo chains, each starting from a random tree. Each chain ran for 2.0 × 107 generations, with samples taken every 100th generation. Phylogenetic trees were visualized by using TreeView version 1.6.6 (33, 34).
Analysis of population structure.
We defined population structure as the presence of more than one distinct genetic population in a group of sequences. To ascertain the presence or absence of population structure in sequences derived from distinct sources, we used two complementary statistical tests. The first test, subsequently referred to as the Slatkin-Maddison test (SM) (43), is implemented in the software package HyPhy (26). This test is a phylogeny-based test that enumerates the minimum number of inferred migration events between two or more populations on the basis of the reconstructed phylogeny. Briefly, each population is assigned a character state, and each sequence derived from that population is labeled with the corresponding character state. The enumeration process begins at the terminal leaves of the tree and moves up, inferring an ancestral character state to each ancestral node. For each character-state mismatch between an ancestral node and one of its descendants, a migration event is inferred. We emphasize that the number of inferred migration events in itself is not meaningful, because the HIV-1 life cycle violates the assumptions of the two-island models in whose context the SM test was originally formulated. Nevertheless, we can use this quantity reliably to test for evidence against incomplete mixing, as explained next.
The SM null hypothesis states that the two (or more) character states are randomly sampled from one large intermixing population. If the null hypothesis is true, then sequences with randomly permuted character states should yield a comparable number of inferred migration events as found in the original data. If the null is false, however, then randomly permuting group assignments should increase the number of inferred migration events. The SM test assumes both the lack of recombination and the lack of selection. Both of these assumptions may be violated in HIV. To address this limitation, we also carried out a genetic-distance-based test of the population structure that does not rely on these assumptions (see below).
We carried out SM tests by adding back all of the duplicate, identical clones to the reconstructed phylogenetic trees, removing the M-group ancestral outgroup, and inputting the resulting trees into HyPhy. The software then enumerated the minimum number of migration events and calculated statistical support against the null hypothesis nonparametrically by randomly permuting group assignments (character states) 1,000 times, recalculating the minimum number of migration events, thus generating the sampling distribution. From this distribution, we estimated the P value by determining the cumulative weight of migration events in the sampling distribution less than or equal to the number of inferred events in the original data. We subsequently subjected the raw P values for all patients to a Benjamini-Hochberg false discovery rate (FDR) correction for multiple significance tests (2).
The second test of population structure involved the analysis of molecular variance (AMOVA) (4, 14, 39). This test is implemented in the software package Arlequin (2). It is a genetic-distance-based test that first calculates Euclidean pairwise distances within and between predefined groups and then partitions covariance components to the respective groups. The test is analogous to a nested analysis of variance, except that the normality assumption is not required. Statistical support for the observed population structure is determined nonparametrically by permuting group assignments 1,000 times and recalculating all statistics to generate their sampling distributions. We again used the FDR method to correct for multiple testing.
We carried out AMOVAs by first formatting aligned sequences into batch files for input into the Arlequin program. Briefly, using a text editor, we partitioned sequences into groups defined by their source, removed the outgroup sequences, and specified the copy number of each sequence. The batch file also contained information regarding the specific population structure to be tested. In all cases, we were testing whether or not sequences from two distinct sources represented a single intermixing population or not. Once the batch files were set up appropriately, they were brought into Arlequin. We let Arlequin compute a distance matrix using Tamura and Nei corrected distances and an empirically defined α shape parameter for gamma-distributed rate variation.
Nucleotide sequence accession numbers.
All newly obtained sequences for the present study are available at GenBank under accession numbers GQ256402 to GQ256627 and GQ261350 to GQ261724.
RESULTS
Proviruses in circulating activated and resting CD4+ T cells belong to one intermixing population.
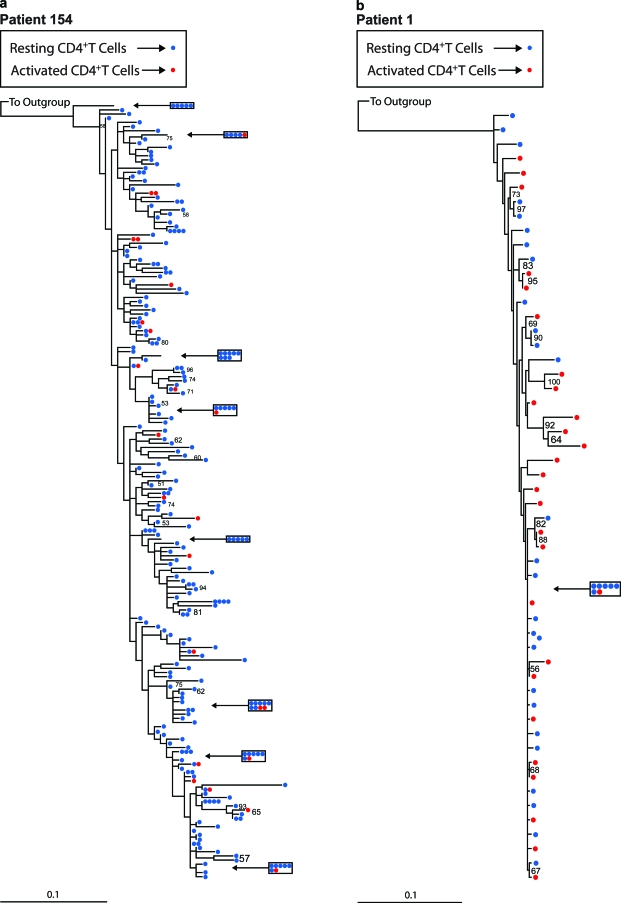
We first tested for the presence of population structure in sets of proviral sequences derived from circulating activated and resting CD4+ T cells. Because activated and resting CD4+ T cells represent a single population of cells at different stages of activation, genetically distinct populations of proviruses are not expected unless activated CD4+ T cells are frequently infected de novo by virus from a distinct origin. To assess population structure, we utilized two complementary tests, the SM test (43) and AMOVA (14), on published (1, 10, 11) and newly obtained datasets. The SM test (43) assesses whether sequences belonging to two or more groups are randomly distributed over the leaves of a phylogenetic tree encompassing all sequences. We used two methods of phylogenetic reconstruction, classical ML and Bayesian inference. In most cases, both methods yielded identical tree topologies, with only minor differences in branch lengths. Figure 1 shows representative ML trees consisting of proviral sequences derived from activated and resting CD4+ T cells from a patient with a defined PPC (1) (Fig. 1a) and a patient without a PPC (Fig. 1b). Both trees show evidence of intermingling between proviral sequences derived from activated and resting CD4+ T cells, devoid of any obvious population level structure. Similar results were obtained for other patients (see Fig. S1a to e and g in the supplemental material).
