Abstract
The function of lentiviral Vif proteins is to neutralize the host antiviral cytidine deaminases APOBEC3G (A3G) and APOBEC3F (A3F). Vif bridges a cullin 5-based E3 ubiquitin ligase with A3G and A3F and mediates their degradation by proteasomes. Recent studies have found that Vif uses different domains to bind to A3G and A3F. A 14DRMR17 domain binds to A3F, 40YRHHY44 binds to A3G, and 69YxxL72 binds to both A3G and A3F. Here, we report another functional domain of Vif. Previously, we demonstrated that human immunodeficiency virus type 1 (HIV-1) Vif failed to mediate A3G proteasomal degradation when all 16 lysines were mutated to arginines. Here, we show that K26, and to a lesser extent K22, is critical for A3G neutralization. K22 and K26 are part of a conserved 21WxSLVK26 (x represents N, K, or H) motif that is found in most primate lentiviruses and that shows species-specific variation. Both K22 and K26 in this motif regulated Vif specificity only for A3G, whereas the SLV residues regulated Vif specificity for both A3F and A3G. Interestingly, SLV and K26 in HIV-1 Vif did not directly mediate Vif interaction with either A3G or A3F. Previously, other groups have reported an important role for W21 in A3F and A3G neutralization. Thus, 21WxSLVK26 is a novel functional domain that regulates Vif activity toward both A3F and A3G and is a potential drug target to inhibit Vif activity and block HIV-1 replication.
The replication of human immunodeficiency virus type 1 (HIV-1) is seriously impaired in human primary lymphocytes when the viral protein Vif is not present (8, 38). The first cellular target of Vif was identified as APOBEC3G (A3G) (34), which belongs to the cytidine deaminase family known as APOBEC (apolipoprotein B mRNA-editing catalytic polypeptide) (14). This family consists of APOBEC1; activation-induced deaminase (AID); APOBEC2; a subgroup of APOBEC3 (A3) proteins, including A3A, A3B, A3C, A3DE, A3F, A3G, and A3H; and APOBEC4 in humans (12). They have one or two copies of a cytidine deaminase domain with a signature motif (HxEx23-28PCx2-4C), and normally only one of the cytidine deaminase domains has deaminase activity.
All seven A3 genes have been shown to inhibit the replication of various types of retroviruses via cytidine deamination-dependent or -independent mechanisms (3). In particular, A3B, A3DE, A3F, and A3G inhibit HIV-1 replication, whereas A3A and A3C do not (1, 6, 7, 19, 34, 42, 50). Recently, it was shown that optimizing A3H expression in cell culture also inhibits HIV-1 replication (4, 10, 25, 39). Among these proteins, A3G and A3F have the most potent anti-HIV-1 activities. A3G and A3F share ∼50% sequence similarity but have different biochemical properties (41) and different target sequence preferences while catalyzing cytidine deamination of viral cDNAs (19).
Nevertheless, HIV-1 is able to elude this defense mechanism and cause human disease for two reasons. First, A3B and A3H are expressed only at low levels in vivo (4, 7, 18, 26). Second, HIV-1 produces Vif, which is expressed in all lentiviruses except equine infectious anemia virus. Vif can destabilize A3DE, A3F, and A3G proteins by targeting them to the proteasomal degradation pathway (6, 22, 35, 37, 50). In addition, Vif may also inhibit A3 activity independently of proteasomal degradation (15, 16, 31).
The action of Vif is highly species specific. Vif from HIV-1 inactivates only A3G from humans, and Vif from simian immunodeficiency virus (SIV) isolated from African green monkeys (AGM) does not inactivate A3G from humans. Nevertheless, Vif from SIV isolated from rhesus macaques (MAC) inactivates A3G from all humans, AGM, and MAC (21). A single residue in A3G at position 128, an aspartic acid in humans versus a lysine in AGM, determines A3G sensitivity to HIV-1 Vif (2, 32, 44). In addition, an N-terminal domain in HIV-1 Vif, 14DRMR17, determines Vif specificity for different A3G proteins (33).
Vif targets A3G to the proteasome by acting as an adaptor protein that bridges A3G with a cullin 5 (Cul5)-based E3 ubiquitin ligase complex, which includes Cul5, elongin B (EloB), and EloC (46). Vif has a BC box motif (144S145L146Q) that binds to EloC (23, 47) and an HCCH motif (114C/133C) that binds to Cul5 (20, 24, 43). It has also been shown that Vif specifically binds to a region from amino acids 126 to 132 of A3G and to amino acids 283 to 300 of A3F (13, 30). It is believed that as a consequence of these interactions, A3G is polyubiquitylated and directed to 26S proteasomes for degradation.
Several domains that determine Vif interactions with A3F and A3G have been identified. Analysis of HIV-1 patient-derived Vif sequences initially found that W11 is essential for A3F recognition and K22, Y40, and E45 are required for A3G recognition (36). The previously identified agmA3G-specific 14DRMR17 domain was also found to determine Vif specificity for A3F (33) by direct binding (29). An A3G-specific binding domain, 40YRHHY44, has also been identified (29), and a 69YxxL72 domain interacts with both A3G and A3F (11, 28, 45).