FIG. 1.
Phylogenetic analysis of proviral sequences derived from activated and resting CD4+ T cells for two representative patients. The trees were generated by using a classical ML approach and were rooted with an M group ancestral sequence. Red circles refer to proviral sequences derived from activated CD4+ T cells, and blue circles correspond to proviral sequences derived from resting CD4+ T cells. Clusters of five or more identical sequences are boxed. Bootstrap values of 50% or greater are shown. (a) ML tree from a patient with a defined predominant plasma sequence (PPC); (b) ML tree from a patient without a PPC.
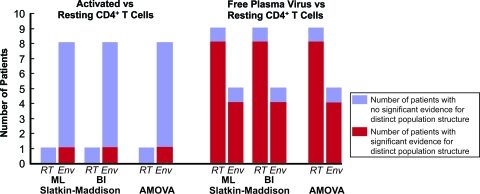
The results from the SM test were consistent with the intermingled nature of the sequences illustrated in the reconstructed phylogenies. In all but one patient, we could not reject the null hypothesis that no population structure existed between the two groups of sequences (Table 1 and Fig. 2). The phylogenetic tree for patient J2, the only patient for whom we could reject the null hypothesis, contained a clade of free plasma virus sequences devoid of any proviral sequences, as well as a clade of proviral sequences devoid of related free plasma sequences (see Fig. S1f in the supplemental material). However, this pattern was atypical. For all of the other patients, the phylogenetic analysis showed a lack of compartmentalized structure. Similar results were obtained when the SM test was performed using phylogenies constructed with the Bayesian inference method (Table 1 and Fig. 2).
TABLE 1.
Analysis of population structure between provirus derived from activated and resting CD4+ T cells
| Patienta | Gene amplified | PPC presentb | No. of independent sequences in cells
|
SM analysisc
|
AMOVA analysis
|
Molecular diversity (π)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Activated cells | Resting cells |
P
|
Adjusted Pd
|
Migration events (ML/BI) | % Variation among or within populations | P | Adjusted P | Activated CD4+ T cells | Resting CD4+ T cells | ||||||
| ML | BI | ML | BI | ||||||||||||
| 154* | RT | Yes | 24 | 242 | 0.130 | 0.140 | 0.170 | 0.209 | 22/22 | -0.25 | 100.25 | 0.602 | 0.63 | 0.019 | 0.020 |
| 1† | Env | NA | 28 | 32 | 0.103 | 0.537 | 0.170 | 0.604 | 17/20 | 1.19 | 98.81 | 0.117 | 0.263 | 0.028 | 0.018 |
| 2† | Env | NA | 23 | 20 | 0.683 | 0.504 | 0.768 | 0.604 | 15/14 | 1.07 | 98.93 | 0.240 | 0.309 | 0.117 | 0.096 |
| 4† | Env | NA | 21 | 22 | 0.106 | 0.043 | 0.170 | 0.097 | 12/11 | 0.65 | 99.35 | 0.186 | 0.297 | 0.090 | 0.107 |
| 5† | Env | NA | 21 | 17 | 0.022 | 0.036 | 0.100 | 0.095 | 9/9 | 10.75 | 89.25 | 0.025 | 0.075 | 0.065 | 0.035 |
| 7† | Env | NA | 20 | 22 | 0.132 | 0.021 | 0.170 | 0.100 | 12/10 | 1.97 | 98.03 | 0.198 | 0.297 | 0.066 | 0.054 |
| 8† | Env | NA | 21 | 17 | 0.871 | 0.871 | 0.871 | 0.871 | 14/14 | -0.94 | 100.94 | 0.633 | 0.633 | 0.034 | 0.038 |
| J2‡ | Env | NA | 22 | 20 | 0.002 | 0.000 | 0.018 | 0.000 | 8/8 | 19.73 | 80.27 | 0.000 | 0.000 | 0.080 | 0.095 |
| J7‡ | Env | NA | 28 | 28 | 0.120 | 0.112 | 0.170 | 0.112 | 16/16 | 4.02 | 95.98 | 0.022 | 0.075 | 0.067 | 0.091 |
*, Patient sequences derived from Bailey et al. (1); †, patient sequences derived from Chun et al. (10); ‡, patient sequences derived from Chun et al. (11).
A PPC was previously defined as being a sequence that represented >50% of all independent sequences from the plasma and <1% of all independent proviral sequences from the resting CD4+ T-cell compartment (1). NA, not applicable (patients could not be evaluated for the presence of a PPC).
ML, phylogenetic trees were generated via a classical ML approach; BI, phylogenetic trees were generated via a Bayesian inference approach.
“Adjusted P” refers to P values obtained for both the SM tests and the AMOVA tests subjected to Benjamini-Hochberg FDR corrections, treating P values obtained by using the ML approach separately from those obtained using BI and both separately from the AMOVA tests (2).
FIG. 2.
Histogram illustrating the proportion of patients studied that exhibit significant evidence for distinct population structure. Blue bars indicate the number of patients with no significant evidence of population structure. Red bars indicate the number of patients with significant evidence of population structure.
Since the SM test relies on phylogenies, which are difficult to estimate in situations where there is low diversity and possible recombination, we also included a phylogeny-independent test of population structure, a genetic-distance-based AMOVA (14). This test analyzes within and among-group molecular variation in a nested analysis-of-variance-like framework. The results of the AMOVA matched those of the SM tests. For all patients but patient J2, the null hypothesis of no population structure could not be rejected (Table 1 and Fig. 2). In other words, proviral sequences derived from circulating resting and activated CD4+ T cells in a typical patient comprise one intermixing genetic population with no significant structure. This result is consistent with the known biology of these cells.
Provirus derived from circulating resting CD4+ T cells and free plasma virus represent distinct genetic populations in most patients.
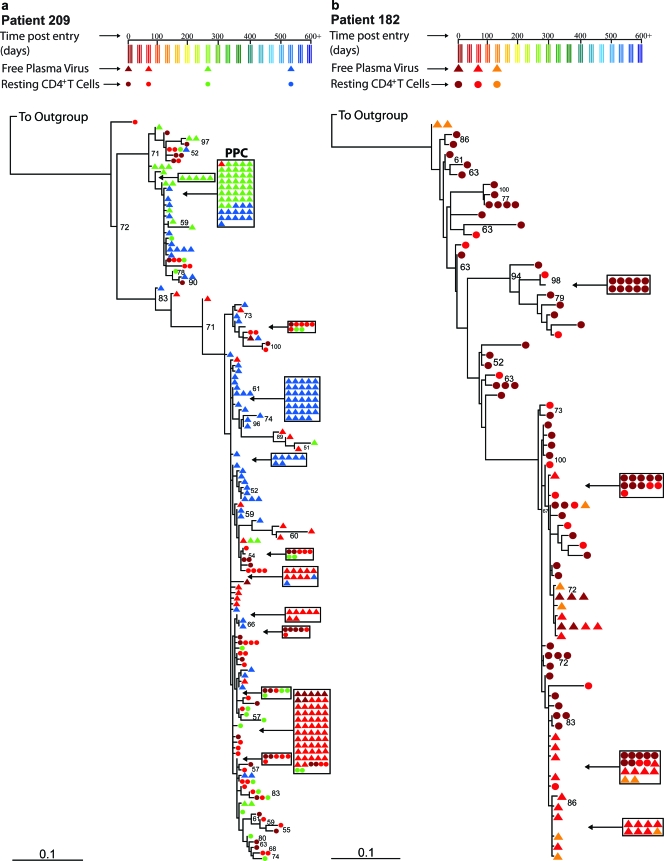
To assess the presence of population structure between free plasma virus and provirus derived from resting CD4+ T cells, we carried out the same tests as described above. Figure 3 shows representative ML trees of proviral sequences derived from resting CD4+ T cells and sequences from free plasma virus for a patient with a defined PPC (1) (Fig. 3a) and a patient without a PPC (Fig. 3b). A PPC was previously defined as a clonal, free-plasma-virus-derived sequence that represented >50% of the total free plasma virus sequences, while also representing <1% of the total proviral sequences obtained from circulating CD4+ T cells. In both cases, some of the plasma sequences were identical to sequences found in the CD4+ reservoir. Thus, at least some of the plasma virus may have been produced by latently infected CD4+ T cells that became activated. However, both trees reveal a tendency for some plasma and reservoir sequences to segregate, in contrast to the completely intermingled pattern observed for provirus in resting and activated CD4+ T cells.
FIG. 3.
Phylogenetic analysis of sequences derived from free plasma virus and proviral sequences derived from resting CD4+ T cells for two representative patients. The trees were generated by using a classical ML approach and were rooted with an M group ancestral sequence. Circles refer to proviral sequences derived from resting CD4+ T cells, and triangles correspond to sequences derived from free plasma virus. Clusters of five or more identical sequences are boxed. The colors of the circles and triangles correspond to the time point at which blood was drawn from the patient. Bootstrap values of 50% or greater are shown. (a) ML tree from a patient with a defined predominant plasma sequence (PPC); (b) ML tree from a patient without a PPC.
The SM analysis revealed significant population structure between the two sources of virus in all but two patients: patient 134 and patient 202 (Table 2 and Fig. 2). For patients 134 and 202, the phylogenetic trees show a pattern of intermingling between proviral sequences and free plasma virus sequences not unlike the pattern we found for proviral sequences derived from activated and resting CD4+ T cells (see Fig. S1k and l in the supplemental material). However, for all of the remaining patients, the phylogenetic trees show segregating proviral and free-plasma sequences (Fig. 3 and see Fig. S1h to j and m to s in the supplemental material). The results were not affected by the method of phylogeny reconstruction (Table 2).
TABLE 2.
Analysis of population structure between free plasma virus and provirus derived from resting CD4+ T cells
| Patienta | Gene amplified | PPC presentb | No. of independent sequences
|
SM analysisc
|
AMOVA analysis
|
Molecular diversity (π)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Free plasma virus | Resting CD4+ T cells |
P
|
Adjusted Pd
|
Migration events (ML/BI) | % Variation among or within populations | P | Adjusted P | Free plasma virus | Resting CD4+ T cells | ||||||
| ML | BI | ML | BI | ||||||||||||
| 113* | RT | Yes | 27 | 24 | 0.005 | 0.008 | 0.006 | 0.009 | 12/12 | 11.53 | 88.47 | 0.000 | 0.000 | 0.007 | 0.013 |
| 135* | RT | Yes | 98 | 151 | 0.000 | 0.000 | 0.000 | 0.000 | 16/22 | 20.45 | 79.55 | 0.000 | 0.000 | 0.013 | 0.024 |
| 139* | RT | Yes | 45 | 83 | 0.000 | 0.000 | 0.000 | 0.000 | 16/16 | 45.43 | 54.57 | 0.000 | 0.000 | 0.018 | 0.021 |
| 148* | RT | Yes | 51 | 138 | 0.000 | 0.000 | 0.000 | 0.000 | 11/11 | 39.23 | 60.77 | 0.000 | 0.000 | 0.006 | 0.020 |
| 154* | RT | Yes | 238 | 242 | 0.000 | 0.000 | 0.000 | 0.000 | 42/44 | 38.43 | 61.57 | 0.000 | 0.000 | 0.011 | 0.020 |
| 099* | RT | No | 105 | 68 | 0.000 | 0.000 | 0.000 | 0.000 | 28/29 | 3.14 | 96.86 | 0.003 | 0.004 | 0.009 | 0.011 |
| 147* | RT | No | 47 | 89 | 0.005 | 0.007 | 0.006 | 0.008 | 32/32 | 2.29 | 97.71 | 0.026 | 0.030 | 0.021 | 0.019 |
| 136* | RT | No | 54 | 66 | 0.000 | 0.000 | 0.000 | 0.000 | 26/27 | 1.84 | 98.16 | 0.007 | 0.009 | 0.004 | 0.004 |
| 134* | RT | No | 12 | 73 | 0.095 | 0.108 | 0.102 | 0.110 | 10/10 | -1.18 | 101.18 | 0.707 | 0.707 | 0.021 | 0.022 |
| 154* | Env | Yes | 67 | 28 | 0.000 | 0.000 | 0.000 | 0.000 | 16/16 | 10.38 | 89.62 | 0.000 | 0.000 | 0.062 | 0.068 |
| 182† | Env | No | 40 | 93 | 0.000 | 0.000 | 0.000 | 0.000 | 9/10 | 17.97 | 82.03 | 0.000 | 0.000 | 0.024 | 0.089 |
| 202† | Env | No | 27 | 18 | 0.113 | 0.110 | 0.113 | 0.110 | 12/12 | -0.76 | 100.76 | 0.525 | 0.565 | 0.064 | 0.077 |
| 206† | Env | No | 24 | 24 | 0.000 | 0.000 | 0.000 | 0.000 | 7/6 | 19.25 | 80.75 | 0.000 | 0.000 | 0.071 | 0.110 |
| 209† | Env | Yes | 262 | 113 | 0.000 | 0.000 | 0.000 | 0.000 | 17/16 | 5.51 | 94.49 | 0.001 | 0.002 | 0.096 | 0.077 |
*, Patient sequences derived from Bailey et al. (1); †, new patients analyzed for this study.