We have previously shown that Vif can mediate A3G proteasomal degradation in the absence of A3G polyubiquitylation and that, unexpectedly, this process is dependent on lysines in Vif (5). Here, we identify two N-terminal lysines that are important for Vif function. We show that these lysines are part of a 21WxSLVK26 motif that is conserved in Vif from primate lentiviruses and that this motif regulates Vif activities against both A3G and A3F via different mechanisms.
MATERIALS AND METHODS
Plasmids.
The HIV-1 proviral constructs pNL4-3Δvif and pNL-LucΔvif and the mammalian expression vectors pcDNA3.1-A3F-V5-6xHis and pcDNA3.1-A3G-V5-6xHis were described previously (6, 50). pNL-LucΔenvΔvif was created by NheI digestion of pNL-LucΔvif, followed by treatment with the large Klenow fragment before T4 ligation. The AGM and MAC A3G cDNAs were from N. Landau (21), and the AGM and MAC A3F cDNAs were from P. Bieniasz (48). These genes were subcloned, using the TOPO cloning strategy (Invitrogen), into pcDNA3.1D/V5-His-TOPO, which expresses a C-terminal V5 tag for protein detection. To create plasmids expressing human A3F or A3G C-terminally fused with a FLAG-hemagglutinin (HA) tag, two oligonucleotides encoding the sense FLAG/HA gene with a 5′-NotI cohesive end or the antisense FLAG/HA gene with a 3′-XbaI cohesive end were synthesized. After being annealed, they were inserted into pcDNA3.1-A3F-V5-6xHis and pcDNA3.1-A3G-V5-6xHis vectors by NotI and XbaI digestion.
The Vif expression vectors pNL-A1, pNL-A1Δvif, pNL-A1SIVmacVif, and pNL-A1SIVagmVif were from K. Strebel. The lysine-free HIV-1 Vif expression vector pNL-A1Vif16K/R was previously described (5). To simplify the cloning and detection of the vif gene, the original pNL-A1 vector was modified. The vif gene was cut out by BssHII and EcoRI digestion and replaced with a linker containing a NotI site and an XbaI site followed by an HA tag. This new vector was named pNL-A1NotI/XbaI/HA. The HIV-1, SIVmac, and SIVagm vif genes were recloned into this vector. The HIV-1 Vif 1-6K/R and 7-16K/R mutants were created by a two-step recombinant PCR method using the wild type (WT) and 16K/R as templates. To make other Vif mutants, vif genes were first cloned into the pCR4-TOPO vector (Invitrogen) and mutated with the QuikChange XL site-directed mutagenesis kit (Stratagene). These vif genes were then cloned back into pNL-A1NotI/XbaI/HA by NotI/XbaI digestion. To express these vif mutants in the replication-competent proviral clone, a modified pNL4-3 vector containing a NotI and an XbaI site in the vif open reading frame, pNL4-3NotI/XbaI, was created (51). This plasmid contains a NotI site at the end of pol and an XbaI site in front of the vpr gene. In addition, the three ATG codons in vif overlapping with pol were silenced without changing the integrase coding sequence. After that, the mutated vif genes were cloned into this vector by NotI and XbaI digestion.
Vif activity assay.
Vif activities were measured by their abilities to rescue HIV-1Δvif virus infectivity in the presence of A3G or A3F. Viruses were produced from 293T cells by a standard calcium phosphate transfection. Typically, 21 μg of plasmid DNAs containing 5 μg pNL-LucΔenvΔvif, 5 μg Vif expression vector, 1 μg vesicular stomatitis virus G (VSV-G) expression vector, and 10 μg A3 expression vector was transfected into 293T cells in a 100-mm culture dish with 20% confluence. The production of HIV-1 was quantified by p24Gag capture enzyme-linked immunosorbent assay (ELISA). Equal numbers of viruses were used to infect GHOST-R3/X4/R5 cells. Thirty-six hours later, the cells were lysed in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 3 mM EDTA (lysis buffer). After the nuclei were removed, the cytosolic fraction was used to determine luciferase activity using a luciferase assay kit (Promega).
HIV-1 growth curve.
The human T-cell lines CEM-SS, H9, Hut78, and PM1, as well as human peripheral blood mononuclear cells (PBMC), were cultured in complete RPMI 1640 culture medium containing 10% fetal bovine serum, 10 μg/ml ampicillin, and 50 μg/ml streptomycin. Human PBMC were isolated from healthy blood donors using a Michigan State University Institutional Review Board-approved protocol and activated by phytohemagglutinin and interleukin 2 before HIV-1 infection. Viruses were produced from 293T cells after transfection with pNL4-3 expressing WT or mutant vif genes. A total of 2 × 105 T cells were infected with HIV-1 containing 150 ng p24Gag for 3 h at 37°C. After being washed, the cells were resuspended in a 24-well culture plate, and culture supernatants were collected every other day for ELISA measurement of viral p24Gag.
Immunoprecipitation.
293T cells were cotransfected with A3-FLAG-HA and Vif expression vectors at a 1:1 ratio. A green fluorescent protein (GFP)-FLAG-HA expression vector was included as a negative control. Twenty-four hours later, the cells were lysed in buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.6, 3 mM EDTA, 1% Triton X-100). The cytosolic fraction was isolated and then rocked with anti-FLAG antibody M2-conjugated beads (Sigma) for 4 h at 4°C. After extensive washing with phosphate-buffered saline containing 500 mM NaCl, the bead-associated proteins were detected by Western blotting.