A PPC was previously defined as being a sequence that represented >50% of all independent sequences from the plasma and <1% of all independent proviral sequences from the resting CD4+ T-cell compartment (1).
ML, phylogenetic trees were generated via a classical ML approach; BI, phylogenetic trees were generated via a Bayesian inference approach.
“Adjusted P” refers to P values obtained for both the SM tests and the AMOVA tests subjected to Benjamini-Hochberg FDR corrections, treating P values obtained using the ML approach separately from those obtained using BI and both separately from the AMOVA tests (2).
The AMOVA results again matched the SM results (Table 2 and Fig. 2). Taken together, these data suggest that the residual viremia is compartmentalized and includes one or more virus populations that are genetically distinct from the proviruses in circulating resting CD4+ T cells.
In a subset of patients on HAART, the residual viremia is dominated by a PPC (1). PPCs were detected in 6 of 13 patients studied (Table 2). The presence of a PPC would be expected to influence the analysis of population structure. In fact, for the RT gene in patients with PPCs, AMOVA found that between 10 and 40% of all molecular variation was among groups (i.e., distinguished proviral from free-plasma sequences), while between 60 and 90% of all molecular variation was shared between the two sequence groups (Table 2). For the RT gene in patients without PPC, on the other hand, the among-group variation was only between 2 and 3%. (In patient 134, the percent variation among populations is negative. Negative percentages can arise in AMOVA [14] but usually coincide with nonsignificant P values.) In contrast, for the Env gene, the absence of a PPC seems to increase rather than decrease the percent variation among populations (Table 2).
To determine to what extent PPCs were influencing the analysis and to determine whether the plasma sequences other than the PPC were genetically distinct from sequences in resting CD4+ T cells, we applied our analyses to datasets stripped of all PPCs (see Table 4). We found that even though PPCs do influence our analyses, in most patients there remains significant population structure even after removal of all PPC sequences. For the SM tests, in all but one patient, patient 113, we can reject the null hypothesis of no population structure after removing the PPCs (see Table 4). The data set for patient 113 had the least number of sequences, and the change of result for this patient may simply reflect loss of statistical power. In the AMOVA analyses, all but two patients, patients 113 and 209, still exhibit a significant difference after removing the PPC. Removal of the PPCs resulted in a reduction of among-population variation of approximately 10 to 30% for RT and 5 to 6% for Env. Surprisingly, for RT, the among-population variation for patients 139, 148, 154 after removal of the PPC still exceeded the among-population variation of any of the patients without PPC. Taking all results together, we find a consistent pattern of free plasma virus and provirus from resting CD4+ T cells showing significant population structure in the majority of patients, regardless of the presence or absence of a PPC and regardless of the gene sequenced.
TABLE 4.
Analysis of population structure between free plasma virus and provirus after removal of all PPCs
| Patient or patient 154 extended-analysis typea | Gene amplified | No. of independent sequences
|
SM analysisb
|
AMOVA analysis
|
Molecular diversity (π)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Free plasma virus | Integrated provirus |
P
|
Adjusted Pc
|
Migration events (ML/BI) | % Variation among or within populations | P | Adjusted P | Free plasma virus | Provirus | |||||
| ML | BI | ML | BI | |||||||||||
| Patient | ||||||||||||||
| 113* | RT | 14 | 24 | 0.432 | 0.219 | 0.432 | 0.229 | 11/10 | -1.45 | 101.45 | 0.704 | 0.704 | 0.009 | 0.013 |
| 135* | RT | 31 | 151 | 0.000 | 0.000 | 0.000 | 0.000 | 16/18 | 2.34 | 97.66 | 0.037 | 0.046 | 0.025 | 0.024 |
| 139* | RT | 15 | 83 | 0.000 | 0.000 | 0.000 | 0.000 | 16/16 | 17.59 | 82.41 | 0.000 | 0.000 | 0.029 | 0.021 |
| 148* | RT | 9 | 138 | 0.006 | 0.008 | 0.008 | 0.011 | 11/11 | 11.01 | 88.99 | 0.005 | 0.008 | 0.021 | 0.020 |
| 154* | RT | 75 | 242 | 0.000 | 0.000 | 0.000 | 0.000 | 42/44 | 6.68 | 93.32 | 0.000 | 0.000 | 0.021 | 0.020 |
| 154* | Env | 46 | 28 | 0.004 | 0.003 | 0.007 | 0.005 | 16/16 | 4.60 | 95.40 | 0.018 | 0.026 | 0.063 | 0.068 |
| 209† | Env | 179 | 113 | 0.000 | 0.000 | 0.000 | 0.000 | 17/16 | 1.66 | 98.34 | 0.055 | 0.061 | 0.067 | 0.077 |
| Patient 154 extended analysis | ||||||||||||||
| Plasma virus vs provirus derived from activated CD4+ T Cells | Env | 75 | 25 | 0.035 | 0.229 | 0.039 | 0.229 | 19/21 | 5.57 | 94.43 | 0.001 | 0.002 | 0.021 | 0.019 |
| Plasma virus vs provirus derived from monocytes | Env | 75 | 17 | 0.005 | 0.020 | 0.007 | 0.025 | 12/13 | 8.13 | 91.87 | 0.001 | 0.002 | 0.021 | 0.020 |
| Plasma virus vs provirus derived from unfractionated PBMC | Env | 75 | 83 | 0.000 | 0.000 | 0.000 | 0.000 | 35/36 | 6.11 | 93.89 | 0.000 | 0.000 | 0.021 | 0.021 |
*, Patient sequences derived from Bailey et al. (1); †, new patient analyzed for this study.