Western blotting.
Horseradish peroxidase (HRP)-conjugated anti-HA antibody (Roche) and HRP-conjugated anti-V5 antibody (Invitrogen) were used to directly detect the expression of GFP, A3F, A3G, or Vif protein. Actin was detected by a polyclonal antibody (C-11) (Santa Cruz Biotechnology). HIV-1 p24Gag and Vif proteins were also detected by two antibodies (no. 3537 and no. 2221) from the NIH AIDS Research and Reference Reagent Program. HRP-conjugated anti-rabbit or -mouse immunoglobulin G secondary antibodies were from Pierce. Detection of the HRP-conjugated antibody was performed using an enhanced chemiluminescence detection kit (Amersham Bioscience).
Motif analysis.
We identified in GenBank approximately 2,800 distinct Vif sequences from HIV-1, HIV-2, SIVcpz, SIVmac, or SIVagm and scanned each sequence for various subsets and variations of the putative motif.
RESULTS
Mapping of critical lysines for HIV-1 Vif function.
Previously, we created a lysine-free HIV-1 Vif mutant from pNL4-3 by replacing all 16 lysines with arginines (16K/R) and found that the mutant lost neutralizing activity against human A3G (5). The sequence of the pNL4-3 vif gene is shown in Fig. 1A. To map lysines critical for Vif function, we created Vif lysine mutants and determined whether they lost activity against human A3G or A3F. Initially, two recombinant vif genes were created from the WT and 16K/R vif genes: 1-6K/R, with the N-terminal 6 lysines (K22, K26, K34, K36, K50, and K63) mutated to arginines, and 7-16K/R, with the C-terminal 10 lysines (K91, K92, K141, K155, K157, K160, K168, K176, K179, and K181) mutated to arginines. These two recombinant genes were cloned into pNL-A1, a pNL4-3-derived HIV-1 subgenomic vector, for vif expression (38). Their activities were determined by measuring their abilities to rescue replication of HIV-1Δvif in the presence of A3G or A3F. In the absence of Vif, the viruses were poorly infectious, indicating potent antiviral activity of both A3G and A3F (Fig. 1B). Consistently, the ΔSLQ mutant, in which the EloC-binding motif 144S145L146Q was mutated to AAA, did not neutralize either A3G or A3F (Fig. 1B). As previously reported, the 16K/R mutant completely lost activity critical to A3G, and we found that it retained only partial activity against A3F (Fig. 1B). The 7-16K/R mutant retained full activity against both A3G and A3F (Fig. 1B), whereas the 1-6K/R mutant completely lost activity against A3G but not A3F (Fig. 1B). This result indicated that the N-terminal six lysines are important for Vif activity against A3G.
FIG. 1.
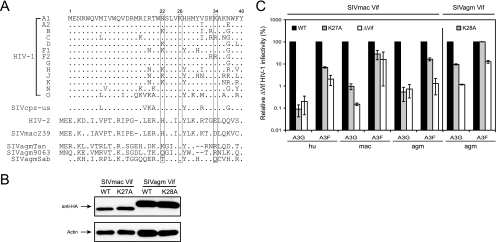
Critical role of the HIV-1 Vif K26 residue in neutralizing A3G, but not A3F. (A) Amino acid sequence alignment of Vif proteins expressed from pNL4-3 and pNL-A1. The lysines are underlined and numbered. (B and C) Mutational analysis of Vif activity against A3G (gray bars) and A3F (open bars). 293T cells were transfected with the HIV-1 reporter construct pNL-LucΔenvΔvif, an indicated Vif expression vector, a VSV-G expression vector, and either an A3F or A3G expression vector. The infectivity of the virus produced from the transfected cells was determined by infection of GHOST cells and quantitation of the luciferase enzyme produced in the GHOST cells after normalization by levels of viral input (p24Gag). Vif activities are presented as relative values of HIV-1Δvif infectivity, with the infectivity in the presence of WT Vif set as 100. The standard errors of the means (error bars) were calculated from three independent experiments. (D and E) A3G or A3F expression in the presence of various Vif mutants. An A3G or A3F expression vector was cotransfected with the indicated Vif expression vector into 293T cells, and protein expression was determined by Western blotting. The A3G and A3F proteins were detected by an anti-V5 antibody, and Vif proteins were detected by an anti-HA antibody.
To identity more critical lysines, we compared Vif protein sequences from pNL4-3 and pNL-A1. Among the N-terminal six lysines in pNL4-3, only three (K22, K26, and K34) exist in pNL-A1 (Fig. 1A). We mutated these three lysines to arginines and created a triple-lysine mutant, 3K/R. The 3K/R mutant, like 1-6K/R, failed to neutralize A3G but retained activity against A3F (Fig. 1B). Thus, K22, K26, and K34 are essential for Vif activity against A3G.