ML, phylogenetic trees were generated via a classical ML approach; BI, phylogenetic trees were generated via a Bayesian inference approach.
“Adjusted P” refers to P values obtained for both the SM tests and the AMOVA tests were subjected to Benjamini-Hochberg FDR corrections, treating P values obtained using the ML approach separately from those obtained using BI and both separately from the AMOVA tests (2).
Previous studies have demonstrated that the total pool of resting CD4+ T cells harboring integrated HIV-1 DNA consists of a mixture of both replication-competent proviruses and defective proviruses, with the former representing only a small fraction of the total (6, 9, 15, 16). Therefore, it may be possible that the population of free plasma viruses appears genetically different from the total population of integrated proviruses, while simultaneously appearing similar to the small fraction of replication-competent viruses. We were able to address this possibility using replication-competent sequences obtained from the extensively characterized patient 154 (1). We found that the two potentially distinct populations (total integrated provirus and replication-competent integrated provirus) appeared to belong to one intermixing population of proviruses (see Table S1 in the supplemental material). Moreover, our results when we compared free plasma virus to the total population of integrated provirus (Table 2) were nearly identical to our results when we compared free plasma virus to only a subset represented by replication-competent virus (see Table S1 in the supplemental material).
In our analysis of population structure between free plasma virus and provirus derived from circulating resting CD4+ T cells, we combined samples obtained from multiple time points. Thus, our analysis of this relationship reflects a time-averaged sampling from these compartments. To determine whether we could consider our data sets as temporally homogeneous, we tested for the presence or absence of temporal population structure in several representative patients. We found no evidence of population structure when we compared proviral populations from different time points (data not shown); however, we found evidence for population structure when we compared populations of free plasma virus from different time points (data not shown).
In addition to comparing free plasma virus to proviruses derived from resting CD4+ T cells, for one patient (patient 154), we were also able to compare free plasma virus to proviruses derived from activated CD4+ T cells, monocytes, and unfractionated PBMC. Our analysis revealed that the residual plasma virus forms a population significantly distinct from all of these cellular sources in this patient (Table 3). Again, to determine the extent to which the PPCs were affecting the results in these analyses, we repeated all analyses with all PPCs removed (Table 4). Table 4 shows that, in all but one scenario (SM test comparing free plasma virus to provirus from activated CD4+ T cells), the results remained significant. The AMOVA analyses found significant population structure for all scenarios (Table 4). Taken together, these results are also largely consistent regardless of the presence or absence of a PPC.
TABLE 3.
Analysis of population structure between free plasma virus and provirus derived from monocytes, PBMC, and activated CD4+ T cells
| Patient 154a analysis | Gene amplified | No. of independent sequences
|
SM analysisb
|
AMOVA analysis
|
Molecular diversity (π)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Free plasma virus | Integrated provirus |
P
|
Adjusted Pc
|
Migration events (ML/BI) | % Variation among or within populations | P | Adjusted P | Free plasma virus | Provirus | |||||
| ML | BI | ML | BI | |||||||||||
| Plasma virus vs provirus derived from activated CD4+ T cells | RT | 238 | 25 | 0.000 | 0.002 | 0.000 | 0.002 | 19/21 | 46.76 | 53.24 | 0.000 | 0.000 | 0.011 | 0.019 |
| Plasma virus vs provirus derived from monocytes | RT | 238 | 17 | 0.000 | 0.000 | 0.000 | 0.000 | 12/13 | 46.64 | 53.36 | 0.000 | 0.000 | 0.011 | 0.020 |
| Plasma virus vs provirus derived from unfractionated PBMC | RT | 238 | 83 | 0.000 | 0.000 | 0.000 | 0.000 | 35/36 | 42.27 | 57.73 | 0.000 | 0.000 | 0.011 | 0.021 |
Patient sequences were derived from Bailey et al. (1).
ML, phylogenetic trees were generated via a classical ML approach; BI, phylogenetic trees were generated via a Bayesian inference approach.
“Adjusted P” refers to P values obtained for both the SM tests and the AMOVA tests were subjected to Benjamini-Hochberg FDR corrections, treating P values obtained using the ML approach separately from those obtained using BI and both separately from the AMOVA tests (2).
DISCUSSION
There is great current interest in the nature of the HIV-1 viruses that persist in patients on HAART and that cause the rebound in viremia that follows cessation of treatment. Previous studies have compared the rebound viremia in patients after interruption of HAART interruption to proviruses in the resting CD4+ T-cell reservoir (6, 7, 22, 50). However, studies of the rebound viremia suffer from the fact that the virus that initially rebounds when treatment is stopped at a particular time point may be different from the virus that would have rebounded if treatment was stopped at a later time point. In other words, the rebound viremia may reflect the stochastic activation of some stable reservoir. In addition, the rebound viremia cannot be attributed to a particular cellular source without extensive sampling of both compartments and rigorous genetic comparisons, features that are missing from previous studies.
A more comprehensive approach to understanding viral persistence is to examine the nature of the free viruses that continue to be produced in patients on HAART and to compare features of this residual viremia with known cellular reservoirs (1, 24, 31). We used population genetics in a statistical framework to systematically analyze the relationship between free plasma virus and the provirus in resting CD4+ T in patients receiving HAART. We found that, in all but two patients, the residual free virus in the plasma was, in general, genetically distinct from proviral sequences in resting CD4+ T cells. In contrast, proviral sequences derived from activated and resting CD4+ T cells comprised one intermixing genetic population. A recently published study (38) found similar results, using the SM test to show significant compartmentalization between plasma and CD4+ T-cell-derived sequences. Here, we addressed a number of issues missing in that study. First, whenever amplifying virus from a patient with low-level viremia, one must be aware of PCR resampling (28), and steps must be made to ensure that this phenomenon does not dominate the results. Our study utilized both patients derived from Bailey et al. (1) and newly enrolled patients, all of which had their samples processed in a manner to avoid PCR resampling. Second, different tests of population structure can yield contradictory results, and a conservative analysis should therefore use at least two complementary tests of compartmentalization (49). For this reason, we used an additional statistical test of population structure, the AMOVA. Third, we addressed the issue of PPCs, which were first described by Bailey et al. (1), and investigated their relationship to the more general phenomenon of compartmentalization.