To compare these three lysines, we created three double-lysine mutants: K22-26/R, with both K22 and K26 changed to arginines; K22-34/R, with K22 and K34 changed to arginines; and K26-34/R, with K26 and K34 changed to arginines. All three mutants were still active against A3F, but K22-26/R and K26-34/R were completely inactive against A3G, while K22-34/R was only partially active against A3G (Fig. 1B). This result suggested that K26 is more critical than K22 and K34 for Vif to neutralize A3G.
We replaced each of these three lysines with alanine, aspartic acid, or arginine and created nine single-lysine mutants (Fig. 1C). Because aspartic acid has a negative charge, alanine has a neutral charge, and both arginine and lysine have positive charges, this mutagenesis allowed us to address whether the charges at these positions affect Vif function. Indeed, these mutants had very different phenotypes. The K22D mutant completely lost activity to neutralize A3G but not A3F, while the K22A and K22R mutants retained almost full activity against both proteins (Fig. 1C). The K34D mutant partially lost activity against both A3G and A3F, while the K34A and K34R mutants retained almost full activity (Fig. 1C). Interestingly, the K26A, K26D, and K26R mutations completely inactivated Vif activity against A3G, but not A3F (Fig. 1C). Because the reduction in Vif activity was more dramatic when mutations were introduced into K22 and K26, we conclude that K22 and K26 play more important roles than K34 in Vif function.
To validate these observations, we determined the levels of A3G and A3F protein expression in the presence of WT Vif or the Vif mutants in viral producer cells. The WT protein effectively reduced both A3G and A3F expression, although its effect on A3G was more potent than that on A3F (Fig. 1D, lanes 1 and 2, and E, lanes 1 and 2). Mutants 16K/R, ΔSLQ, 3K/R, 1-6K/R, K22D, K26A, K26D, and K26R completely failed to reduce A3G protein expression; mutants K22-26/R, K26-34/R, and K34D partially reduced A3G protein expression; and mutants 7-16K/R, K22-34/R, K22A, K22R, K34A, and K34R fully reduced A3G expression, like the WT protein (Fig. 1D). In addition, mutants 16K/R, ΔSLQ, and K34D failed to reduce A3F expression, and the other mutants reduced A3F expression either as effectively as or more effectively than the WT Vif protein (Fig. 1E). In general, these results are in agreement with the previous results, further highlighting the important roles of K22 and K26 in Vif function.
Identification of a lysine critical for SIV Vif function.
To further explore the functions of K22 and K26, we compared the N-terminal amino acid sequences of Vif proteins from different primate lentiviruses. Vif sequences from all subtypes of HIV-1, SIVcpz, HIV-2, SIVmac239, and three different SIVagm strains, SIVagmTan, SIVagm9063, and SIVagmSab, were initially selected (Fig. 2A). It was found that both K22 and K26 were located in a highly conserved 21WKSLVK26 motif in HIV-1 Vif (22WHSLIK27 in HIV-2 and SIVmac239 and 23WxxxVK28 in SIVagm). Further analyses indicated that a consensus motif, W[N/K/H]SL[V/I]K, is present in >90% of 2,742 Vif sequences from HIV-1, HIV-2, and SIVmac, and it is present in ∼75% of 113 Vif sequences from SIVcpz (Table 1). Clear species-specific motifs were observed for HIV-1 (W[N/K]SLVK), HIV-2 (WHSL[V/I]K), SIVmac (WHSLIK), and SIVcpz (W[N/H]SL[I/V]K), while SIVagm showed more variation (Table 1). We therefore summarized these sequences as a general consensus motif, WxSLVK.
FIG. 2.
Critical roles of SIVmac Vif K27 and SIVagm Vif K28 residues in neutralizing A3G, but not A3F. (A) N-terminal amino acid sequence alignment of Vif proteins from HIV-1, HIV-2, SIVcpz, SIVmac239, and three SIVagm isolates. Three lysines (K22, K26, and K34) are indicated. (B) Expression of SIVmac Vif K27A and SIVagm Vif K28A mutants. SIV Vif proteins were expressed from the HIV-1 subgenomic expression vector pNL-A1, and protein expression was determined in 293T cells by Western blotting with an anti-HA antibody. (C) Activities of SIV Vif K27A and K28A mutants. 293T cells were transfected with pNL-LucΔenvΔvif, an indicated SIV Vif expression vector, a VSV-G expression vector, and either an A3F or A3G expression vector. Vif activities were determined by measuring the infectivity of HIV-1Δvif virus as for Fig. 1B. The standard errors of the means (error bars) were calculated from three independent experiments.
TABLE 1.
Motif sequences in HIV and SIV strainsa
| Unique Vif sequence | % that contain each motif variant
|
||||
|---|---|---|---|---|---|
| HIV-1 (2,683) | HIV-2 (27) | SIVcpz (113) | SIVmac (32) | SIVagm (9) | |
| W K S L V K | 43.6 | 0.0 | 0.9 | 0.0 | 0.0 |
| W N S L V K | 47.4 | 0.0 | 20.4 | 0.0 | 0.0 |
| W H S L V K | 4.3 | 74.1 | 6.2 | 0.0 | 0.0 |
| W H S L I K | 0.0 | 22.2 | 40.7 | 90.6 | 11.1 |
| W x S L V K | 1.0 | 0.0 | 2.7 | 0.0 | 11.1 |
| W Q G I V K | 0.0 | 0.0 | 0.9 | 0.0 | 11.1 |
| W x G I V K | 0.0 | 0.0 | 0.9 | 0.0 | 11.1 |
| W x G I V R | 0.0 | 0.0 | 0.9 | 0.0 | 11.1 |
| W x G I V x | 0.0 | 0.0 | 0.9 | 0.0 | 11.1 |
| Total | 96.3 | 96.3 | 74.5 | 90.6 | 66.7 |
Vif sequences from the indicated virus strains were identified in GenBank and analyzed for the presence of the motifs as shown.
Since the K26 (or K27 in SIVmac and K28 in SIVagm) residue is highly conserved, we decided to continue to study the role of this residue in SIV Vif function. A K27A or K28A mutation was introduced into the SIVmac239 or SIVagm9063 vif gene, respectively, and the genes were subsequently cloned into the pNL-A1 vector for expression. Both SIVmac Vif K27A and SIVagm Vif K28A were expressed at levels comparable to those of the WT proteins after transfection into 293T cells (Fig. 2B). SIVmac Vif K27A almost completely lost activity against human A3G, macA3G, and agmA3G and partially lost activity against human A3F and agmA3F (Fig. 2C). As previously reported, macA3F had very low antiviral activity (48), and it was impossible to compare the abilities of WT and K27A Vifs to neutralize macA3F (Fig. 2C). The SIVagm Vif K28A mutant partially lost activity against agmA3G and completely lost activity against agmA3F (Fig. 2C). In general, the K27A or K28A mutation in SIV Vif tended to selectively inactivate the A3G protein. Thus, we conclude that, like the K26 in HIV-1 Vif, the K27 or K28 in SIV Vif is more important in protecting against A3G than A3F.
Role of SLV in Vif function.
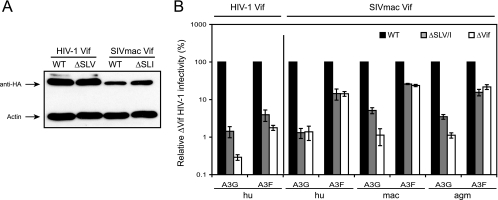
We found that SL[V/I] residues are also conserved, except in SIVagm (Fig. 2A and Table 1). We next addressed how they could regulate Vif function. All three SLV residues in HIV-1 Vif (or SLI in SIVmac) were mutated to alanines, creating a ΔSLV mutant for HIV-1 Vif and ΔSLI for SIVmac Vif. These mutants were expressed at levels comparable to those of their WT proteins after transfection into 293T cells (Fig. 3B). However, ΔSLV and ΔSLI failed to neutralize both A3G and A3F proteins from humans, and ΔSLI failed to neutralize both A3G and A3F proteins from humans, MAC, and AGM (Fig. 3B). Thus, we conclude that, unlike K22 and K26, the SLV residues are critical for Vif neutralization of both A3G and A3F.
FIG. 3.
Critical roles of SLV/I residues in Vif function. (A) Expression of HIV-1 Vif ΔSLV and SIVmacVif ΔSLI mutants in 293T cells. (B) Activities of ΔSLV and ΔSLI mutants determined as for Fig. 2C. The standard errors of the means (error bars) were calculated from three independent experiments.
Actions of K22, SLV, and K26 in Vif activity.
How do the K22, SLV, and K26 residues regulate HIV-1 Vif function? One important question was whether they could regulate A3G or A3F binding and degradation. As noted above, the 14DRMR17 domain of Vif binds to A3F and the 69YxxL72 domain of Vif binds to both A3G and A3F. We introduced an M16A or Y69A mutation into HIV-1 Vif to disrupt the DRMR or YxxL motif, respectively. We had confirmed that the M16A mutant failed to neutralize A3F and Y69A failed to neutralize both A3F and A3G (data not presented). These two mutants were used as controls in Vif/A3 protein binding experiments.
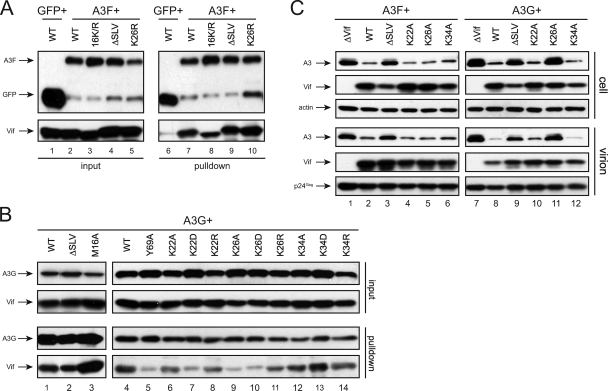
A3F or A3G fusion proteins with a FLAG-HA tag were coexpressed with different Vif mutants in 293T cells, and a similar GFP expression vector was included as an additional negative control. The proteins were pulled down by the anti-FLAG antibody-conjugated beads, and bead-associated proteins were determined by Western blotting. GFP did not bind to the WT Vif proteins, whereas both A3F and A3G bound to the WT Vif proteins (Fig. 4A, lanes 6 and 7, and B, lane 1). As expected, the M16A mutant bound to A3G, whereas the Y69A mutant bound poorly to A3G (Fig. 4B, lanes 3 and 5). We previously reported that the binding of the 16K/R mutant to A3G was slightly reduced (5), and we found that its binding to A3F was also slightly reduced (Fig. 4A, lane 8). The ΔSLV mutant bound to A3F and A3G as efficiently as did the WT Vif (Fig. 4A, lane 9, and B, lane 2), and the K26R mutant still bound to A3F (Fig. 4A, lane 10). We further compared how the mutations at amino acid position 22, 26, or 34 affected Vif binding to A3G. Mutants K22D, K26A, and K26D completely lost binding ability to A3G, whereas mutants K22A, K22R, K26R, K34A, K34D, and K34R still bound to A3G (Fig. 4B, lanes 6 to 14). In general, these binding results were consistent with those from Vif neutralization assays, although it was unclear why ΔSLV and K26R failed to neutralize A3G or A3F.