Our study also addressed the relationship between proviral sequences derived from activated and resting CD4+ T cells. A previous study (10, 11) derived asymmetric migration rates of HIV-1 sequences between activated and resting CD4+ T cells, implicitly assuming compartmentalization between these two cell types. Since in the majority of patients we could not reject the null hypothesis of no compartmentalization, we conclude that the migration rates calculated earlier (10, 11) have no meaningful interpretation.
Although HAART can halt ongoing replication, memory CD4+ T cells harboring replication-competent HIV-1 provirus can still produce progeny virus after reactivation. Thus, the latent reservoir likely contributes to the residual viremia in patients on HAART. However, we show here that at the population level, proviruses derived from resting CD4+ T cells and free viruses in the plasma exist as two distinct genetic populations. There are several possible explanations. Because we only sampled the circulating, resting CD4+ T-cell reservoir, one explanation is that resting CD4+ T cells sequestered in various lymphoid tissues could be the source of the free viruses observed in the plasma of these patients. The assumption that sampling from the periphery can yield a good representation of the archived quasispecies present in the CD4+ T-cell reservoir, while a potential limitation of the present study, is not uncommon in this area of research. Another potential explanation is that other cell types or anatomical compartments could also function as long-term stable reservoirs (1, 3, 31, 32, 45). Recent studies identifying PPCs provide additional evidence suggesting that an alternate reservoir for HIV-1 may be responsible for some of the observed residual viremia (1). We suggest that the presence of PPCs may be a manifestation of a more general phenomenon in which a major source of the free virus in the plasma is some cellular compartment severely underrepresented in the peripheral blood.
Whether we found population structure between free plasma virus and provirus from resting CD4+ T cells did not depend on the presence of a PPC. However, the percentage of molecular variation among (i.e., not shared by) these two groups of sequences was strongly influenced by the PPCs. We studied the impact of PPCs by reanalyzing all datasets after removing all PPCs. We found that removal of the PPC reduced the percent variation among groups, but in most cases significant population structure remained even after removal of the PPC.
Our analysis encompassed two genes: RT and Env. Our finding of population structure between free plasma virus and provirus derived from resting CD4+ T cells did not seem to depend on the gene under study. However, the percentage of molecular variation among groups, as calculated by AMOVA, was different for the two genes, as was the effect of the presence of PPC sequences or removal thereof. The observed difference in the percent variation among groups for RT and Env may be explained by the difference in the overall diversity found in these two genes. AMOVA is based on molecular diversity, which is much lower in RT than in Env. Since the initial HIV-1 infection of a patient was likely established by a single clone (23, 51), two distinct subpopulations can display little among-population variation if the time to accumulate diversity has been short, and this effect would be stronger in RT than in Env. Under this scenario, we can conclude that there is a major source of residual viremia other than reactivated CD4+ T cells but that this source becomes clearly visible in the RT gene only when it produces a PPC. Alternatively, if we reject the notion that the Env gene is significantly more diverse than RT, we can explain the correlation between the presence or absence of a PPC and the percent variation among populations by a model where the majority of plasma virus originates from resting CD4+ T cells but a subset (exemplified in the PPC) originates from an as-yet-unidentified source. Based on our analyses of patients involving Env sequences, we feel that the former model explaining the source of residual viremia best fits the data. We did not have the data to analyze both the RT and Env genes for each patient, but we did have sufficient data for one patient (patient 154). For this patient, we found that our results were comparable for the two genes.
In our analysis of population structure between free plasma virus and provirus derived from resting CD4+ T cells, we used samples derived from multiple time points. Thus, our analysis reflects a time-averaged sampling from the compartments under study. Temporal sampling is a logistical necessity when carrying out analyses of free plasma virus in patients on suppressive HAART, because of the very small numbers of sequences obtainable at any given blood draw. We found no evidence for temporal variation in proviral populations from different time points. We did, however, observe that plasma viruses isolated at different time points seemed to be sampled from distinct subpopulations. This observation lends further support to our overall conclusion that free plasma virus does not exclusively derive from circulating CD4+ T cells. Our analysis of longitudinally sampled plasma viral sequences is in agreement with previously published work by Joos et al. (22), who found temporal variation but no evidence of continued evolution in free plasma virus sampled at different time points.
It is known that the total pool of resting T-cell-derived HIV-1 provirus contains a mixture of both replication-competent and -incompetent species, with the former representing only a small fraction of the total (6, 9, 15, 16). Thus, it is possible that the free plasma virus sampled could be genetically similar to a subset of the total proviral pool, represented by the replication-competent population, while appearing genetically distinct compared to the total population. Methods developed to isolate replication-competent provirus in patients are technically challenging and often yield too few sequences to be of any use for a detailed phylogenetic analysis (30). Thus, we only had sufficient data from replication-competent proviruses to carry out an analysis in one extensively characterized patient (patient 154). Our results showed that replication-competent provirus and total integrated provirus comprise one intermixing genetic compartment. Furthermore, when we compared free plasma virus to replication-competent provirus, we found that these two populations form distinct genetic compartments. Although limited to one patient, our data support the hypothesis that infected T cells isolated from the peripheral circulation represent a single compartment. Therefore, mutations or other events resulting in replication incompetence are occurring within the context of one intermingling genetic population, and thus, on a population level, there should be little overall genetic difference.
We have also shown that proviruses derived from activated and resting CD4+ T cells form one intermixing genetic population. We can explain this observation in the context of basic T-cell biology. Activated and resting CD4+ T cells represent the same population of cells but fixed at different stages of activation. Therefore, proviruses derived from activated or resting CD4+ T cells come from the same cellular compartment and should form one intermixing genetic population. This will only be true, however, if activated CD4+ T cells are not newly infected by free plasma virus. If there were ongoing replication, one might expect discordance between proviruses derived from activated and resting CD4+ T cells and might expect little evidence for population structure between free plasma virus and provirus derived from activated CD4+ T cells. We had the data to test the latter hypothesis for only one patient, patient 154. In this patient, free virus was significantly different from provirus derived from both activated and resting CD4+ T cells. In contrast, provirus derived from activated and resting CD4+ T cells showed no evidence of population structure, a finding consistent with the overall pattern we found in the present study.
Our study did not address the phylogenetic relationship between plasma virus and virus in resting CD4+ T cells of patients who have active viral replication. In patients with active viral replication, most of the plasma virus is derived from recently infected cells that turnover very rapidly (20, 47). The dominant viral variants in the plasma are typically those that are the most fit under the existing condition. In contrast, the latent reservoir in resting CD4+ T cells harbors a stable archive of preexisting viral variants. For example, we have previously shown that in patients failing therapy, the plasma contains drug-resistant variants, while the latent reservoir harbors the original wild-type form and earlier drug-resistant variants (30). Thus, in viremic patients, it is expected that there will be differences in the viral quasispecies detected in the plasma and the latent reservoir. However, these reflect differences between active replication versus production from stable reservoirs rather than differences between stable reservoirs. Unfortunately, technical difficulties currently preclude a detailed analysis of this problem. Stevenson and coworkers showed that most of the HIV-1 DNA in resting CD4+ T cells of viremic patients is a labile unintegrated form in recently infected cells (5). This unintegrated HIV-1 DNA greatly complicates the detection of the much rarer integrated form. We have developed an experimental approach to isolate integrated HIV-1 DNA (30), but the number of sequences that can be obtained by this approach is generally too limited for a detailed phylogenetic analysis. It is, however, sufficient to confirm the general impression that in viremic patients there are substantial differences between the actively replicating pool of viruses observed in the plasma and the stable archival pool of integrated proviruses in resting CD4+ T cells (30).
That a major part of free plasma virus may be derived from some as of yet unidentified cellular source has several important clinical implications with respect to HAART regimen management, virologic failure, rebound viremia associated with treatment interruption, and strategies aimed at eradication. Numerous laboratories are actively pursuing various eradication strategies, most of which involve some aspect of targeting and purging the latent reservoir in resting memory CD4+ T cells. If much of the residual viremia of patients undergoing HAART comes from another reservoir or compartment as suggested here, then eradication strategies will have to include ways to target and purge this additional reservoir to be successful.
Supplementary Material
Acknowledgments
We thank D. C. Nickle and T. W. Chun for providing the sequences for patients 1 to 5, 7, 8, J2, and J7.
This research was supported by NIH grant AI43222, the Doris Duke Charitable Foundation, and the Howard Hughes Medical Institute (R.F.S.) and by NIH grant AI065960 (C.O.W.).
Footnotes
Published ahead of print on 17 June 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bailey, J. R., A. R. Sedaghat, T. Kieffer, T. Brennan, P. K. Lee, M. Wind-Rotolo, C. M. Haggerty, A. R. Kamireddi, Y. Liu, J. Lee, D. Persaud, J. E. Gallant, J. Cofrancesco, Jr., T. C. Quinn, C. O. Wilke, S. C. Ray, J. D. Siliciano, R. E. Nettles, and R. F. Siliciano. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 806441-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57289-300. [Google Scholar]
- 3.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53557-593. [DOI] [PubMed] [Google Scholar]
- 4.Borderia, A. V., F. M. Codoner, and R. Sanjuan. 2007. Selection promotes organ compartmentalization in HIV-1: evidence from gag and pol genes. Evolution 61272-279. [DOI] [PubMed] [Google Scholar]
- 5.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387183-188. [DOI] [PubMed] [Google Scholar]
- 7.Chun, T. W., R. T. Davey, Jr., M. Ostrowski, J. Shawn Justement, D. Engel, J. I. Mullins, and A. S. Fauci. 2000. Relationship between preexisting viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6757-761. [DOI] [PubMed] [Google Scholar]
- 8.Chun, T. W., D. Engel, S. B. Mizell, C. W. Hallahan, M. Fischette, S. Park, R. T. Davey, Jr., M. Dybul, J. A. Kovacs, J. A. Metcalf, J. M. Mican, M. M. Berrey, L. Corey, H. C. Lane, and A. S. Fauci. 1999. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat. Med. 5651-655. [DOI] [PubMed] [Google Scholar]
- 9.Chun, T. W., D. Finzi, J. Margolick, K. Chadwick, D. Schwartz, and R. F. Siliciano. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 11284-1290. [DOI] [PubMed] [Google Scholar]
- 10.Chun, T. W., D. C. Nickle, J. S. Justement, D. Large, A. Semerjian, M. E. Curlin, M. A. O'Shea, C. W. Hallahan, M. Daucher, D. J. Ward, S. Moir, J. I. Mullins, C. Kovacs, and A. S. Fauci. 2005. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J. Clin. Investig. 1153250-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun, T. W., D. C. Nickle, J. S. Justement, J. H. Meyers, G. Roby, C. W. Hallahan, S. Kottilil, S. Moir, J. M. Mican, J. I. Mullins, D. J. Ward, J. A. Kovacs, P. J. Mannon, and A. S. Fauci. 2008. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 197714-720. [DOI] [PubMed] [Google Scholar]
- 12.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 9413193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dornadula, G., H. Zhang, B. VanUitert, J. Stern, L. Livornese, Jr., M. J. Ingerman, J. Witek, R. J. Kedanis, J. Natkin, J. DeSimone, and R. J. Pomerantz. 1999. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 2821627-1632. [DOI] [PubMed] [Google Scholar]
- 14.Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5512-517. [DOI] [PubMed] [Google Scholar]
- 16.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 2781295-1300. [DOI] [PubMed] [Google Scholar]
- 17.Frenkel, L. M., Y. Wang, G. H. Learn, J. L. McKernan, G. M. Ellis, K. M. Mohan, S. E. Holte, S. M. De Vange, D. M. Pawluk, A. J. Melvin, P. F. Lewis, L. M. Heath, I. A. Beck, M. Mahalanabis, W. E. Naugler, N. H. Tobin, and J. I. Mullins. 2003. Multiple viral genetic analyses detect low-level human immunodeficiency virus type 1 replication during effective highly active antiretroviral therapy. J. Virol. 775721-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. Eron, Jr., J. E. Feinberg, H. H. Balfour, Jr., L. R. Deyton, J. A. Chodakewitz, M. A. Fischl, et al. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N. Engl. J. Med. 337725-733. [DOI] [PubMed] [Google Scholar]
- 19.Havlir, D. V., M. C. Strain, M. Clerici, C. Ignacio, D. Trabattoni, P. Ferrante, and J. K. Wong. 2003. Productive infection maintains a dynamic steady state of residual viremia in human immunodeficiency virus type 1-infected persons treated with suppressive antiretroviral therapy for five years. J. Virol. 7711212-11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373123-126. [DOI] [PubMed] [Google Scholar]