FIG. 4.
Mechanism of K22, SLV, K26, and K34 residues in regulating Vif function. (A and B) Interaction of Vif mutants with A3F or A3G. The indicated Vif proteins were coexpressed with GFP, A3F, or A3G protein with a FLAG-HA tag in 293T cells. The proteins were pulled down by anti-FLAG antibody-conjugated beads and analyzed by Western blotting. Membranes were first incubated with a polyclonal anti-Vif antibody to detect Vif proteins and then an anti-HA antibody to detect GFP, A3F, and A3G proteins. (C) A3F and A3G expression in virus producer cells and virions. The indicated HIV-1 strains bearing vif mutations were produced from 293T cells in the presence of A3F or A3G protein expression. Protein expression in cells and purified virions was determined by Western blotting. A3F and A3G were detected by an anti-V5 antibody, Vif was detected by a polyclonal antibody, and Gag was detected by a monoclonal antibody.
Next, we determined whether the mutations affect the degradation of A3F and A3G. We determined A3F and A3G protein levels in both viral producer cells and virions in the presence of Vif mutants. HIV-1 proviral clones containing a ΔSLV, K22A, K26A, or K34A mutation in the vif gene were coexpressed with A3F or A3G proteins in 293T cells; viral particles were collected and purified from these cell cultures; and A3F and A3G levels were determined by Western blotting. The WT, K22A, and K34A Vif proteins effectively decreased A3F and A3G levels in both cells and virions (Fig. 4C, lanes 2, 4, 6, 8, 10, and 12); the ΔSLV mutant did not decrease A3G and A3F expression in cells or virions (Fig. 4C, lanes 3 and 9); and the K26A mutant could decrease only A3F, but not A3G, expression in cells and virions (Fig. 4C, lanes 5 and 11). These results are consistent with those from viral replication assays (Fig. 1C) and confirmed that these residues regulate A3F and A3G neutralization via protein degradation and virion exclusion.
Replication of HIV-1 with K26R and ΔSLV mutations in human T-cell lines.
To further evaluate the functions of these residues, human T cells were infected with HIV-1 bearing either a K26R or a ΔSLV mutation in the vif gene, and viral growth curves were determined. As a comparison, viruses with either an M16A or a Y69A mutation in vif were also included. One permissive cell line (CEM-SS) and three nonpermissive cell lines (H9, Hut-78, and PM1), as well as human PBMC, were selected. These cells were infected with equal amounts of each virus, and viral replication was determined by measuring the p24Gag concentration in the supernatant using an ELISA kit. As expected, in CEM-SS cells, the WT and vif-defective viruses and four vif mutant viruses (M16A, K26R, ΔSLV, and Y69A) all grew well, because viruses do not need a functional vif gene in this cell line (Fig. 5A). Moreover, we were able to detect all these different Vif proteins in CEM-SS cells by Western blotting on days 6 and 9 postinfection, indicating that they were well expressed during viral replication (Fig. 5A). In contrast, in Hut-78, H9, and PM1 cells and PBMC, only the WT virus replicated well, whereas the ΔVif virus showed approximately a 2-log-unit reduction in viral production, because viruses need a functional vif gene in these cell lines (Fig. 5B). In addition, viral replication was inhibited by both M16A and K26R mutations, and the K26R mutation caused a more profound inhibition than the M16A mutation. Because the M16A and K26R mutants did not neutralize A3F or A3G, respectively, this result confirmed that A3G has stronger antiviral activity than A3F. The replication of HIV-1 with a Y69A or ΔSLV mutation was further reduced to the Δvif HIV-1 levels, confirming that these two vif genes failed to neutralize both A3F and A3G. Thus, these results were consistent with those from viral replication assays (Fig. 1C) and further confirmed the importance of SLV and K26 in Vif function.
FIG. 5.
Replication of HIV-1 with mutations in the vif gene in human T-cell lines. (A) Replication in a permissive cell line. CEM-SS cells were infected with equal amounts of six different HIV-1 strains (WT, ΔVif, M16A, K26R, ΔSLV, and Y69A). Viral growth was monitored for 8 days by p24Gag ELISA. In addition, Vif protein expressions in CEM-SS cells was determined by Western blotting on the third, sixth, and ninth days postinfection using a polyclonal antibody. (B) Replication in nonpermissive cell lines. The indicated nonpermissive cells were infected with the same six different viruses as for panel A, and viral replication was determined similarly.