- 21.Huelsenbeck, J. P., B. Larget, R. E. Miller, and F. Ronquist. 2002. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst. Biol. 51673-688. [DOI] [PubMed] [Google Scholar]
- 22.Joos, B., M. Fischer, H. Kuster, S. K. Pillai, J. K. Wong, J. Boni, B. Hirschel, R. Weber, A. Trkola, H. F. Gunthard, et al. 2008. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. USA 10516725-16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 1057552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieffer, T. L., M. M. Finucane, R. E. Nettles, T. C. Quinn, K. W. Broman, S. C. Ray, D. Persaud, and R. F. Siliciano. 2004. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J. Infect. Dis. 1891452-1465. [DOI] [PubMed] [Google Scholar]
- 25.Korin, Y. D., D. G. Brooks, S. Brown, A. Korotzer, and J. A. Zack. 2002. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J. Virol. 768118-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosakovsky Pond, S. L., S. D. W. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21676-679. [DOI] [PubMed] [Google Scholar]
- 27.Kulkosky, J., G. Nunnari, M. Otero, S. Calarota, G. Dornadula, H. Zhang, A. Malin, J. Sullivan, Y. Xu, J. DeSimone, T. Babinchak, J. Stern, W. Cavert, A. Haase, and R. J. Pomerantz. 2002. Intensification and stimulation therapy for human immunodeficiency virus type 1 reservoirs in infected persons receiving virally suppressive highly active antiretroviral therapy. J. Infect. Dis. 1861403-1411. [DOI] [PubMed] [Google Scholar]
- 28.Liu, S. L., A. G. Rodrigo, R. Shankarappa, G. H. Learn, L. Hsu, O. Davidov, L. P. Zhao, and J. I. Mullins. 1996. HIV quasispecies and resampling. Science 273415-416. [DOI] [PubMed] [Google Scholar]
- 29.Liu, S. L., T. Schacker, L. Musey, D. Shriner, M. J. McElrath, L. Corey, and J. I. Mullins. 1997. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 714284-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monie, D., R. P. Simmons, R. E. Nettles, T. L. Kieffer, Y. Zhou, H. Zhang, S. Karmon, R. Ingersoll, K. Chadwick, H. Zhang, J. B. Margolick, T. C. Quinn, S. C. Ray, M. Wind-Rotolo, M. Miller, D. Persaud, and R. F. Siliciano. 2005. A novel assay allows genotyping of the latent reservoir for human immunodeficiency virus type 1 in the resting CD4+ T cells of viremic patients. J. Virol. 795185-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nettles, R. E., T. L. Kieffer, P. Kwon, D. Monie, Y. Han, T. Parsons, J. Cofrancesco, Jr., J. E. Gallant, T. C. Quinn, B. Jackson, C. Flexner, K. Carson, S. Ray, D. Persaud, and R. F. Siliciano. 2005. Intermittent HIV-1 viremia (Blips) and drug resistance in patients receiving HAART. JAMA 293817-829. [DOI] [PubMed] [Google Scholar]
- 32.Nickle, D. C., M. A. Jensen, D. Shriner, S. J. Brodie, L. M. Frenkel, J. E. Mittler, and J. I. Mullins. 2003. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J. Virol. 775540-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page, R. D. 2002. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinformatics 6.2. http://www.currentprotocols.com/protocol/bi0602. [DOI] [PubMed]
- 34.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 35.Palmer, S., A. P. Wiegand, F. Maldarelli, H. Bazmi, J. M. Mican, M. Polis, R. L. Dewar, A. Planta, S. Liu, J. A. Metcalf, J. W. Mellors, and J. M. Coffin. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 414531-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387188-191. [DOI] [PubMed] [Google Scholar]
- 37.Ramratnam, B., J. E. Mittler, L. Zhang, D. Boden, A. Hurley, F. Fang, C. A. Macken, A. S. Perelson, M. Markowitz, and D. D. Ho. 2000. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med. 682-85. [DOI] [PubMed] [Google Scholar]
- 38.Sahu, G. K., D. Paar, S. D. Frost, M. M. Smith, S. Weaver, and M. W. Cloyd. 2009. Low-level plasma HIVs in patients on prolonged suppressive highly active antiretroviral therapy are produced mostly by cells other than CD4 T cells. J. Med. Virol. 819-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanjuan, R., F. M. Codoner, A. Moya, and S. F. Elena. 2004. Natural selection and the organ-specific differentiation of HIV-1 V3 hypervariable region. Evolution 581185-1194. [DOI] [PubMed] [Google Scholar]
- 40.Sedaghat, A. R., J. D. Siliciano, T. P. Brennan, C. O. Wilke, and R. F. Siliciano. 2007. Limits on replenishment of the resting CD4+ T-cell reservoir for HIV in patients on HAART. PLoS Pathog. 3e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sedaghat, A. R., R. F. Siliciano, and C. O. Wilke. 2008. Low-level HIV-1 replication and the dynamics of the resting CD4+ T-cell reservoir for HIV-1 in the setting of HAART. BMC Infect. Dis. 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9727-728. [DOI] [PubMed] [Google Scholar]
- 43.Slatkin, M., and W. P. Maddison. 1989. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123603-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 222688-2690. [DOI] [PubMed] [Google Scholar]
- 45.Sundstrom, J. B., J. E. Ellis, G. A. Hair, A. S. Kirshenbaum, D. D. Metcalfe, H. Yi, A. C. Cardona, M. K. Lindsay, and A. A. Ansari. 2007. Human tissue mast cells are an inducible reservoir of persistent HIV infection. Blood 1095293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tobin, N. H., G. H. Learn, S. E. Holte, Y. Wang, A. J. Melvin, J. L. McKernan, D. M. Pawluk, K. M. Mohan, P. F. Lewis, J. I. Mullins, and L. M. Frenkel. 2005. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J. Virol. 799625-9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, and B. H. Hahn. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373117-122. [DOI] [PubMed] [Google Scholar]
- 48.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 2781291-1295. [DOI] [PubMed] [Google Scholar]
- 49.Zarate, S., S. L. Pond, P. Shapshak, and S. D. Frost. 2007. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J. Virol. 816643-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, L., C. Chung, B. S. Hu, T. He, Y. Guo, A. J. Kim, E. Skulsky, X. Jin, A. Hurley, B. Ramratnam, M. Markowitz, and D. D. Ho. 2000. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J. Clin. Investig. 106839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 2611179-1181. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, T., D. Muthui, S. Holte, D. Nickle, F. Feng, S. Brodie, Y. Hwangbo, J. I. Mullins, and L. Corey. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14+ monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76707-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.