DISCUSSION
In this report, we identified a conserved WxSLVK motif in Vif proteins from diverse primate lentiviruses. Within this motif, SL[V/I] residues regulate Vif specificity for both A3F and A3G, whereas K22 and K26 regulate Vif specificity only for A3G.
We found that this motif was very sensitive to mutagenesis and could not tolerate even conservative substitutions. When K26 was replaced with arginine (conserving a positive charge), Vif completely lost the ability to neutralize A3G, and HIV-1 bearing the same mutation replicated poorly in nonpermissive cell lines. In addition, even though N22 is commonly found in WT HIV-1 Vif (Fig. 2A and and Table 1), mutating this position to an aspartic acid (D) also selectively disrupted Vif activity against A3G. These results are consistent with previous observations made by Khamsri et al. that a K26R viral mutant showed a delayed growth curve in H9 cells (17) and by Simon et al. that a K22E Vif mutant selectively lost activity against A3G (36). Although K22 and K26 regulate Vif activity similarly, different mechanisms are involved. Position 22 is more variable in natural Vif sequences, with N, K, and H all being common, and in this study, only replacement of K22 with negatively charged aspartic acid disrupted Vif activity, whereas any substitution in the highly conserved K26 made Vif inactive. Similarly, only replacement of K34 with a negatively charged residue (D) partially disrupted Vif activity. These results suggest that charge plays an important role in Vif function. Moreover, we showed that mutation of SLV to AAA completely disrupted Vif function. Previously, when Yamashita et al. introduced a single L24A mutation within KSLVK, the activity of Vif to neutralize A3G and A3F did not change (45). These results support the concept that all three SLV residues have to be mutated simultaneously to disable Vif function. Furthermore, the W21 at the N termini of Vif proteins is also highly conserved. When Tian et al. made a W21A mutation, Vif failed to neutralize both A3G and A3F (40). Yamashita et al. not only confirmed this observation, but also further demonstrated that the W21A mutant failed to bind to A3G (45). Moreover, Vif neutralization of A3 proteins is highly species specific, in that Vif more effectively neutralizes the A3 proteins from the same species. This specificity is also detectable in the variants of the basic W[N/K]SL[V/I]K motif. The variant positions tend to be highly conserved within each lentivirus strain (Table 1), suggesting that this motif may be important in determining the species specificity of Vif. Taken together, these results demonstrate that WxSLVK represents a functional motif and plays a critical role in regulating Vif neutralizing activity against A3G and A3F.
It should be noted that the ability of Vif to bind to A3 proteins does not always correlate with its capability to neutralize these proteins. We found that although ΔSLV and K26R mutants failed to neutralize A3G or A3F, they still bound to these two proteins very efficiently (Fig. 4A and 4B). This observation is not unprecedented. He et al. reported that the Vif L72S mutant still bound to A3G but failed to neutralize it (11). In addition, several groups have reported that although WT Vif can bind to several other A3 proteins, including the human A3A YFW mutant (9), A3A and A3C chimeric mutants (49), an A3G C97A mutant (27), the N-terminal half of A3G (9), and human A3H (4), it did not initiate their destruction. Similarly, the A3G D128K mutant seems to still bind to HIV-1 Vif without being degraded (29). These observations indicate that, in addition to assembly of the A3, Vif, and Cul5 E3 ligase protein complex, there must be other, secondary determinants on both A3 and Vif proteins that are critical in mediating A3 protein degradation.
We found that K22D and other K26 mutants selectively lost activity against A3G, whereas ΔSLV and K34D mutants lost activity against both A3F and A3G. Thus, the functions of K22 and K26 resemble that of the YRHHY motif, and those of W21, SLV, and K34 resemble that of the YxxL motif. It is interesting that residues in the WxSLVK motif can be functionally differentiated. Although the DRMR, YRHHY, and YxxL motifs directly bind to A3F and/or A3G, the WxSLVK motif does not appear to affect Vif activity solely via this interaction, because the inactive ΔSLV and K26R mutants still bind to A3F and A3G (Fig. 4A and B). It is possible that the WxSLVK motif contributes to a secondary signal that is critical for A3F and A3G neutralization. Indeed, such secondary signals have been implicated in previous studies (29, 49). Thus, the WxSLVK motif provides an important tool to further study these unknown mechanisms. It also emerges as a novel target for pharmaceutical inhibition of Vif function to prevent HIV-1 infection.
Acknowledgments
We thank X. F. Yu for helpful discussions. We thank N. Landau, P. Bieniasz, K. Strebel, and the NIH AIDS Research and Reference Reagent Program for various reagents.
Y.-H.Z was supported by grants AI063944 and AI080225 from the National Institutes of Health.
Footnotes
Published ahead of print on 17 June 2009.
REFERENCES
- 1.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 141392-1396. [DOI] [PubMed] [Google Scholar]
- 2.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 1013770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26317-353. [DOI] [PubMed] [Google Scholar]
- 4.Dang, Y., L. M. Siew, X. Wang, Y. Han, R. Lampen, and Y. H. Zheng. 2008. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 28311606-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang, Y., L. M. Siew, and Y. H. Zheng. 2008. APOBEC3G is degraded by the proteasomal pathway in a Vif-dependent manner without being polyubiquitylated. J. Biol. Chem. 28313124-13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang, Y., X. Wang, W. J. Esselman, and Y. H. Zheng. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 8010522-10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339281-288. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, A. G., B. Ensoli, L. Ivanoff, M. Chamberlain, S. Petteway, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237888-893. [DOI] [PubMed] [Google Scholar]
- 9.Gooch, B. D., and B. R. Cullen. 2008. Functional domain organization of human APOBEC3G. Virology 379118-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harari, A., M. Ooms, L. C. Mulder, and V. Simon. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, Z., W. Zhang, G. Chen, R. Xu, and X. F. Yu. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 3811000-1011. [DOI] [PubMed] [Google Scholar]
- 12.Holmes, R. K., M. H. Malim, and K. N. Bishop. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32118-128. [DOI] [PubMed] [Google Scholar]
- 13.Huthoff, H., and M. H. Malim. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 813807-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79285-296. [DOI] [PubMed] [Google Scholar]
- 15.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 7711398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao, S., E. Miyagi, M. A. Khan, H. Takeuchi, S. Opi, R. Goila-Gaur, and K. Strebel. 2004. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khamsri, B., M. Fujita, K. Kamada, A. Piroozmand, T. Yamashita, T. Uchiyama, and A. Adachi. 2006. Effects of lysine to arginine mutations in HIV-1 Vif on its expression and viral infectivity. Int. J. Mol. Med. 18679-683. [PubMed] [Google Scholar]
- 18.Kidd, J. M., T. L. Newman, E. Tuzun, R. Kaul, and E. E. Eichler. 2007. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 3e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 141385-1391. [DOI] [PubMed] [Google Scholar]
- 20.Luo, K., Z. Xiao, E. Ehrlich, Y. Yu, B. Liu, S. Zheng, and X. F. Yu. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. USA 10211444-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 11421-31. [DOI] [PubMed] [Google Scholar]
- 22.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 91398-1403. [DOI] [PubMed] [Google Scholar]
- 23.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 182861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehle, A., E. R. Thomas, K. S. Rajendran, and D. Gabuzda. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 28117259-17265. [DOI] [PubMed] [Google Scholar]
- 25.OhAinle, M., J. A. Kerns, M. M. Li, H. S. Malik, and M. Emerman. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4249-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.OhAinle, M., J. A. Kerns, H. S. Malik, and M. Emerman. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 803853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opi, S., S. Kao, R. Goila-Gaur, M. A. Khan, E. Miyagi, H. Takeuchi, and K. Strebel. 2007. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 818236-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pery, E., K. S. Rajendran, A. J. Brazier, and D. Gabuzda. 2009. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J. Virol. 832374-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell, R. A., and V. K. Pathak. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 818201-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell, R. A., J. Smith, R. Barr, D. Bhattacharyya, and V. K. Pathak. 2009. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J. Virol. 831992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santa-Marta, M., F. A. da Silva, A. M. Fonseca, and J. Goncalves. 2005. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J. Biol. Chem. 2808765-8775. [DOI] [PubMed] [Google Scholar]
- 32.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 1013927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrofelbauer, B., T. Senger, G. Manning, and N. R. Landau. 2006. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 805984-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 35.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 91404-1407. [DOI] [PubMed] [Google Scholar]
- 36.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12591-601. [DOI] [PubMed] [Google Scholar]
- 38.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature 328728-730. [DOI] [PubMed] [Google Scholar]
- 39.Tan, L., P. T. Sarkis, T. Wang, C. Tian, and X. F. Yu. 2008. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 23279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian, C., X. Yu, W. Zhang, T. Wang, R. Xu, and X. F. Yu. 2006. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 803112-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, X., P. T. Dolan, Y. Dang, and Y. H. Zheng. 2007. Biochemical differentiation of APOBEC3F and APOBEC3G proteins associated with HIV-1 life cycle. J. Biol. Chem. 2821585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 232451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao, Z., E. Ehrlich, Y. Yu, K. Luo, T. Wang, C. Tian, and X. F. Yu. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349290-299. [DOI] [PubMed] [Google Scholar]
- 44.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 1015652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita, T., K. Kamada, K. Hatcho, A. Adachi, and M. Nomaguchi. 2008. Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes Infect. 101142-1149. [DOI] [PubMed] [Google Scholar]
- 46.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]
- 47.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 182867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zennou, V., and P. D. Bieniasz. 2006. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology 34931-40. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, W., M. Huang, T. Wang, L. Tan, C. Tian, X. Yu, W. Kong, and X. F. Yu. 2008. Conserved and non-conserved features of HIV-1 and SIVagm Vif mediated suppression of APOBEC3 cytidine deaminases. Cell Microbiol. 101662-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 786073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, T., Y. Han, Y. Dang, X. Wang, and Y. H. Zheng. 2009. A novel HIV-1 restriction factor that is biologically distinct from APOBEC3 cytidine deaminases in a human T cell line CEM.NKR. Retrovirology 631. [DOI] [PMC free article] [PubMed] [Google Scholar]